Abstract
GWAS-identified 10q22.3 loci with lead SNP rs704017 are significantly associated with CRC risk in both Asian and European populations. However, the functional mechanism of this region is unclear. In this study, we performed a fine-mapping analysis to identify the causal SNPs. To identify potential functional SNPs in linkage disequilibrium with the lead SNP, we searched for the potential target genes using a Hi-C database and an RNA interfering-based on-chip approach. The results indicated that rs12263636 (r2 = 0.41) showed the highest potential to be functional. It resided in a region with enhancer markers and a topologically associating domain. We found that RPS24 was the only gene that significantly promoted the proliferation rate of CRC cells and might have promoter–enhancer interaction with rs12263636. Dual-luciferase reporter assays confirmed that the risk alleles of two variants (rs3740253 and rs7071351) in RPS24 promoter could increase the expression of luciferase. Case control study consisting of 1134 cases and 2039 health controls confirmed that both the two variants were associated with risk of CRC (rs3740253: P = 0.0079, OR = 1.15, 95% CI 1.04–1.28; rs7071351: P = 0.0085, OR = 1.15, 95% CI 1.04–1.28). And plasmid containing mutant haplotypes containing all the three mutations (rs12263636 or rs3740253 and rs7071351) could most significantly increase luciferase expression, compared with any haplotype of the three mutations. The study explained the functional mechanism for the 10q22.3 loci and provided new insights into the prevention and treatment of CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third most common type of cancer among worldwide population in both genders (Siegel et al. 2018). The incidence and mortality of CRC keep increasing in China these years with age-standardized incidence rates of 376.3 per 100,000 and mortality rate of 191.0 per 100,000 in 2015 (Chen et al. 2016; Torre et al. 2015). Evidently, CRC remains an important public health concern, it is of great urgency to explore and develop more efficient biomarkers and targets to facilitate the diagnosis and treatment of CRC.
Population-based studies had suggested a positive association between CRC and genetic background. Researchers have identified more than 100 genome-wide association studies (GWAS) loci associated with risk of CRC. Several sites are found in both European and Asian descent cases (Jia et al. 2013; Zeng et al. 2016; Zhang et al. 2014a, b). Though molecular mechanisms underlying many of the genetic loci have not been well characterized (Chang et al. 2018; Gundert et al. 2019), it has been recognized that the majority of the GWAS-identified SNPs reside in non-coding gene regions indicating that they might execute their effects through regulation of gene expression (Zhu et al. 2016; Zou et al. 2018). Promoter–enhancer interaction is one of the key gene regulation mechanisms (Ron et al. 2017). Public databases of several high-throughput experimental methods like Hi-C(Rao et al. 2015) provided great opportunities to understand the long-range interaction mechanism of three-dimensional chromatin organization. And variants residing in different regulatory elements might have interactive effects and strengthen the transcriptional activity of genes and lead to worse result (Chen et al. 2018; Tian et al. 2019; Visser et al. 2012). However, studies on SNP interactions are limited.
Here, we performed fine mapping analysis in GWAS-identified 10q22.3 region. This region is significantly associated with CRC risk in both Asian and European populations (Zhang et al. 2014a), but little is known about the underlying functional mechanism for this signal. We identified that rs12263636 with enhancer markers was the most potential functional variant. By prioritizing the candidate target genes by performing an on-chip RNA interfere assay, we successfully identified a risk gene RPS24. The expression of RPS24 might be upregulated by two SNPs (rs3740253 and rs7071351) with effect on promoter function. And we further identified that these two SNPs together with rs12263636 might increase the expression of RPS24 through enhancer–promoter interaction. These three SNPs may serve as novel biomarkers for early detection and prevention of CRC.
