Abstract
The chb operon of Escherichia coli is involved in the utilization of chitooligosaccharides. While acquisition of two classes of mutations leading to altered regulation of the chb operon is necessary to confer the ability to utilize the glucose disaccharide cellobiose to wild-type strains of E. coli, in the closely related organism Shigella sonnei, Cel+ mutants arise relatively faster, requiring only a single mutational event. In Type I mutants, the insertion of IS600 at −21 leads to ChbR regulator-independent, constitutive expression of the operon. In Type II mutants, the insertion of IS2/600 within the distal binding site of the negative regulator NagC leads to ChbR-dependent cellobiose-inducible expression of the operon. These studies underscore the significance of strain background, specifically the diversity of transposable elements, in the evolution of novel metabolic functions. Constitutive expression of the chb operon also enables utilization of the aromatic β-glucosides arbutin and salicin, implying that the chb structural genes are inherently promiscuous.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacteria are versatile organisms that exist in a variety of ecological niches. Among the different types of stress that they face in their natural habitats, nutrient scarcity is very prominent. When grown in the presence of novel substrates, bacteria often evolve the ability to utilize them through modifications of preexisting genetic systems, primarily at the regulatory level (Patrick et al. 2007; Wright 2004). Recent studies have demonstrated this in the case of the naturally occurring aromatic β-glucosides salicin, arbutin, and esculin (Zangoui et al. 2015).
Cellobiose, a cellulose-derived abundant β-glucoside in nature, cannot be utilized by many wild-type strains of Escherichia coli as well as several members of Enterobacteriaceae. However, spontaneous Cel+ mutants can arise from Cel− E. coli K12 strains as papillae on MacConkey cellobiose medium after prolonged exposure of 20–30 days (Kachroo et al. 2007; Kricker and Hall 1984; Parker and Hall 1990a, b). The Cel+ phenotype of these mutants was attributed to the cel operon, originally believed to be a cryptic genetic system which required activating mutations to enable cellobiose utilization (Parker and Hall 1990b). The operon was later renamed as chb operon when studies showed that the wild-type operon is involved in the utilization of chitobiose and chitotriose, which are products of chitin degradation (Keyhani and Roseman 1997).
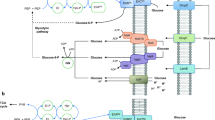
The chb operon is an inducible system consisting of six genes with specific roles in transport, hydrolysis, modification of the substrate, and regulation of the operon (Fig. 1a). The chbB, chbC, and chbA genes together constitute the permease that transports chitobiose across the inner membrane and concomitantly phosphorylates it, resulting in the accumulation of intracellular chitobiose-6-phosphate (Keyhani et al. 2000a, b, c). The chbF gene codes for a phospho-β-glucosidase required for hydrolysis of the substrate (Thompson et al. 1999). The chbR gene codes for an AraC family dual-function repressor/activator involved in the transcriptional regulation of the operon (Plumbridge and Pellegrini 2004). The chbG gene encodes a monodeacetylase that removes the acetyl group from the non-phosphorylated end of chitobiose, which is essential for induction of the operon and hydrolysis of the sugar (Verma and Mahadevan 2012).
a Organization of the chb operon. b Nucleotide sequence of the wild-type chb promoter. The positions of all regulator binding sites with respect to the transcription start site (TSS) are marked. The inherent −10 and −35 elements of the promoter are shown in boxes. c Location of IS element insertions within the chb promoter of Cel+ mutants Type I mutants harbor IS600 insertion at −21 while Type II mutants have IS2 insertion at −113 or IS600 insertion at −114
Transcription of the chb operon is regulated by the three transcription factors NagC, ChbR, and CRP (Fig. 1b, c). In the absence of chitobiose, NagC and ChbR together lead to negative regulation of the operon. In the presence of chitobiose, NagC repression is removed and ChbR along with CRP activates the operon. Hydrolysis of chitobiose-6-phosphate, but not of cellobiose-6-phosphate, results in the production of N-acetyl glucosamine-6-phosphate, which acts as an inducer for relieving NagC repression (Plumbridge and Pellegrini 2004).
