Abstract
This study aimed to evaluate the short-term effects of irrigation with diluted fish-processing effluents on soil pH, electrical conductivity, nitrification rate and abundance of ammonia oxidizers. To accomplish that, we constructed microcosms of soil from an undisturbed arid ecosystem of Patagonia, and irrigated them for 2 months with diluted effluents from a fish-processing factory or with water as control. In the initial soil sample, and along the experiment, we determined soil pH, electrical conductivity, and the concentration of inorganic nitrogen forms, which we used to calculate the net nitrification rate. We further estimated the abundances of ammonia-oxidizing archaea and bacteria in the initial soil sample and at the end of the experiment, by qPCR of amoA genes. Soil pH decreased and electrical conductivity increased in both irrigation treatments, although the effect was higher in effluent-irrigated microcosms. Soil nitrate + nitrite concentration, and thus the nitrification rate, was higher in effluent than in water-irrigated microcosms. The abundance of archaeal amoA genes was higher under effluent than water-irrigation, but that of bacterial amoA genes did not vary significantly between treatments. Neither ammonia-oxidizing archaea nor bacteria were influenced by the changes in soil pH and electrical conductivity induced by effluent irrigation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The water crisis is among the 10 most likely proposed global risks and among the 5 risks with a higher global impact, according to the World Economic Forum (World Economic Forum 2020). Currently, 17 countries that are home to one quarter of the world’s population face extremely high freshwater scarcity issues (World Resources Institute 2019). Irrigated agriculture is responsible for nearly 70% of the world's freshwater consumption (FAO 2014); therefore, wastewater reuse for irrigation has been adopted by many countries, particularly from arid and semiarid regions, in an attempt to reduce their consumption of freshwater. Wastewater reuse for irrigation was proposed to have additional advantages in nutrient-poor soils, such as benefits on soil fertility due to increased nutrient supplies; though negative effects, e.g. on soil salinity and pH were also reported (Chen et al. 2017).
Fish-processing factories consume on average 11–15 m3 of water per ton of processed product, and the estimated amount of fish production worldwide is 179 million tons; leading to a rough estimate of global water consumption by the fishery industry of 2.0–2.7 billion m3 (de Melo Ribeiro et al. 2020; FAO 2020). This large water consumption is directly related to the amount of produced wastewater; part of which may be reused in some industrial applications to reduce the factory consumption of freshwater (Guimarães et al. 2018). The remaining amount, after a proper treatment to fulfill the local regulations, is usually discharged into water bodies. However, resources contained in fish-processing effluents have a high potential to be reused in agriculture, e.g. in irrigation or as liquid fertilizers (Gwon and Kim 2012; Cristóvão et al. 2015; Santoyo Figueroa et al. 2015; Jung and Kim 2016). Nevertheless, there has still been little discussion on the effects of soil irrigation with effluents from the fish industry on the soil microbial community.
Fish-processing effluents are saline if seawater or saline groundwater is used in the process, and have a high content of organic matter, of which proteins represent ca. 70% (Veiga et al. 1994; Mishra et al. 2015). As a result of protein degradation, they also have high contents of ammonium. Besides changes in the overall microbial community (Vallejos et al. 2020); this high content of ammonium and ammonifiable organic matter, together with the potential changes in soil pH and salinity, may particularly affect a specific group of soil prokaryotes, i.e., the ammonia oxidizers. To the best of our knowledge, there is no evidence of nitrification inhibitory molecules in fish-processing wastewaters. The ammonia oxidizers perform the rate-limiting step of nitrification under oxic conditions and are represented by archaea of the phylum Thaumarchaeota (AOA), and bacteria of the β- and γ-proteobacteria (AOB). Nitrifiers are critical players in the soil nitrogen cycle, as this process controls the total inorganic nitrogen available in soil (Prosser 2011). In addition, since ammonia oxidizers are susceptible to different kinds of soil perturbations, they have been previously reported as possible bioindicators for soil monitoring (Wessén and Hallin 2011; Zabaloy et al. 2017). In other arid regions, despite having no significant effects on the composition of the total soil bacterial community, and having only a minor effect on total bacterial activities, treated municipal wastewater stimulated soil nitrification rates and the relative abundance of OTUs related to Nitrosococcus, Nitrosovibro, and Nitrospira in short- and mid-term studies (≤ 2 years), whilst the impact on AOA was not analyzed (Frenk et al. 2015; Ibekwe et al. 2018). In addition, shifts in the composition of AOB from Nitrosospira-like to Nitrosomonas-like dominated populations were observed in response to soil irrigation with treated urban effluents (Oved et al. 2001; Frenk et al. 2015). In contrast, the use of sewage sludge compost tea as soil fertilizer had no effect on either AOB or AOA abundances (Vela-Cano et al. 2018). It is still unknown how irrigation with effluents from fish-processing industries could affect soil nitrifiers; however, in accordance with previous studies based on irrigation with municipal wastewater (Frenk et al. 2015; Ibekwe et al. 2018), we hypothesize that through their input of ammonium, fish-processing effluents may stimulate nitrification and the abundances of AOB and AOA, despite potential changes in soil pH and salinity. Therefore, the aims of this study were (i) to evaluate the short-term effects of irrigation with diluted fish-processing effluents on soil salinity and pH in contrast with soils irrigated with freshwater; and (ii) to analyze the effects of irrigation with diluted fish-processing effluents or fresh water on soil ammonium (NH4+) and nitrate + nitrite (NO3− + NO2−) concentrations, nitrification rate, and on the abundance of AOB and AOA populations.