Methods
Study subjects participated in this study
This study consists of 1134 cases and 2039 healthy controls. There were 61.2% males in cases and 61.9% in controls. The average age of cases and controls was 55.95 and 61.33 (Table S1). CRC patients were enrolled from Tongji hospital in Wuhan, China. Health controls were selected from a community cancer screening program for early detection conducted in the same region during the same period as cases were collected. Some of whom were included in our previous studies (Gong et al. 2018; Li et al. 2018). All participants were unrelated Han Chinese descent, and the inclusion criteria included histopathologically confirmed primary CRC, without any radiotherapy or chemotherapy treatment prior to blood samples collected. The informed consent was obtained from every participant at recruitment and peripheral blood samples and demographic characteristics such as sex, age, smoking status, drinking status and ethnicity were collected by interviewers. This study was conducted under the approval of the institutional review boards of the Tongji medical college, Huazhong University of science and technology. SNPs were genotyped by Taqman SNP Genotyping Assay (Applied Biosystem).
Identification of functional SNPs in LD with tag SNP
SNP in linkage disequilibrium (LD) with the tag SNP (r2 > 0.2) was made as function annotation using Haploreg4.1. The LD was calculated based on 1000 Genome Phase 3 population of Asian. For the physical interaction prediction of the promoter and enhancer region, we used an online database 3D Genome Browser (http://promoter.bx.psu.edu/hi-c/). Hi-C data were generated from HCT116_RAD21-mAC_no_auxin from Rao SSP (Rao et al. 2017).
Candidate gene selection and RNA interfering-based on-chip assays
We first found the potential target gene of rs12263636 according to Hi-C data. Since rs12263636 resided in enhancer region and a topologically associating domain as well, all of the genes within the boundaries of the domains were predicted to have enhancer–promoter interaction with rs12263636. We reviewed the gene functional annotation in NCBI database and excluded genes encoding small nuclear RNAs or pseudo genes. The remaining genes were chosen for the function interrogation on CRC cell proliferation rate. An RNA interfering-based on-chip approach was used to screen out the potential target genes that may affect the proliferation rate of CRC cells. The CCK-8 assay was used to evaluate the cell proliferation of cell lines after transfection with siRNAs. The GAPDH expression was used as control for siRNA knock down experiment. Both criteria of P < 0.05 and fold change > 1.2 or < 0.8 were selected as the threshold of significance. Data were generated from three independent experiments and each independent experiment has three replications. We used GTEx (https://www.gtexportal.org/home/) to extract eQTLs of the functional genes identified from RNA interfering experiment. The gene expression data were generated from GEPIA (http://gepia.cancer-pku.cn/).
Luciferase reporter assays
The mut-type DNA sequence centering on rs3740253 and rs7071351 was inferred to affect the promoter function of RPS24. We first constructed PGL3 plasmid (Promega, Madison, WI, USA) with a 1.5 kb region containing the promoter of RPS24 and both rs3740253 and rs7071351 polymorphic sites (C at rs3740253 and C at rs7071351) as control plasmid. And then T-A haplotype (T at rs3740253 and A at rs7071351) plasmid was generated based on the control plasmid. To examine whether the rs12263636 variant might affect RPS24 enhancer activity, a 750 bp region centered on rs12263636 was synthesized and added to the above-described vector with T-A or C-C haplotype of rs3740253 and rs7071351. This region was inferred as enhancer of RPS24 and constructed to the downstream region of the luciferase gene to mimic the real structure. Additionally, we further identified the function of rs3740253, rs7071351 and rs12263636 separately. Each of the three SNPs was site-specifically mutated. The plasmid was co-transfected with pRL-SV40 into SW480 and LoVo cells using Lipofectamine 3000 (Invitrogen, Waltham, MA, USA).
Cell lines
LoVo, SW480 and HCT116 cell lines were obtained from the China Center for Type Culture Collection (Wuhan, PR China). Cells were tested for the absence of mycoplasma contamination (MycoAlert, Lonza Rockland, ME, USA) and were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NJ, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% antibiotics (100 U/ml penicillin and 0.1 mg/ml streptomycin) at 37 °C in a humidified atmosphere of 5% CO2. All cell lines were tested and authenticated by DNA sequencing using the AmpF/STR method (Applied Biosystems).