The Cel+ phenotype of E. coli K12 is contributed by two classes of mutations that alter the regulation of the chb operon. The first class of mutations either within the nagC locus or at the NagC binding site within the chb promoter abrogates NagC repression. The second class consisting of gain-of-function mutations in chbR enables the enhanced recognition of cellobiose as an inducer by ChbR resulting in the activation of the operon. Acquisition of both classes of mutations is necessary and sufficient for the manifestation of a Cel+ phenotype in E. coli (Kachroo et al. 2007), partly explaining the prolonged exposure on cellobiose necessary for the appearance of the Cel+ mutants. This was contrary to an earlier report which suggested that Cel+ phenotype in E. coli arises due to either of these mutational events (Parker and Hall 1990a). However, the study failed to explain the necessity for prolonged exposure on cellobiose to obtain the Cel+ mutants. Additionally, there were many differences in the nucleotide sequence of the chb operon of the strain used in the study (Parker and Hall 1990b) and the E. coli K12 genome sequence published subsequently. These inherent differences between the strains used could have led to the differences in the results reported in the two studies (Kachroo et al. 2007).
Though Shigella and Escherichia genuses are phylogenetically very close, marked differences have been observed in the genetic systems involved in the metabolism of other β-glucosides like salicin and arbutin (Desai et al. 2010; Kharat and Mahadevan 2000). In an attempt to understand the mechanism of cellobiose utilization in Shigella sonnei in comparison with E. coli, Cel+ mutants were isolated from a Cel− wild-type strain of S. sonnei. Interestingly, Cel+ papillae appeared on MacConkey cellobiose agar relatively faster in the case of S. sonnei compared to E. coli. Deletion of the chb operon or the phospho-β-glucosidase gene, chbF, in these Cel+ mutants resulted in the loss of the Cel+ phenotype, confirming the role of the chb operon in these mutants. These strains were seen to harbor insertions either between the −10 and −35 sequences of the chb promoter leading to ChbR- and cellobiose-independent expression of the chb operon (Type I) or within the strong NagC binding site resulting in ChbR- and cellobiose-dependent expression of the chb operon (Type II). The mechanism of chb operon activation in both types of mutants has been characterized, and these results are described below. These studies also reveal that the transport and catabolic functions encoded by the chb operon can enable the catabolism of salicin and arbutin once the regulation of the chb genes is bypassed.
Materials and methods
Bacterial strains and plasmids
The strains used in the study are listed in Table 1. The plasmids used are listed in Table 2.
Media and growth conditions
Bacterial strains were normally grown in Luria-Bertani (LB) broth or agar (1.5 %) at 37 °C. Beta-glucoside utilization was checked by growth on MacConkey indicator agar media supplemented with 1 % cellobiose, arbutin, or salicin. Strains that could utilize the sugar formed bright red colonies while the strains that could not utilize the sugar formed yellow colonies. Growth on cellobiose was also tested using M9 minimal agar containing 0.4 % cellobiose as sole carbon source. M9 minimal agar with 0.4 % glycerol was used as a positive control. Antibiotic concentrations used were as follows: ampicillin −100 µg/ml, chloramphenicol −15 µg/ml, tetracycline −10 µg/ml.
Construction of plasmids
For cloning chbR, chbF, and chbRF into pACDH vector, these sequences were amplified from the S. sonnei strain AK1 using flanking primers (Online resource 1 Table S1) and ligated with pTZ57R/T vector using InsTAclone PCR cloning kit (Thermo Fisher Scientific). The identity of the clones was confirmed by sequencing the complete insert. The positive clones containing chbR, chbF, or chbRF were digested with Sac1/EcoR1 enzyme pair and ligated with pACDH vector digested with the same enzyme pair. For cloning chbR from S. sonnei and E. coli into pBR322, the chbR sequences were amplified from strains AK1 and MG1655, respectively. The purified PCR products were digested with EcoR1/BamH1 enzyme pair and ligated with pBR322 vector digested with the same enzyme pair.
DNA sequencing
DNA sequencing was carried out commercially at Macrogen Inc., South Korea. Both strands were sequenced in all cases. All available sequences were retrieved from Ecocyc and NCBI databases, and alignments were carried out using NCBI nucleotide BLAST.
Generation of chbR, chbF, and chbBCARF knockout strains
Knockout strains were constructed in appropriate mutant background using λ red gam recombination method as described previously (Datsenko and Wanner 2000). Briefly, hybrid cassettes used for replacement were generated by PCR amplification of the chloramphenicol resistance gene from pKD3 plasmid using specific primers with flanking regions corresponding to the genes to be deleted. These hybrid cassettes were electroporated into strains expressing λ red gam genes and plated on chloramphenicol-containing plates to select for resistant colonies. The knockout strains were confirmed using the corresponding confirmatory primers (Online resource 1 Table S1).