Materials and methods
Soil sampling and characterization
Soil samples were collected in a field near a cluster of fish-processing industries in Puerto Madryn city, Chubut Province, Argentina (42° 43′ S; 65° 02′ W). This is an arid-climate location (mean annual temperature: 13.4 ± 0.1 °C, mean annual precipitation: 184.4 ± 10.5 mm, 1970–2019 time series INTA SIPAS, http://sipas.inta.gob.ar/) in the southern part of the Monte Phytogeographic Province (Patagonian Monte) were vegetation corresponds to a shrubland of Larrea divaricata Cav., with perennial grasses (León et al. 1998). Soils are a complex of Typic Torriorthents (Pereyra and Bouza 2019), and vegetation is heterogeneously distributed in plant patches surrounded by bare soil areas (Bisigato and Bertiller 1997). Soil samples were collected from the bare soil areas (to minimize the interaction with vegetal organic matter) using cores (0–10 cm depth and 10 cm in diameter), and immediately transported to the laboratory at 4 °C. Thereafter, samples were pooled, homogenized and sieved through a 2 mm mesh to have a homogeneous soil sample for microcosm construction.
Air-dried sub-samples were characterized according to their physicochemical properties. Soil moisture was determined gravimetrically (105 °C, 48 h), and all the results were expressed on the basis of dry soil weight. Soil pH, electrical conductivity (EC), and sodium adsorption ratio (SAR) were assessed in soil saturation extracts, as described in Allison and Richards (1954). Soil texture (sand, silt and clay percentages) was assessed by the Bouyoucos Hydrometer method (Bouyoucos 1962). Total soil carbon (C) and nitrogen (N) concentrations were measured using a CN628 Carbon/Nitrogen Determinator (LECO Corporation, USA). The concentration of inorganic C was assessed gravimetrically, after removing soil carbonates with 3 N HCl (Allison and Moodie 1965). The concentration of soil organic C was assessed by wet combustion, and was used to calculate soil C/N ratio. The concentration of NH4+ in soil sample extracts was assessed according to Keeney and Nelson (1982), and that of NO3− + NO2− as described by Shand et al. (2008). All soil analyses were performed in triplicate, and are reported in Table 1. Soil physicochemical properties were similar to previously reported values in uncultivated soils from this and other arid regions (Xie et al. 2001; Li et al. 2006; Vallejos et al. 2020).
Fish-processing effluent sampling and characterization
Fish-processing effluents were collected from the outgoing of a fishing-factory treatment plant in Puerto Madryn city. The effluent processing in this plant consisted of a primary treatment, a secondary treatment by activated sludge, and a final chlorination step. Effluent samples were collected according to method 1060 of the American Public Health Association (APHA et al. 2017), and immediately transported to the laboratory at 4 °C, where sub-samples were characterized according to their physicochemical properties. The effluent was 16-fold diluted in tap water, aliquoted in 500 ml bottles and stored in a freezer at − 20 °C until its use for microcosm irrigation (Ching and Redzwan 2017). The 16-fold dilution was chosen so that the effluent salinity was lower than 0.45 g total dissolved solids (TDS)/l, which is the limit value for effluent reuse in irrigation with no restrictions, according to Chubut Province guidelines (adapted from Ayers and Westcot 1994).
The EC, TDS and temperature of the fish-processing effluents were measured with a Hanna HI 98192 conductivity meter, while pH was measured using a Hanna pH 211 meter (Hanna Instruments, USA). The organoleptic characteristics of the effluents were assessed according to methods 2150 B (odor) and 2130 B (turbidity) of the APHA et al. (2017). The effluent SAR was determined as described in Allison and Richards (1954). Chemical oxygen demand (COD) was measured using a Hanna HI83099 photometer (Hanna Instruments, USA), (method EPA 410.4-adapted; US EPA 1993). In addition, effluent alkalinity (concentration of bicarbonates; method 2320 B), oils and greases (method 5520 B), and NH4+ concentration (method 4500-NH3 F) were carried out according to APHA et al. (2017). The concentration of NO3− in the effluents was assessed following EPA method 352.1 (Keith 1996). All effluent analyses were performed in triplicate.
Microcosm experimental setup
Soil microcosms were prepared in plastic pots containing 350 g of fresh soil, and incubated in a greenhouse at room temperature for 61 days. Microcosms were irrigated daily to constant 15% soil moisture (corresponding to the usual water content of these soils in wet periods) with either tap water (W) as a control, or fish-processing effluent diluted 16-fold in water (DFE). Twelve replicates were prepared per irrigation treatment, which were destructively sampled in quadruplicates on days 8, 19 and 61 to determine soil pH, EC, and the concentrations of NH4+ and NO3− + NO2−, as described above. The net nitrification rate in soil microcosms was calculated as the increase in NO3− + NO2− concentration after 61 days of incubation (Drury et al. 2007).