Statistical analyses
Unconditional multivariate logistic regression analysis adjusted for age, sex, smoking and drinking status was used to assess the associations between two functional polymorphisms and CRC risk. All statistical analyses were performed using R (3.6.0) with a significance level of 0.05 and all tests were two-sided. Genotype data in 1 Mb flanking the tag SNP were downloaded from 1000 genome (http://grch37.ensembl.org/index.html) and LD analysis was performed using PLINK. We imputed rs3740253 and rs7071351 for all CRC samples from TCGA with IMPUTE2, and we used 1000 Genomes Phase 3 as the reference panel.
Results
The rs12263636 variant was predicted to have enhancer function
We performed function annotation for 22 SNPs in high LD (r2 > 0.2) with the lead SNP rs704017 in the 10q22.3 region. The results suggested that most of them resided in the region with enhancer histone markers (Table S2). None of these SNPs had cis-eQTL hit in the colon tissues from GTEx database. The rs12263636 (LD r2 = 0.41, D’ = 0.86) showed the highest functional potential with enhancer histone markers in 23 tissues and DNase in 22 tissues. Chromatin immunoprecipitation sequencing (ChIP-seq) data from the CistromDB generated in different CRC cell lines showed that rs12263636 resided in a region with strong signal of enhancer markers (Fig. 1a). Therefore, we focused on this SNP in the following study.
rs12263636 with enhancer markers resided in a TAD region. a Chromatin modification of H3K4me1, H3K27ac at the region of interest which contains rs12263636. The position of rs12263636 was indicated by dashed lines. Data were downloaded from CistromDB. b Hi-C data generated in HCT116_RAD21-mAC_no_auxin from Rao SSP indicated that promoters of several genes might interact with rs12263636 resided in the intron of ZMIZ1 (the black triangle show two instances of the candidate genes)
Functional interrogation identified RPS24 as the susceptibility gene
To identify tumor-related genes regulated by rs12263636, we first used a Hi-C database to find gene promoters which might have interaction with rs12263636. After excluding genes encoding small nuclear RNAs or pseudo genes, we finally selected 15 genes including 12 protein-coding genes and 3 large intergenic non-coding RNAs (Fig. 1b). The siRNA sequences specific to the candidate genes are shown in Table S3 and the knock down efficiency for each gene was shown in Fig S1. Four genes were not effectively knocked down due to low expression in CRC cells (Fig S2). We then used an RNA interfering-based on-chip approach to screen out potential target genes that might affect the proliferation rate of CRC cells. The result showed that only RPS24 had an enhancer–promoter interaction with rs12263636 and was significantly associated with cell proliferation rate in both HCT116 and SW480 cell lines. When knocking down RPS24, the proliferation rate of HCT116 and SW480 cell lines were significantly enhanced with P values of 0.0010 and 0.0078, respectively (Fig. 2 and Table S4).