β-galactosidase assay
Assays for lacZ reporter gene activity were carried out as described by Miller (Miller 1972). Cells were grown in M9 minimal media containing 0.4 % glycerol as carbon source, in the absence or presence of 10 mM cellobiose (Sigma-Aldrich). For each assay, a minimum of three independent measurements were used to calculate β-galactosidase activity expressed in Millers units.
Total RNA isolation and quantitative RT-PCR (qRT-PCR)
For total RNA isolation, the strains were grown in LB broth in the absence or presence of 10 mM cellobiose for 8 h. RNA was isolated from these cultures using the acid phenol method as previously described (Singh et al. 1995) and was quantified using a Nanodrop 1000 spectrophotometer. After DNase treatment to remove genomic DNA contamination, 2 µg of RNA was used for cDNA synthesis using RevertAid First-Strand cDNA Synthesis Kit (Thermo Scientific) as per manufacturer’s instructions. cDNA equivalent to 10 ng RNA was used for qRT-PCR using Maxima SYBR Green qPCR Kit (Thermo Scientific) according to the manufacturer’s instructions on an Applied Biosystems StepOne-Plus real-time PCR system (version 2.2.3). The primers used are listed in Online resource 1 Table S1. All reactions were carried out in technical triplicates for two biological replicates. Transcript levels of chb operon were normalized to rrnC transcript levels (16S rRNA), and fold change was calculated as previously described (Schmittgen and Livak 2008). Data were analyzed, and statistical significance was calculated using Bonferroni’s multiple comparison test following one-way ANOVA or unpaired t test in GraphPad Prism 5. P values ≤0.05 were considered significant.
Results
Isolation of Cel+ mutants from S. sonnei
The wild-type S. sonnei strain AK1 is unable to utilize arbutin, salicin, and cellobiose and forms yellow colonies on MacConkey medium containing any of these β-glucosides. Previous studies from the laboratory have shown that the Arb− Sal− Cel− strain AK1 gives rise to Arb+ Sal− Cel− mutants in two days by activation of the silent bgl operon (Kharat and Mahadevan 2000). The S. sonnei phospho-β-glucosidase gene bglB is inactive and cannot enable the utilization of arbutin or salicin. Arbutin utilization in these mutants is facilitated by transport function provided by BglF and hydrolysis by BglA, an unlinked phospho-β-glucosidase specific for arbutin. The Arb+ Sal− Cel− mutant gives rise to Arb+ Sal+ Cel− mutants in five days through activation of another phospho-β-glucosidase SSO1595, which is absent in most commensal strains of E. coli including K12. Thus, salicin utilization in these mutants is the result of the combined action of BglF for transport and SSO1595 for hydrolysis (Desai et al. 2010; Kharat and Mahadevan 2000). It was observed that the Arb+ Sal+ Cel− strain can mutate to Arb+ Sal+ Cel+ in two days. Further characterization of one of the mutants, RsC3, showed that the Cel+ phenotype in this mutant was contributed by the partial activation of the chb operon leading to transport of cellobiose by ChbBCA and hydrolysis of cellobiose by SSO1595. The partial activation of the chb operon was caused by an A to G transition at position 550 within chbR, resulting in a single amino acid change, K184E in ChbR (Sonowal and Mahadevan unpublished). This mutation was one of the effective gain-of-function mutations reported among the Cel+ mutants of E. coli (Kachroo et al. 2007).
Cel+ mutants could also be isolated directly from AK1 as red papillae on MacConkey cellobiose agar within 5–10 days of incubation. This was a relatively shorter period compared to the E. coli Cel+ mutants, which required 20–30 days of incubation. All Cel+ mutants isolated from S. sonnei could grow on minimal media containing cellobiose as the sole carbon source. Surprisingly, the frequency of Cel+ mutants arising from S. sonnei was an order of magnitude lower than that from E. coli, and no papillae appeared outside the time window of 5–10 days. Five single-step Cel+ mutants of S. sonnei, isolated from four independent experiments, were used for all further characterization.