Soil DNA extraction and qPCR of amoA genes
Total DNA was extracted from ca. 0.5 g of the soil used for microcosms set up (day 0) and of soil microcosms at the end of the irrigation experiment (day 61) using the FastDNA® SPIN Kit for Soil (MP Biomedicals, USA) and following the manufacturer’s instructions. The DNA extraction protocol included a washing step with 5.5 M guanidine thiocyanate solution, to remove PCR inhibitors (Tournier et al. 2015). DNA was quantified using a Quantus™ Fluorometer and the QuantiFluor® dsDNA Dye System (Promega Corporation, USA). Real-time PCR amplifications of amoA genes were performed on a 7500 Real-Time PCR System (Applied Biosystems, USA), using the 7500 System Software (Applied Biosystems, USA) for the setting of qPCR parameters, and the visualization and analyses of the amplicons. The amplifications were conducted using the SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad Laboratories, USA), and the amoA-1F/amoA-2R (Rotthauwe et al. 1997; amplicon size 491 bp) and Arch-amoAF/Arch-amoAR (Francis et al. 2005; amplicon size 635 bp) primer sets for AOB and AOA, respectively. All qPCRs were performed using 10 ng of template DNA, except for control reactions where DNA was replaced by ultrapure water. Primer concentrations were 0.3 µM and 0.2 µM for amoA-1F and amoA-2R (AOB), respectively; and 0.4 µM for both Arch-amoAF and Arch-amoAR (AOA). The amplification program for bacterial amoA genes was: 5 min at 95 °C and then 44 cycles of 30 s at 95 °C, 1 min at 55 °C, 30 s at 72 °C and a final step before fluorescence read of 40 s at 81 °C. In addition, the program for the quantification of archaeal amoA genes was: 5 min at 95 °C and then 45 cycles of 30 s at 95 °C, 1 min at 53 °C, 1 min at 72 °C and a final step of 40 s at 80 °C before fluorescence read. Melting curves were run at the end of the amplification program to verify the specificity of the amplified DNA fragments. Standard curves were constructed by performing 1:10 serial dilutions of linearized plasmids containing the amoA gene from Nitrosomonas europaea or from an uncultured archaeon (clone E2), in the range of 107 – 102 amoA gene copies/µl (r2 > 0.99). qPCR efficiencies were 93% and 92% for bacterial and archaeal amoA genes, respectively.
Statistical analyses
The statistical significance of differences in soil physicochemical properties between irrigation treatments at each incubation time was tested with Student’s t tests, and that among incubation times within each irrigation treatment was tested with one-way analysis of variance (ANOVA) followed by the Tukey test when data accomplished the assumptions of normality and homoscedasticity, or by the Tamhane test otherwise. The significance in the (log-transformed) amoA gene abundances between incubation day 0 and day 61 within each irrigation treatment, and between irrigation treatments on day 61 were analyzed with Student’s t tests. The relationships among soil properties were analyzed by Spearman rank-order correlation tests. The relationship between amoA gene abundances and nitrification rates was tested with regression analyses. Normality and homoscedasticity assumptions were tested before performing all parametric tests. Significance levels were set at 0.05.
Results and discussion
Fish-processing effluent characteristics
Fish-processing effluent physicochemical characteristics are reported in Table 2. Considering a potential irrigation reuse, one of the main threats of the fish-processing effluent was its salinity. According to the Chubut Province Standards, EC and TDS effluent values implied severe irrigation restrictions (Table 2), whereby the effluent was diluted to a concentration of approximately 0.45 g TDS/l for microcosm irrigation. Effluent dilution also contributed to reducing the values of other parameters such as SAR and bicarbonate concentration; therefore, the diluted effluent fell within the limits of irrigation without restrictions (Table 2). Moreover, the pH value of the diluted effluent was circumneutral and within the accepted values for irrigation. On the other hand, even though effluent dilution led to a decrease in NH4+ concentration to approximately 17 mg/l, this value is still close to the NH4+ concentrations of culture media used for enrichment and isolation of ammonia-oxidizers (Achuthan et al. 2006; Koops et al. 2006).
Irrigation effects on soil pH and salinity
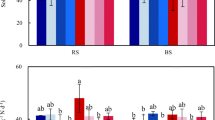
Irrigation with fish-processing effluents produced a significant decrease in soil pH of 1.3 pH units by the end of the experiment (Fig. 1a). Previous studies showed that pH may decrease in response to soil irrigation with wastewater, probably as a consequence of ammonia absorption by plants or nitrification, or due to leaching of base cations, such as Na+, Ca2+, K+, and Mg2+, that in turn reduce the alkaline reserves of the soil (Tarchouna et al. 2010; Bedbabis et al. 2014; Ganjegunte et al. 2017). In this study, irrigation with W also caused soil pH to decrease at the end of the experiment (day 61), although the extent of the pH drop was lower than in DFE. The rewetting of soil may cause pH to either increase or decrease as a result of the balance between two mechanisms that pursue the N mineralization induced by wetting, i.e., ammonification and nitrification (Haynes and Swift 1989). When ammonification exceeds nitrification, ammonium may accumulate leading to an increase in soil pH; whilst when ammonification is followed by nitrification -as seems to be the case in this study-, nitrate may accumulate producing a decrease in soil pH (Haynes and Swift 1989).