Knock down of RPS24 with siRNA inhibits the cell proliferation in both two colorectal cancer cell lines. CCK8 assay was used to evaluate the cell proliferation of HCT116 and SW480 cell lines after transfection with siRNAs for 11 genes. Data were from three independent experiments and each have three replications. log2 transformed fold change and log10 transformed P values for each gene were showed in x and y axis, respectively
Two variants were predicted to affect the promoter activity of RPS24
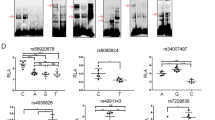
We then explored whether there were SNPs affecting the promoter activity of RPS24 and function together with rs12263636. Cis-eQTL data of 169 human Colon-Transverse tissues were obtained from the GTEx database (Carithers et al. 2015) (downloaded at November, 22ed, 2016). The result showed that the RPS24 expression was correlated with 15 SNPs with P values ranging from 2.9 × 10−6 to 3.4 × 10−5 (Table S5 and Fig. 3a). These variants were all in a LD block (Fig S3). We then predicted whether these variants have effect on RPS24 expression according to chromatin modification data from the HaploReg 4.1 database. Among these 15 variants, two SNPs (rs3740253 and rs7071351) in perfect LD were strongly predicted to affect the promoter function of RPS24, and data of 24 different tissues suggested that transcription factors motif changed at both two sites (Table S6). eQTL analysis of the association between rs3740253/rs7071351 and the expression of RPS24 using the genotype and expression data from TCGA also showed that samples with the T/A allele of rs3740253/rs7071351 had significantly higher expression of RPS24 than samples with the C allele (Fig S4). ChIP-seq data from the CistromDB also showed that rs3740253 and rs7071351 resided in a region with strong signal of H3K4me3 (Fig. 3b). GATA1 was predicted to bind to the sequence with the A allele at rs3740253 according to the JASPAR database online prediction (Fig. 3c), and a GATA1 ChIP-seq peak was also found in the SNP region (Fig. 3b). Similarly, IRF2 was predicted to bind to the sequence with the T allele at rs7071351 (Fig. 3d) and IRF2 ChIP-seq peak was also found in the SNP region (Fig. 3b). These results suggested that these two SNPs might influence transcription factor binding at the promoter region of RPS24 and then affect RPS24 expression.
rs3740253 within intron region of RPS24 was predicted to have promoter function with alteration of GATA family transcription factors binding. a Box plot shows the correlation between rs3740253 and rs7071351 with RPS24 expression in colon tissues. Data were from GTEx database. b Chromatin modification of H3K4me3, GATA1 and IRF2 transcription factor binding sites at the region of interest which contains rs3740253 and rs7071351. c The predicted binding sites sequence of rs3740253 and rs7071351 were shown and the risk allele of rs3740253 creates a stronger GATA1 motif while the risk allele of rs3740253 creates a stronger IRF2 motif
The rs3740253 and rs7071351 variant affected the promoter activity of RPS24
We then tested the function of these two promoter variants through a dual-luciferase reporter assay. As these two variants were in perfect LD, we constructed four different luciferase reporter gene constructs including plasmids with C-C haplotype, T-A haplotype (T at rs3740253 and A at rs7071351), C>T transition at rs3740253 and C>A transition at rs7071351. We observed a significantly twofold higher level of promoter activity derived from transfection of the construct containing T-A haplotype compared with those of the construct containing C-C haplotype (Fig. 4a, both P < 0.0001 for LoVo and SW480 cells). Intriguingly, either C>T transition at rs3740253 or C>A transition at rs7071351 displayed higher promoter activity and significantly increased the expression of luciferase in both CRC cell lines (Fig. 4b).
Function annotation of rs3740253 and rs7071351 by dual-luciferase reporter assay. a The T-A haplotype of rs3740253 and rs7071351 increases the relative luciferase activity in both LoVo and SW480 cell lines. Data are from 3 independent transfection experiments with assays conducted in 3 replications. Comparisons were conducted by unpaired Wilcoxon rank-sum test. b rs3740253 and rs7071351 was then site-specifically mutated in the template sequence and the results indicated that either C>T transition at rs3740253 or C>A transition at rs7071351 significantly increased the expression of luciferase in both two CRC cell lines. Data are from three independent transfection experiments with assays conducted in three replications. Comparisons were conducted by unpaired t test
Both rs3740253 and rs7071351 are associated with increased risk of CRC
We further tested whether rs3740253 and rs7071351 were associated with susceptibility to CRC through a case–control study consisting of 1134 cases and 2039 health controls. Through a logistic regression analysis, we identified that rs3740253 was significantly correlated with increased risk of CRC with odds ratio (OR) being 1.15 in an additive model (95% CI 1.04–1.28, P = 0.0079). Compared with CC genotype carriers, CT genotype and TT genotype carriers were associated with increased risk of CRC with OR being 1.24 (95% CI 1.04–1.48, P = 0.0187) and 1.31 (95% CI 1.06–1.61, P = 0.0134), respectively. Significant associations were also observed for rs7071351. The results are shown in Table 1. Then we performed a stratified analysis to determine the association between the two SNPs (rs3740253 and rs7071351) and CRC risk (Fig S5). Significant association between the SNPs and risk of CRC was observed for subgroup patients of female, age < 60, non-smoker and non-drinker.