Cellobiose utilization in the S. sonnei Cel+ mutants is linked to the chb operon
The Cel+ phenotype in S. sonnei could be achieved through activation of either the chb operon, as in E. coli, or any other unknown genetic systems unique to S. sonnei. In order to confirm the role of the chb operon, we deleted the chbBCARF genes from the S. sonnei Cel+ mutants AK104 and AM1. The deletion resulted in loss of the Cel+ phenotype in these mutants confirming their role in conferring the phenotype (Online resource 1 Fig. S1a).
One possibility for the Cel+ phenotype is the partial activation of the chb operon resulting in the expression of low levels of the permease, which allows transport of cellobiose into the cell, and hydrolysis of cellobiose by an unlinked enzyme, as seen in the case of RsC3, the three-step activated Cel+ mutant of S. sonnei. However, deletion of chbF also led to the loss of Cel+ phenotype (Online resource 1 Fig. S1b), suggesting that ChbF is the enzyme responsible for the hydrolysis function in these mutants. These results indicate that in the one-step activated Cel+ mutants of S. sonnei, the chb operon contributes to both transport and hydrolysis of cellobiose.
The chbR and nagC loci of S. sonnei Cel+ mutants do not show any genetic alteration
All Cel+ mutants of E. coli carry gain-of-function mutations in the chbR locus, and these mutations were necessary for conferring the Cel+ phenotype in E. coli (Kachroo et al. 2007). Sequencing of the complete chbR locus from S. sonnei Cel+ mutants showed that the chbR loci from all the mutants were identical to that of the wild-type strain AK1. Additionally, the nagC locus of these mutants also remained identical to that of the wild-type strain, suggesting that the abrogation of NagC repression in all these mutants occurred through a different mechanism.
The Cel+ mutants of S. sonnei harbor insertions in the chb promoter region
One possible mechanism of activation of the chb operon is by rearrangements of the promoter. To test this possibility, the chb promoter region was PCR-amplified using specific flanking primers. While the amplicon corresponding to the wild-type chb promoter was ~350 bp long, the promoter fragments of all Cel+ mutants were close to 1.6 kb long, indicating the presence of insertions within the chb promoter of all mutants. Sequencing of these promoters confirmed the presence of ~1.2-kb-long IS elements within the promoter. Three of the five mutants referred to as Type I mutants (AM1, AM3, and AM4) had identical insertion of IS600 at position −21. The other two mutants referred to as Type II mutants had IS2 (in AK104) or IS600 (in AM2) insertion within the distal but strong NagC binding site, at positions −113 and −114, respectively (Fig. 1c).
Insertion of IS600 in Type I mutants leads to constitutive activation of the chb operon
One possible consequence of IS600 insertion at −21 within the chb promoter in Type I mutants is the creation of a new −35 element which is closer to the consensus −35 sequence (Fig. 2). The insertion also distances the ChbR, CRP, and strong NagC binding sites by ~1.2 kb from the transcription start site, which could make the promoter independent of these regulators. Activation could be a consequence of both events. Surprisingly, when chbR was deleted from AM1 (Type I mutant), the Cel+ phenotype was lost, but it could not be restored on transformation with a plasmid expressing wild-type chbR. Interestingly, expression of chbF immediately downstream to chbR could rescue the phenotype in the chbR knockout strain of AM1 (Online resource 1 Fig. S2). These results indicate that replacement of chbR with the antibiotic cassette exerted a polar effect on the expression of the downstream gene chbF, which is indispensible for the manifestation of the Cel+ phenotype. The observation that the loss of the Cel+ phenotype caused by deletion of chbR could be restored only with expression of chbF and not with chbR indicates that chb transcription in Type I mutants is independent of ChbR.
To check the impact of the IS600 insertion on chb transcription, we analyzed chb mRNA levels in Type I mutants in the presence or absence of the inducer cellobiose. The chb transcript levels were significantly higher in Type I mutants regardless of the presence or absence of cellobiose compared to the wild-type strain (Fig. 3a), indicating that the chb operon is constitutively active in Type I mutants. These results are consistent with the predicted role of the IS600 element in activation by providing a stronger −35 element as well as delinking the regulatory elements from the chb promoter.