Irrigation effects on soil pH (a) and electrical conductivity (EC; b). Mean values ± Standard errors (n = 4). Soil(0): soil initial sample (before microcosms irrigation), W: water-irrigated microcosms, DFE: diluted fish effluent-irrigated microcosms. Different uppercase letters indicate significant differences (p < 0.05) in soil pH and EC of microcosms irrigated with W among incubation times, whereas lowercase letters refer to the same comparisons in microcosms irrigated with DFE. The results of pairwise comparisons between irrigation treatments within each date are indicated with asterisks (** p < 0.01) or with “ns” (not significant, p > 0.05)
Contrarily to soil pH, EC significantly increased along the experiment in both treatments, with a significantly higher effect in DFE- than in W-irrigated microcosms (Fig. 1b). This result was expected, as soil salinization is a commonly reported adverse effect of wastewater irrigation (Becerra-Castro et al. 2015; Ibekwe et al. 2018). However, the microcosm study probably had higher evapotranspiration and lower lixiviation conditions than those in the field, contributing to a higher accumulation of soil salts than that expected under field conditions. Accordingly, the exposure of Patagonian arid soils to fish-processing effluents of higher EC (2.8 mS/cm) in a field study showed only a mild increase in soil EC, and the soil neither became saline nor sodic after exposure to the effluents (Vallejos et al. 2020). Nevertheless, regular soil monitoring should be performed if effluents are to be reused for irrigation to prevent soil salinization.
Irrigation effects on inorganic nitrogen forms, nitrification rates, and abundances of AOB and AOA
Ammonium concentration in DFE irrigated soils increased at the beginning, and then decreased to the initial NH4+ values at the end of the experiment (Fig. 2a), reflecting the input of NH4+ supplied by the effluent and, possibly, its later consumption by soil microorganisms (e.g., through nitrification). In contrast, this variable did not change significantly along the experiment in soil microcosms irrigated with W. Soil NO2− + NO3− concentration increased significantly after irrigation with DFE compared to the initial soil sample, whilst that in soil irrigated with W, in general, did not change significantly along the experiment (Fig. 2b). However, a slight but significant increase in NO2− + NO3− concentration in W was observed after 8 days of irrigation despite no external input of NH4+ (Fig. 2b). This could be due to a rapid stimulation of N mineralization of the soil organic matter induced by dry soil rewetting, followed by the NH4+ depletion through nitrification and microbial consumption for growth (Schimel 2018). At the end of the experiment (day 61), NO2− + NO3− concentration in DFE was significantly higher than in W. Furthermore, soil NO2− + NO3− concentration was used to calculate the net nitrification rate, which after 61 days of irrigation was significantly higher in DFE than W irrigated microcosms (W: 0.13 ± 0.03 µg NO3− + NO2−/g dry soil × day, DFE: 0.25 ± 0.02 µg NO3− + NO2−/g dry soil × day, p < 0.05).
Effects of irrigation with diluted fish-processing effluents (DFE) or freshwater (W) on soil microcosms inorganic nitrogen forms and nitrifying populations. a, NH4+ concentration (µg/g soil); b, NO3− + NO2− concentration (µg/g soil); c, bacterial and archaeal amoA gene abundances (copy numbers/µg DNA); d, nitrification rate as a function of amoA gene abundance. Soil(0): soil initial sample (before microcosms irrigation). In panels a and b, different uppercase letters indicate significant differences (p < 0.05) in soil NH4+ or NO3− + NO2− of microcosms irrigated with W among incubation times, whereas lowercase letters refer to the same comparisons in microcosms irrigated with DFE. In panel c, uppercase and lowercase letters indicate the significance of differences in bacterial and archaeal amoA genes, respectively. The results of pairwise comparisons between irrigation treatments within each date (panels a and b) or between bacterial and archaeal genes within each irrigation condition (panel c) are indicated with asterisks (**p < 0.01, *p < 0.05) or with “ns” (not significant, p > 0.05). Only the regression adjustment between nitrification rate and archaeal amoA genes is shown in panel d since that between nitrification rate and bacterial amoA genes was not significant (p > 0.05)
We quantified the abundance of bacterial and archaeal amoA genes in the initial soil sample and in soil from microcosms at the end of the experiment (day 61). A predominance of AOA over AOB has been reported in several soils (Leininger et al. 2006); particularly from arid regions, since AOA are highly resistant to the unfavorable environmental conditions of drylands (Delgado-Baquerizo et al. 2013). However, there is also evidence of AOB prevailing in drylands (Banning et al. 2015; Marcos et al. 2016); which could indicate that soil site-specific effects (e.g., degree of aridity, soil conditions and plant litter input) may modulate AOB or AOA predominance (Delgado-Baquerizo et al. 2016; Trivedi et al. 2019). In this study, the abundance of archaeal amoA genes in the initial soil sample was significantly higher than that of AOB (Fig. 2c). In agreement, previous analyses of 16S rRNA gene sequences showed that ammonia-oxidizing archaea of the family Nitrososphaeraceae were dominant among the archaeal communities of Patagonian arid soils (reaching abundances as high as 99.5% of the archaeal sequences, Marcos et al. 2019; Vallejos et al. 2020); suggesting that they may be stable and highly resistant members of the community in this arid ecosystem. Moreover, we detected a positive linear relationship between the net nitrification rate and the log10 abundance of the amoA gene of AOA (linear regression, R2 = 0.786, p = 0.003, Fig. 2d), but not with that of AOB (p = 0.819); reflecting that under the conditions of this study the former gene could be considered as an estimator of nitrification. Nevertheless, also other microorganisms not targeted in this study (like the complete ammonia oxidizers, i.e. comammox, or heterotrophic nitrifiers; Daims et al. 2015; van Kessel et al. 2015; Li et al. 2018) could be contributing to nitrification in the analyzed soils from this arid region.