The rs12263636 variant interacts with rs3740253 and rs7071351 in the promoter of RPS24
We constructed plasmids containing all the three variants including rs12263636 (G>A), rs3740253 (C>T) and rs70701351 (C>A) (Fig. 5a). The plasmids containing the C-C-A haplotype (P = 0.0369 in SW480, P = 0.0003 in LoVo), the T-A-G haplotype (P = 0.0014 in SW480, P < 0.0001 in LoVo), and the T-A-A haplotype (P < 0.0001 in SW480, P < 0.0001 in LoVo) significantly increased the activity of luciferase, compared with the C-C-G haplotype. The plasmid with all mutant types for the three SNPs (T-A-A) showed the highest luciferase activity (Fig. 5b).
Function annotation of rs12263636 by dual-luciferase reporter assay. a DNA fragments containing rs12263636 [G/A] and RPS24 promoter containing rs3740253 [C/T] and rs70701351 [C/A] were cloned into pGL3-Basic vector. b The T-A-A haplotype increases the relative luciferase activity in both LoVo and SW480 cell lines. Data are from three independent transfection experiments with assays conducted in three replications. Comparison was conducted by unpaired t test
Discussion
GWAS has so far identified numbers of susceptibility loci related to complex traits (Karaderi et al. 2015; Patel and Ye 2011). However, most of the identified variants are tag SNPs, with a limitation in elucidation of the disease-related genes and the biological relevance of those susceptibility loci (Wang and Chatterjee 2017). In the present study, by fine-mapping analysis of GWAS loci 10q22.3, we identified that the function of rs12263636 was exerted by enhancer–promoter interaction. The rs12263636, rs3740253, and rs7071351 variants function together to influence the expression of RPS24 and were associated with CRC risk.
The 10q22.3 region is one of the CRC GWAS loci identified in East Asians and is also associated with CRC risk in individuals of European descent (Zhang et al. 2014a). This region is tagged by the rs704017 which resides in the 3rd intron of ZMIZ1-AS1 gene. However, ZMIZ1-AS1 is a miscellaneous RNA gene with unknown function and both ZMIZ1 and ZMIZ1-AS1 failed to affect cell growth in our functional interrogation. More experiments are needed to find out more of the function of those genes.
In this study, we found rs12263636 might work together with two variants (rs3740253 and rs7071351) to increase RPS24 gene expression, and contributed to CRC risk. Previous studies have reported that RPS24 was a multiple cancer-related gene (Choesmel et al. 2008; Inoue et al. 2015). RPS24 is known to encode a ribosomal protein that is a component of the 40S ribosomal subunits and co-expressed with family of ribosomal proteins (Fig S6). Mutations in ribosomal protein genes might cause cellular stress and activate cell cycle inhibitors. It has been reported that knockdown of RPS24 could significantly inhibit cell proliferation, colony formation, cell migration in CRC cell lines HCT116 and HT-29 cells by arresting cells in S phase (Wang et al. 2015). RPS24-deficient fibroblasts cells showed increased levels of the cell cycle inhibitor p21 and a seemingly opposing increase of p21 target genes in Cyclin-E, CDK4 and CDK6 (Badhai et al. 2009) and which might explain the reduced growth. In this study, we successfully replicated the previous results and further demonstrated that RPS24 was an important susceptibility gene for CRC.
By integrating Hi-C data and eQTL analysis, we successfully determined the potential cis-regulated genes that physically interacted with predicted functional SNPs in GWAS loci. Our results demonstrated that the Hi-C data along with eQTL will facilitate identification of functional variants and candidate genes responsible for CRC risk.