Expression of the chb operon in Type I and Type II mutants. Steady-state levels of chb mRNA from AK1 (wild type), AM1 (Type I) (a) and AK104 (Type II) (b) strains in the absence or presence of 10 mM cellobiose measured using real-time RT-PCR. The y-axis represents the fold change in chb transcript expression relative to the Cel− wild-type strain in the absence of cellobiose. Error bars represent standard deviations obtained from a minimum of two independent biological replicates, each carried out in technical triplicates
The insertion of IS elements in Type II mutants leads to cellobiose-inducible activation
The Type II mutants (AK104 and AM2) had IS element insertions within the distal, strong NagC binding site (Fig. 1c). AK104 harbors an IS2 insertion at −113, while AM2 harbors an IS600 insertion at −114 with respect to the transcription start site. This could lead to disruption of NagC binding to the chb promoter, thus relieving the promoter from NagC-mediated repression. Nevertheless, the CRP and ChbR binding sites are intact in these mutants, suggesting that activation of the operon remains ChbR-dependent. Deletion of chbR resulted in the loss of the Cel+ phenotype, which could not be restored with expression of chbR or chbF alone, indicating that the disruption of chbF expression due to polar effect of chbR deletion is not the only reason for the loss of phenotype. To circumvent this problem, we cloned the chbRF open reading frames from S. sonnei into the low copy number vector pACDH. When the strain AK104∆chbR was transformed with this plasmid construct enabling simultaneous expression of chbR and chbF, the Cel+ phenotype could be regained (Online resource 1 Fig. S3). These results indicate that Type II Cel+ mutants are dependent on ChbR for chb operon activation and ChbF is involved in cellobiose hydrolysis.
To examine the inducibility of the chb promoter in these mutants, we analyzed chb transcript levels in the presence or absence of the inducer, cellobiose. Compared to the wild-type strain, the basal transcription from the chb promoter is significantly higher in Type II mutants. This increase is further amplified in the presence of cellobiose (Fig. 3b), indicating that in Type II mutants, the chb promoter remains ChbR-dependent and cellobiose-inducible.
The activation of the chb operon in Type II mutants is caused by a combination of NagC derepression and a direct effect of IS element insertion on transcription
Previous work from our laboratory has shown that gain-of-function mutation in chbR is essential to give rise to Cel+ phenotype in E. coli. The wild-type ChbR allele of S. sonnei is different from that of E. coli by a single amino acid (A197T). To check whether wild-type S. sonnei chbR can induce transcription more efficiently from the chb promoter compared to wild-type E. coli ChbR in combination with NagC derepression, an E. coli reporter strain JMchb22, with a chromosomal copy of a chb-lacZ fusion and deletion of both nagC and chbR loci (Plumbridge and Pellegrini 2004) was used. Transcriptional activity of the chb promoter in the strain was measured as β-galactosidase activity after introduction of the vector plasmid (pBR322), plasmid containing the wild-type chbR allele from E. coli (pBR322-ECchbR), wild-type chbR allele from S. sonnei (pBR322-SSchbR), or the chbRN238 allele (pBR322-chbRN238S). The chbRN238S allele was one of the predominant alleles identified from E. coli Cel+ mutants (Kachroo et al. 2007). While the presence of chbRN238S allele led to substantial induction of transcription from the chb promoter, the wild-type alleles from both E. coli and S. sonnei failed to induce transcription beyond the basal levels in the presence of cellobiose (Fig. 4). Further, consistent with the previous results (Kachroo et al. 2007), transformation of JMchb22 strain with chbRN238S allele resulted in Cel+ colonies, while that with wild-type chbR alleles from both E. coli and S. sonnei resulted in Cel− colonies on MacConkey cellobiose plates. These observations indicate that the wild-type Shigella sonnei chbR allele cannot function similar to chbR alleles of E. coli that carry gain-of-function mutations in the absence of NagC repression in E. coli.