Archaeal but not bacterial amoA genes were more abundant in DFE than in the W irrigated control (Fig. 2c). Such AOA difference could be not only associated with its prevalence in the soils under study, but also with the moderate ammonium concentration in the effluent used for irrigation. The dilution of the fish-processing effluent to meet the local guidelines for irrigation led to a final ammonium concentration of 17 mg/l (ca. 1 mM), which does not represent an excessive input of ammonium, as is within the range of culture media for the isolation of ammonia oxidizers adapted to low ammonia concentrations, and considerably below traditional media (with up to 20 mM ammonium) used for enrichment or isolation of AOB (Koops et al. 2006; Bollmann et al. 2011). The ammonium concentration in the diluted effluent is also below the inhibitory ammonium concentration reported for both AOA and AOB (Lehtovirta-Morley et al. 2016). In addition, while in general AOA have been associated with low nutrient soils (Delgado-Baquerizo et al. 2013); members of the Nitrososphaeraceae—as those previously detected at high abundance in soils from Patagonian arid regions—have also shown a mixotrophic or even heterotrophic metabolism, with a capability to use organic carbon compounds (Sauder et al. 2017). Most of the COD of fish-processing effluents is represented by proteins, which when degraded produce not only ammonia but also organic acids (Hwang and Hansen 1998); that could have benefited the growth of this group of AOA.
Neither archaeal nor bacterial amoA genes were influenced by the decrease in pH or the increase in soil EC induced by irrigation with DFE (Spearman RhoAOA-pH: 0.132, p = 0.699; Spearman RhoAOA-EC: − 0.100, p = 0.769; Spearman RhoAOB-pH: − 0.591, p = 0.056; Spearman RhoAOB-EC: 0.509, p = 0.110). AOA are known to prevail in acidic soils (pH < 5.5), but both groups of ammonia oxidizers are found in neutral and alkaline soils (Prosser and Nicol 2012). Even though DFE caused pH to decrease, it still remained within the pH range tolerated by both groups of ammonia oxidizers. Regarding salinity, previous studies of irrigation with saline waters showed contrasting results. Guo et al. (2020) found that the irrigation of desert soils with saline water inhibited nitrification, and reduced the abundance of the amoA gene of AOA and AOB. In contrast, other studies found that irrigation with high-salinity wastewater increased nitrification and promoted an enrichment of OTUs assigned to AOB (Frenk et al. 2015; Ibekwe et al. 2018). In addition, the AOA Candidatus Nitrosocosmicus exaquare, Candidatus Nitrosocosmicus franklandus, and Candidatus Nitrososphaera gargensis (all members of the Nitrososphaeraceae) were predominantly active in microcosms treating highly saline municipal wastewater (30 g/l) (Pan et al. 2019). Despite the fact that these AOA are thought to be non-halophilic, the authors proposed that they may be able to survive salinity stresses and be highly active after a lag period. Interestingly, one of the most abundant OTUs previously found in soils of the same study site (accounting in average 13% of the archaeal sequences) had its closest match in the NCBI database (98.9% identity) with Nitrosocosmicus franklandus (Vallejos et al. 2020); thus, the high archaeal amoA gene abundances in DFE could be associated with the presence of these AOA adapted to high soil salinity.
Conclusions
Irrigation with diluted effluents from a fish-processing factory promoted nitrification and stimulated the ammonia-oxidizing archaea inhabiting soils of an arid region of Patagonia compared to the water-irrigated control. In contrast, ammonia-oxidizing bacteria were not affected by the irrigation treatment. Irrigation with diluted fish-processing effluents also provoked changes in soil pH and EC, however, neither of those changes affected the abundances of AOA or AOB negatively. Nevertheless, although these effluents have potential as water resource and liquid fertilizers, regular soil monitoring should be considered if reused for irrigation purposes, to avoid the risk of soil salinization. Further studies should be conducted to analyze the diversity of the ammonia-oxidizers inhabiting these soils, as they seem highly resistant to salinity stress.