There are still some limitations in our study. First, we only selected genes in a restricted region for functional interrogation. And those genes have no effect on CRC cell proliferation might have other functions and might also affect the susceptibility of CRC. Second, the results of case–control study are needed to be replicated in larger samples in the future. Third, the SNP identification was only conducted in the Chinese, and Chinese populations may be different from other populations, such as Caucasian, in terms of genetic predisposition to cancer. Thus, whether rs3740253/rs7071351 were risk SNPs in other populations needs to be replicated in the future.
In conclusion, we provided a feasible method to identify potentially functional SNPs to distally regulate their target gene underlying the pathogenesis of CRC using bioinformatics data analyses and experimental validation. We first identified a functional SNP rs12263636 at GWAS-identified 10q22.3 region. We further found that two variants (rs3740253 and rs7071351) together with rs12263636 might upregulate RPS24 expression via enhancer–promoter interaction. Knocking RPS24 down could significantly inhibit proliferation in both LoVo and SW480 cells which suggested the important role of RPS24 in CRC. These results expanded our understanding of the etiology of CRC and might be used in the precise prevention of CRC in the future.
References
Badhai J, Frojmark AS, Davey JE, Schuster J, Dahl N (2009) Ribosomal protein S19 and S24 insufficiency cause distinct cell cycle defects in Diamond-Blackfan anemia. Biochim Biophys Acta 1792(10):1036–1042. https://doi.org/10.1016/j.bbadis.2009.08.002
Carithers LJ, Ardlie K, Barcus M et al (2015) A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank 13(5):311–319. https://doi.org/10.1089/bio.2015.0032
Chang J, Tian J, Yang Y et al (2018) A rare missense variant in TCF7L2 associates with colorectal cancer risk by interacting with a GWAS-identified regulatory variant in the MYC enhancer. Cancer Res 78(17):5164–5172. https://doi.org/10.1158/0008-5472.CAN-18-0910
Chen W, Zheng R, Baade PD et al (2016) Cancer statistics in China, 2015. CA 66(2):115–132. https://doi.org/10.3322/caac.21338(A Cancer Journal for Clinicians)
Chen XF, Zhu DL, Yang M et al (2018) An osteoporosis risk SNP at 1p36.12 acts as an allele-specific enhancer to modulate LINC00339 expression via long-range loop formation. Am J Hum Genet 102(5):776–793. https://doi.org/10.1016/j.ajhg.2018.03.001
Choesmel V, Fribourg S, Aguissa-Toure AH et al (2008) Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet 17(9):1253–1263. https://doi.org/10.1093/hmg/ddn015
Gong J, Tian J, Lou J et al (2018) A polymorphic MYC response element in KBTBD11 influences colorectal cancer risk, especially in interaction with an MYC-regulated SNP rs6983267. Ann Oncol 29(3):632–639. https://doi.org/10.1093/annonc/mdx789
Gundert M, Edelmann D, Benner A et al (2019) Genome-wide DNA methylation analysis reveals a prognostic classifier for non-metastatic colorectal cancer (ProMCol classifier). Gut 68(1):101–110. https://doi.org/10.1136/gutjnl-2017-314711
Inoue E, Watanabe Y, Egawa J et al (2015) Rare heterozygous truncating variations and risk of autism spectrum disorder: whole-exome sequencing of a multiplex family and follow-up study in a Japanese population. Psychiatry Clin Neurosci 69(8):472–476. https://doi.org/10.1111/pcn.12274
Jia WH, Zhang B, Matsuo K et al (2013) Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat Genet 45(2):191–196. https://doi.org/10.1038/ng.2505
Karaderi T, Drong AW, Lindgren CM (2015) Insights into the genetic susceptibility to type 2 diabetes from genome-wide association studies of obesity-related traits. Curr Diabetes Rep. https://doi.org/10.1007/s11892-015-0648-8