Induction of the chb promoter in the presence of chbR variants in E. coli. Activation of the chb promoter in the reporter strain JMchb22 (∆chbR ∆nagC) in the presence of wild-type chbR allele from E. coli, wild-type chbR allele from S. sonnei or the chbRN238S allele, measured as β-galactosidase activity. Error bars represent standard deviations obtained from a minimum of three independent biological replicates
To elucidate the exact mechanism by which the Type II mutants of S. sonnei acquire Cel+ phenotype by insertion of IS elements within the NagC binding site, we checked whether NagC derepression alone was sufficient to cause the Cel+ phenotype in S. sonnei. This was achieved by knocking out the complete nagC locus in the S. sonnei wild-type strain AK1. Deletion of the nagC locus was not sufficient to give a Cel+ phenotype (Online resource 1 Fig. S4), indicating that in addition to relieving NagC-mediated repression, the IS element also exerts a cis-effect on chb operon activation in Type II mutants. This was further confirmed by monitoring chb transcript levels in AK1 (wild type), AK1∆nagC (wild-type strain with a deletion of the nagC locus), AK104 and AM2 (Type II mutants), both in the absence and presence of the inducer cellobiose. While the basal level of chb transcription in AK1∆nagC was moderately higher than AK1, basal transcription from the chb promoter in Type II mutants was significantly higher than both AK1 and AK1∆nagC (Fig. 5). While the AK1∆nagC strain showed a twofold induction in the presence of cellobiose, there was a dramatic increase in transcription in the mutant strains AK104 and AM2 in the presence of cellobiose. These observations indicate that the Cel+ phenotype of the Type II mutants is a combined effect of the moderate enhancement of transcription from the chb promoter due to the loss of NagC binding as well as the augmentation of transcription brought about by the insertion event. This augmentation could be by providing additional promoter elements or by altering the sequence context of the existing promoter.
Comparison of the expression of the chb operon in AK1 (wild type), AK1∆nagC (NagC derrepressed), AK104 and AM2 (Type II) strains. Steady-state levels of chb mRNA in the absence or presence of 10 mM cellobiose were measured using real-time RT-PCR. The y-axis represents the fold change in chb expression relative to the Cel− wild-type strain in the absence of cellobiose. Error bars represent standard deviations obtained from a minimum of two independent biological replicates, each carried out in technical triplicates
Strains carrying deletion of nagC can acquire Cel+ phenotype by accumulating gain-of-function mutations in chbR
Interestingly, the strain AK1∆nagC could give rise to multiple Cel+ papillae after 2–4 days of incubation on MacConkey cellobiose agar, indicating the need for additional mutations for cellobiose utilization in the absence of NagC repression. Sequencing analysis of the chbR locus from ten Cel+ papillae isolated from AK1∆nagC confirmed the presence of gain-of-function mutations in these mutants (Table 3). These mutations were similar to the ones previously reported in E. coli Cel+ mutants (Kachroo et al. 2007). Though N238S was the most prominent mutation reported in E. coli, variants of this mutation where aspargine (N) was replaced by either lysine or threonine were also seen in S. sonnei. Y30C has been reported both in E. coli and in a Cel+ natural isolate whose chb locus resembles E. coli O157:H7 (Kachroo et al. 2007). K184E was seen in both E. coli Cel+ mutants and S. sonnei three-step activated Cel+ mutant, RsC3.
Salicin and arbutin utilization by S. sonnei Cel+ mutants
In an attempt to verify whether the Cel+ mutants have an expanded ability to respond to the aromatic β-glucosides arbutin and salicin, the mutants were scored on MacConkey arbutin/salicin media in the absence of cellobiose. All Type I Cel+ mutants of S. sonnei with insertion between −10 and −35 elements of the promoter resulting in constitutive chb expression showed an Arb+ Sal+ phenotype. Interestingly, Type II mutants that show inducible chb expression failed to show an Arb+ Sal+ phenotype in the absence of cellobiose. Furthermore, chbBCA and chbF expressed from a heterologous promoter in the wild-type strain AK1 resulted in an Arb+ Sal+ phenotype (Online resource 1 Fig. S5). Therefore, constitutive activation of the chb operon can lead to utilization of arbutin and salicin without additional mutations in other genetic systems, indicating that the transport and hydrolytic functions encoded by the chb operon are promiscuous and can function on a variety of β-glucosides.