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
Achuthan C, Rejish Kumar VJ, Manju NJ et al (2006) Development of nitrifying bacterial consortia for immobilizing in nitrifying bioreactors designed for penaeid and non-penaeid larval rearing systems in the tropics. Indian J Mar Sci 35:240–248
Allison LE, Moodie CD (1965) Carbonate. In: Norman AG (ed) Methods of soil analysis: part 2 chemical and microbiological properties. American Society of Agronomy Inc, Madison, pp 1379–1396
Allison LE, Richards LA (1954) Diagnosis and improvement of saline and alkali soils. No. 60. Soil and Water Conservative Research Branch, Agricultural Research Service, US Department of Agriculture, Washington D.C., USA
APHA, AWWA, WEF (2017) Standard methods for the examination of water and wastewater, 23rd edn. American Public Health Association, American Water Works Association, Water Environment Federation, Washington
Ayers RS, Westcot DW (1994) Water quality for agriculture. FAO. Irrigation and drainage paper 29. Rome
Banning NC, Maccarone LD, Fisk LM, Murphy DV (2015) Ammonia-oxidising bacteria not archaea dominate nitrification activity in semi-arid agricultural soil. Sci Rep 5:11146. https://doi.org/10.1038/srep11146
Becerra-Castro C, Lopes AR, Vaz-Moreira I et al (2015) Wastewater reuse in irrigation: a microbiological perspective on implications in soil fertility and human and environmental health. Environ Int 75:117–135. https://doi.org/10.1016/j.envint.2014.11.001
Bedbabis S, Ben Rouina B, Boukhris M, Ferrara G (2014) Effect of irrigation with treated wastewater on soil chemical properties and infiltration rate. J Environ Manag 133:45–50. https://doi.org/10.1016/j.jenvman.2013.11.007
Bisigato AJ, Bertiller MB (1997) Grazing effects on patchy dryland vegetation in northern Patagonia. J Arid Environ 36:639–653. https://doi.org/10.1006/jare.1996.0247
Blumenthal UJ, Duncan Mara D, Peasey A et al (2000) Guidelines for the microbiological quality of treated wastewater used in agriculture: recommendations for revising WHO guidelines. B World Health Organ 78:1104–1116. https://doi.org/10.1590/S0042-96862000000900006
Bollmann A, French E, Laanbroek HJ (2011) Isolation, cultivation, and characterization of ammonia-oxidizing bacteria and archaea adapted to low ammonium concentrations. In: Klotz MG (ed.). Methods in enzymology, Vol. 486. Research on nitrification and related processes, part A. pp 55–88
Bouyoucos GJ (1962) Hydrometer method improved for making particle size analyses of soils. Agron J 54:464–465. https://doi.org/10.2134/agronj1962.00021962005400050028x
Chen L, Feng Q, Li C et al (2017) Impacts of aquaculture wastewater irrigation on soil microbial functional diversity and community structure in arid regions. Sci Rep 7:11193. https://doi.org/10.1038/s41598-017-11678-z
Ching YC, Redzwan G (2017) Biological treatment of fish processing saline wastewater for reuse as liquid fertilizer. Sustainability 9:1062. https://doi.org/10.3390/su9071062
Cristóvão RO, Botelho CM, Martins RJE et al (2015) Fish canning industry wastewater treatment for water reuse—a case study. J Clean Prod 87:603–612. https://doi.org/10.1016/j.jclepro.2014.10.076
Daims H, Lebedeva EV, Pjevac P et al (2015) Complete nitrification by Nitrospira bacteria. Nature 528:504–509. https://doi.org/10.1038/nature16461
de Melo Ribeiro FH, dos Santos Silva Y, Pena Naval L (2020) Auxiliary strategies for water management in industries: minimization of water use and possibility of recycling and/or reuse of effluent. In: Sabban A (ed.). Innovation in global green technologies 2020. IntechOpen. pp 79–93
Delgado-Baquerizo M, Gallardo A, Wallenstein MD, Maestre FT (2013) Vascular plants mediate the effects of aridity and soil properties on ammonia-oxidizing bacteria and archaea. FEMS Microbiol Ecol 85:273–282. https://doi.org/10.1111/1574-6941.12119
Delgado-Baquerizo M, Maestre FT, Eldridge DJ, Singh BK (2016) Microsite differentiation drives the abundance of soil ammonia oxidizing bacteria along aridity gradients. Front Microbiol 7:505. https://doi.org/10.3389/fmicb.2016.00505
Drury CF, Hart SC, Yang XM (2007) Nitrification techniques for soils. In: Carter MR, Gregorich EG (eds) Soil sampling and methods of analysis, 2nd edn. CRC Press, Boca Raton, pp 495–513
FAO (2014) FAO statistical yearbook 2014. Latin America and the Caribbean food and agriculture. Santiago
FAO (2020) The state of world fisheries and aquaculture 2020. Sustainability in action. Rome
Francis CA, Roberts KJ, Beman JM et al (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102:14683–14688. https://doi.org/10.1073/pnas.0506625102