Li J, Chang J, Tian J et al (2018) A rare variant P507L in TPP1 interrupts TPP1-TIN2 interaction, influences telomere length, and confers colorectal cancer risk in Chinese population. Cancer Epidemiol Biomarkers Prev 27(9):1029–1035. https://doi.org/10.1158/1055-9965.EPI-18-0099(A Publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology)
Patel RS, Ye S (2011) Genetic determinants of coronary heart disease: new discoveries and insights from genome-wide association studies. Heart 97(18):1463–1473. https://doi.org/10.1136/hrt.2010.219675
Rao SSP, Huntley MH, Durand NC et al (2015) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 162(3):687–688. https://doi.org/10.1016/j.cell.2015.07.024
Rao SSP, Huang SC, St Hilaire BG et al (2017) Cohesin loss eliminates all loop domains. Cell 171(2):305. https://doi.org/10.1016/j.cell.2017.09.026
Ron G, Globerson Y, Moran D, Kaplan T (2017) Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat Commun. https://doi.org/10.1038/S41467-017-023863
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA 68(1):7–30. https://doi.org/10.3322/caac.21442(A Cancer Journal for Clinicians)
Tian J, Chang J, Gong J et al (2019) Systematic functional interrogation of genes in GWAS loci identified ATF1 as a key driver in colorectal cancer modulated by a promoter-enhancer interaction. Am J Hum Genet 105(1):29–47. https://doi.org/10.1016/j.ajhg.2019.05.004
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA 65(2):87–108. https://doi.org/10.3322/caac.21262(A Cancer Journal for Clinicians)
Visser M, Kayser M, Palstra RJ (2012) HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res 22(3):446–455. https://doi.org/10.1101/gr.128652.111
Wang Z, Chatterjee N (2017) Increasing mapping precision of genome-wide association studies: to genotype and impute, sequence, or both? Genome Biol 18(1):118. https://doi.org/10.1186/s13059-017-1255-6
Wang Y, Sui JK, Li X et al (2015) RPS24 knockdown inhibits colorectal cancer cell migration and proliferation in vitro. Gene 571(2):286–291. https://doi.org/10.1016/j.gene.2015.06.084
Zeng C, Matsuda K, Jia WH et al (2016) Identification of susceptibility loci and genes for colorectal cancer risk. Gastroenterology 150(7):1633–1645. https://doi.org/10.1053/j.gastro.2016.02.076
Zhang B, Jia WH, Matsuda K et al (2014a) Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet 46(6):533–542. https://doi.org/10.1038/ng.2985
Zhang B, Jia WH, Matsuo K et al (2014b) Genome-wide association study identifies a new SMAD7 risk variant associated with colorectal cancer risk in East Asians. Int J Cancer 135(4):948–955. https://doi.org/10.1002/ijc.28733
Zhu Z, Zhang F, Hu H et al (2016) Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48(5):481–487. https://doi.org/10.1038/ng.3538
Zou D, Lou J, Ke J et al (2018) Integrative expression quantitative trait locus-based analysis of colorectal cancer identified a functional polymorphism regulating SLC22A5 expression. Eur J Cancer 93:1–9. https://doi.org/10.1016/j.ejca.2018.01.065
Acknowledgements
We gratefully acknowledge the members of the Miao lab for the suggestions and contributions to this work. This work was supported by the National Natural Science Foundation of China [81502875 to J.C.]; the National Key Research and Development Plan Program [2016YFC1302702, 2016YFC1302703 to X.M.]; National Program for Support of Top-notch Young Professionals, National Natural Science Foundation of China [81171878, 81222038 to X.M.]; the National Science Fund for Distinguished Young Scholars of China and the Fok Ying Tung Foundation for Young Teachers in the Higher Education Institutions of China [131038 to X.M.].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zou, D., Zhang, H., Ke, J. et al. Three functional variants were identified to affect RPS24 expression and significantly associated with risk of colorectal cancer. Arch Toxicol 94, 295–303 (2020). https://doi.org/10.1007/s00204-019-02600-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-019-02600-9