Discussion
Though the Shigella genus has been grouped separately from E. coli due to its pathogenic significance, the genus status of Shigella is quite debated and many studies show that Shigella falls within the Escherichia genus. It is believed that Shigella species were derived from E. coli through multiple independent origins, which subsequently over a period of time underwent convergent evolution to form a single pathovar with enteroinvasive E. coli (EIEC) (Lan and Reeves 2002; Peng et al. 2009). It has been proposed that the three main clusters comprising most strains of S. boydii, S. flexneri, and S. dysenteriae evolved independently from different E. coli ancestors 35,000–270,000 years ago (Pupo et al. 2000), while S. sonnei emerged around 10,000 years ago, as a human pathogenic clone of E. coli (Shepherd et al. 2000). In spite of genetic similarities, they show major differences in catabolic functions like utilization of lactate and mucate. Shigella species have lost many gene functions, including several catabolic pathways, befitting their pathogenic lifestyle (Lan and Reeves 2002). One of the striking features of Shigella spp. is the presence of hundreds of IS elements within their genomes. These IS elements are responsible for causing many DNA rearrangements like translocations, deletions, and insertions. It is speculated that the high density of IS elements is a reason for the DNA rearrangements which partly reshaped the Shigella genome through the course of evolution (Yang et al. 2005).
Previous studies from our laboratory have shown that the mechanism of activation of genetic systems responsible for β-glucoside utilization is markedly different between E. coli and a sewage isolate of S. sonnei, AK1 (Desai et al. 2010; Kharat and Mahadevan 2000). The experiments described in this study were aimed at understanding the differences and similarities in chb operon activation for cellobiose utilization between these two closely related organisms. One of the significant observations of the present study is the relatively shorter duration taken for S. sonnei Cel+ mutants to appear (5–10 days), compared to E. coli. This may be related to the fact that while E. coli requires two mutational events, S. sonnei needs only a single insertion event to acquire a Cel+ phenotype. In E. coli, NagC derepression acquired either through loss-of-function mutations at the nagC locus or insertions within the NagC binding site could provide a low-level growth advantage to these mutants, facilitating gain-of-function mutations accumulating in the chbR locus (Kachroo et al. 2007). In S. sonnei, this mechanism of activation involving two mutational steps seems to be absent, though the chb open reading frames are very similar, and the chb promoter is identical to that of E. coli, including all the regulator binding sites. All Cel+ mutants of S. sonnei had insertions within the chb regulatory region at very specific locations. In E. coli, NagC derepression of only a very small proportion of Cel+ mutants was caused by insertions within the chb regulatory region, while the remaining Cel+ mutants harbored loss-of-function mutations in the nagC locus (Kachroo et al. 2007). None of the one-step activated mutants of S. sonnei isolated in this study harbored loss-of-function mutations in the nagC locus and/or gain-of-function mutations in the chbR locus. The specificity of the mechanism employed by S. sonnei in activating the chb operon could possibly explain the reduced number of Cel+ mutants arising from this strain compared to E. coli. The non-appearance of Cel+ mutants in S. sonnei beyond 5–10 days could be related to the poorer viability of the S. sonnei strain, especially during prolonged exposure under nutrient limitation necessary for the accumulation of the two classes of mutations needed to confer a Cel+ phenotype. This is consistent with the observation that in a strain that already carries a deletion of the nagC locus, gain-of-function mutations in chbR similar to those seen in E. coli accumulate in 2–4 days.
Gene derepression, leading to constitutive or altered expression of a previously regulated enzyme, is one of the underpinning mechanisms for evolution of new metabolic functions under environmental stress (Wright 2004). The Cel+ phenotype of S. sonnei is contributed by such alterations in the chb promoter. The mechanism of activation of chb operon in Type I mutants is very precise. The IS600 insertion occurring at a very specific location close to the chb transcription start site is very likely enhancing transcription by bringing in a new −35 and distancing all the regulatory elements (Figs. 1c, 2). This is expected to result in constitutive activation of the operon independent of ChbR, consistent with the transcription data (Fig. 3a). This type of insertion has not been reported in E. coli K12 strains (Kachroo et al. 2007; Parker and Hall 1990a) which possess only an attenuated copy of IS600, making it unlikely for such type of activation events to occur, highlighting the importance of the strain background in determining the evolutionary path taken for acquiring a Cel+ phenotype.
In Type II mutants, the IS element insertions occurring at the distal NagC binding site without the disruption of other regulatory elements within the chb promoter lead to a ChbR-dependent and cellobiose-inducible activation of the chb operon (Fig. 3b). Since the basal transcription seen in the mutants is higher than that seen in a ΔnagC strain, (Fig. 5), the insertion is also augmenting chb transcription most likely by altering the sequence context of the promoter. In the presence of this enhanced transcription providing elevated levels of chbR, the induction mediated by wild-type ChbR in the presence of cellobiose is necessary and sufficient for conferring a Cel+ phenotype. While activation of genes mediated by insertion elements has been reported previously, their impact in most cases is to disrupt the normal regulation of the downstream gene as seen in the Type I mutants. The observations on Type II mutants presented above indicate that they can also facilitate enhanced induction of transcription.