Frenk S, Dag A, Yermiyahu U et al (2015) Seasonal effect and anthropogenic impact on the composition of the active bacterial community in Mediterranean orchard soil. FEMS Microbiol Ecol 91:fiv096. https://doi.org/10.1093/femsec/fiv096
Ganjegunte G, Ulery A, Niu G, Wu Y (2017) Effects of treated municipal wastewater irrigation on soil properties, switchgrass biomass production and quality under arid climate. Ind Crops Prod 99:60–69. https://doi.org/10.1016/j.indcrop.2017.01.038
Guimarães JT, Souza ALM, Brígida AIS et al (2018) Quantification and characterization of effluents from the seafood processing industry aiming at water reuse: a pilot study. J Water Process Eng 26:138–145. https://doi.org/10.1016/j.jwpe.2018.10.006
Guo H, Ma L, Liang Y et al (2020) Response of ammonia-oxidizing Bacteria and Archaea to long-term saline water irrigation in alluvial grey desert soils. Sci Rep 10:489. https://doi.org/10.1038/s41598-019-57402-x
Gwon BG, Kim JK (2012) Feasibility study on production of liquid fertilizer in a 1 m3 reactor using fishmeal wastewater for commercialization. Environ Eng Res 17:3–8. https://doi.org/10.4491/eer.2012.17.1.003
Haynes RJ, Swift RS (1989) Effect of rewetting air-dried soils on pH and accumulation of mineral nitrogen. J Soil Sci 40:341–347. https://doi.org/10.1111/j.1365-2389.1989.tb01278.x
Hwang S, Hansen CL (1998) Formation of organic acids and ammonia during acidogenesis of trout-processing wastewater. Trans Am Soc Agric Eng 41:151–156. https://doi.org/10.13031/2013.17139
Ibekwe AM, Gonzalez-Rubio A, Suarez DL (2018) Impact of treated wastewater for irrigation on soil microbial communities. Sci Total Environ 622–623:1603–1610. https://doi.org/10.1016/j.scitotenv.2017.10.039
Jung HY, Kim JK (2016) Eco-friendly waste management of mackerel wastewater and enhancement of its reutilization value. Int Biodeterior Biodegrad 111:1–13. https://doi.org/10.1016/j.ibiod.2016.04.002
Keeney DR, Nelson DW (1982) Nitrogen—Inorganic forms. In: Page AL (ed.). Methods of soil analysis, Part 2. Chemical and microbiological properties. American society of agronomy, Inc., and Soil science society of America Inc., Madison, WI, pp 643–698
Keith LH (1996) Compilation of EPA´s sampling and analysis methods, 2nd edn. Lewis publishers, Boca Raton
Koops HP, Purkhold U, Pommerening-Röser A et al (2006) The lithoautotrophic ammonia-oxidizing bacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes. Springer, New York, pp 778–811
Lehtovirta-Morley LE, Ross J, Hink L et al (2016) Isolation of “Candidatus Nitrosocosmicus franklandus”, a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol Ecol 92:fiw057. https://doi.org/10.1093/femsec/fiw057
Leininger S, Urich T, Schloter M et al (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. https://doi.org/10.1038/nature04983
León RJC, Bran D, Collantes M et al (1998) Grandes unidades de vegetación de la Patagonia extra andina. Ecol Aust 8:125–144
Li XG, Li FM, Rengel Z et al (2006) Cultivation effects on temporal changes of organic carbon and aggregate stability in desert soils of Hexi Corridor region in China. Soil Tillage Res 91:22–29. https://doi.org/10.1016/j.still.2005.10.004
Li Y, Chapman SJ, Nicol GW, Yao H (2018) Nitrification and nitrifiers in acidic soils. Soil Biol Biochem 116:290–301. https://doi.org/10.1016/j.soilbio.2017.10.023
Marcos MS, Bertiller MB, Saraví Cisneros H, Olivera NL (2016) Nitrification and ammonia-oxidizing bacteria shift in response to soil moisture and plant litter quality in arid soils from the Patagonian Monte. Pedobiol J Soil Ecol 59:1–10. https://doi.org/10.1016/j.pedobi.2015.11.002
Marcos MS, Bertiller MB, Olivera NL (2019) Microbial community composition and network analyses in arid soils of the Patagonian Monte under grazing disturbance reveal an important response of the community to soil particle size. Appl Soil Ecol 138:223–232. https://doi.org/10.1016/j.apsoil.2019.03.001
Mishra SS, Markande AR, Keluskar RP et al (2015) Simultaneous nitrification and denitrification by novel heterotrophs in remediation of fish processing effluent. J Basic Microbiol 55:772–779. https://doi.org/10.1002/jobm.201400783
Oved T, Shaviv A, Goldrath T et al (2001) Influence of effluent irrigation on community composition and function of ammonia-oxidizing bacteria in soil. Appl Environ Microbiol 67:3426–3433. https://doi.org/10.1128/AEM.67.8.3426-3433.2001
Pan K-L, Gao J-F, Li D-C, Fan X-Y (2019) The dominance of non-halophilic archaea in autotrophic ammonia oxidation of activated sludge under salt stress: a DNA-based stable isotope probing study. Bioresour Technol 291:121914. https://doi.org/10.1016/j.biortech.2019.121914
Pereyra FX, Bouza P (2019) Soils from the Patagonian region. In: Rubio G, Lavado RS, Pereyra FX (eds) The soils of Argentina. Springer, Madison, pp 101–121
Prosser JI (2011) Soil nitrifiers and nitrification. In: Ward BB, Arp DJ, Klotz MG (eds) Nitrification. ASM Press, Washington, pp 347–383
Prosser JI, Nicol GW (2012) Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20:523–531. https://doi.org/10.1016/j.tim.2012.08.001
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Santoyo Figueroa JG, Jung HY, Jeong G-T, Kim JK (2015) The high reutilization value potential of high-salinity anchovy fishmeal wastewater through microbial degradation. World J Microbiol Biotechnol 31:1575–1586. https://doi.org/10.1007/s11274-015-1906-2
Sauder LA, Albertsen M, Engel K et al (2017) Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J 11:1142–1157. https://doi.org/10.1038/ismej.2016.192
Schimel JP (2018) Life in dry soils: effects of drought on soil microbial communities and processes. Annu Rev Ecol Evol Syst 49:409–432. https://doi.org/10.1146/annurev-ecolsys-110617-062614
Shand CA, Williams BL, Coutts G (2008) Determination of N-species in soil extracts using microplate techniques. Talanta 74:648–654. https://doi.org/10.1016/j.talanta.2007.06.039
Tarchouna LG, Merdy P, Raynaud M et al (2010) Effects of long-term irrigation with treated wastewater. Part I: evolution of soil physico-chemical properties. Appl Geochem 25:1703–1710. https://doi.org/10.1016/j.apgeochem.2010.08.018
Tournier E, Amenc L, Pablo AL et al (2015) Modification of a commercial DNA extraction kit for safe and rapid recovery of DNA and RNA simultaneously from soil, without the use of harmful solvents. MethodsX 2:182–191. https://doi.org/10.1016/j.mex.2015.03.007
Trivedi C, Reich PB, Maestre FT et al (2019) Plant-driven niche differentiation of ammonia-oxidizing bacteria and archaea in global drylands. ISME J 13:2727–2736. https://doi.org/10.1038/s41396-019-0465-1
US EPA (1993) Method 410.4. The determination of chemical oxygen demand by semi-automated colorimetry. Cincinnati, OH
Vallejos MB, Marcos MS, Barrionuevo C, Olivera NL (2020) Fish-processing effluent discharges influenced physicochemical properties and prokaryotic community structure in arid soils from Patagonia. Sci Total Environ 714:136882. https://doi.org/10.1016/j.scitotenv.2020.136882
van Kessel MAHJ, Speth DR, Albertsen M et al (2015) Complete nitrification by a single microorganism. Nature 528:555–559. https://doi.org/10.1038/nature16459
Veiga MC, Méndez R, Lema JM (1994) Waste water treatment for fisheries operations. In: Martin AM (ed) Fisheries processing. Springer, Boston, pp 344–369
Vela-Cano M, Gómez-Brandón M, Pesciaroli C et al (2018) Study of total bacteria and ammonia-oxidizing bacteria and ammonia-oxidizing archaea in response to irrigation with sewage sludge compost tea in agricultural soil. Compost Sci Util 26:145–155. https://doi.org/10.1080/1065657X.2018.1432429
Wessén E, Hallin S (2011) Abundance of archaeal and bacterial ammonia oxidizers—Possible bioindicator for soil monitoring. Ecol Indic 11:1696–1698. https://doi.org/10.1016/j.ecolind.2011.04.018
World Economic Forum (2020) The global risks report 2020
World Resources Institute (2019) RELEASE: update global water risk atlas reveals top water-stressed countries and states. Washington D.C., USA
Xie G, Lahav I, Barness G, Steinberger Y (2001) Dynamics of soil nitrogen along a topoclimatic gradient in the Judean desert. Arid Land Res Manag 15:135–146. https://doi.org/10.1080/15324980151062760
Zabaloy MC, Allegrini M, Tebbe DA et al (2017) Nitrifying bacteria and archaea withstanding glyphosate in fertilized soil microcosms. Appl Soil Ecol 117–118:88–95. https://doi.org/10.1016/j.apsoil.2017.04.012
Funding
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica [grant numbers PICT 2013-1505, 2015-1689, and 2019-01480] and the Consejo Nacional de Investigaciones Científicas y Técnicas [PUE IPEEC N° 22920160100044].
Author information
Authors and Affiliations
Contributions
Designed the study, acquired financial support, provided reagents/materials/laboratory supplies, supervised the research activities, and wrote the paper: MSM and NLO; collected samples and performed experiments: MSM, MCG, MBV, CGB, and NLO; analyzed data: MSM and MCG; prepared graphical arts: MSM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Marcos, M.S., González, M.C., Vallejos, M.B. et al. Impact of irrigation with fish-processing effluents on nitrification and ammonia-oxidizer abundances in Patagonian arid soils. Arch Microbiol 203, 3945–3953 (2021). https://doi.org/10.1007/s00203-021-02358-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02358-8