Similar to E. coli, NagC derepression alone brought about by deletion of nagC was insufficient to give rise to Cel+ mutants in S. sonnei (Online resource 1 Fig. S4). This was consistent with the observation that the Cel+ papillae that arose from AK1∆nagC strain harbored chbR gain-of-function mutations very similar to the ones reported previously in E. coli. We also observed that in the strain AK104∆chbR that has lost the Cel+ phenotype in the absence of chbR, introduction of the wild-type E. coli chbRF open reading frame resulted in a Cel+ phenotype after 48 h (Online resource 1 Fig. S6), indicating that the wild-type E. coli chbR is able to function in S. sonnei. This observation further strengthens the assumption that in Type II mutants, the insertion element provides a distinct enhancement in transcription which bypasses the need for additional mutations in chbR for complete activation of the chb operon. The specificity and location of the insertion could play a significant role in imparting the cis-effect shown by the insertion event in S. sonnei. When compared to the nonpathogenic laboratory E. coli strain MG1655, Shigella strains not only possess higher copy numbers of IS elements, but also harbor additional IS species including IS1N, IS600, and IS629 (Yang et al. 2005). Though we have only tested the S. sonnei strain AK1 in this study, the high abundance and variety of IS elements in Shigella raises the probability of such insertion events happening in other Shigella strains as well leading to a Cel+ phenotype.
The Type I mutants with constitutive activation of the chb promoter also show an Arb+ Sal+ phenotype, while the Type II mutants with inducible promoter are Arb− Sal− in the absence of cellobiose. The constitutive expression of the permease ChbBCA and phospho-β-glucosidase ChbF in the wild-type strain AK1 also results in an Arb+ Sal+ phenotype. These results suggest that the chb operon can enable arbutin and salicin utilization if its expression can bypass the chbR-mediated regulation. Though multiple studies have proposed the contribution of the chb operon for arbutin and salicin utilization, none of the Cel+ mutants isolated from E. coli in our previous study showed an arbutin- or salicin-positive phenotype (Kachroo et al. 2007). This is most probably due to the inability of these substrates to activate transcription from the inducible promoters that are dependent on ChbR and cellobiose.
Consistent with previous reports, our study demonstrates that the utilization of novel substrates by bacteria is facilitated by altering the regulation of a preexisting genetic system, while maintaining the integrity of the downstream genes. Such an ability of bacteria to evolve novel metabolic functions is expected to provide a distinct selective advantage under specific environmental conditions. These results indicate that the diversity of insertion elements carried by an organism within its genome can have a significant impact on its evolutionary trajectory to facilitate utilization of a broader range of substrates.
References
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Desai SK, Nandimath K, Mahadevan S (2010) Diverse pathways for salicin utilization in Shigella sonnei and Escherichia coli carrying an impaired bgl operon. Arch Microbiol 192:821–833
Kachroo AH, Kancherla AK, Singh NS, Varshney U, Mahadevan S (2007) Mutations that alter the regulation of the chb operon of Escherichia coli allow utilization of cellobiose. Mol Microbiol 66:1382–1395
Keyhani NO, Roseman S (1997) Wild-type Escherichia coli grows on the chitin disaccharide, N, N′-diacetylchitobiose, by expressing the cel operon. Proc Natl Acad Sci USA 94:14367–14371
Keyhani NO, Bacia K, Roseman S (2000a) The transport/phosphorylation of N, N′-diacetylchitobiose in Escherichia coli. Characterization of phospho-IIB(Chb) and of a potential transition state analogue in the phosphotransfer reaction between the proteins IIA(Chb) AND IIB(Chb). J Biol Chem 275:33102–33109
Keyhani NO, Boudker O, Roseman S (2000b) Isolation and characterization of IIAChb, a soluble protein of the enzyme II complex required for the transport/phosphorylation of N, N′-diacetylchitobiose in Escherichia coli. J Biol Chem 275:33091–33101
Keyhani NO, Wang LX, Lee YC, Roseman S (2000c) The chitin disaccharide, N, N′-diacetylchitobiose, is catabolized by Escherichia coli and is transported/phosphorylated by the phosphoenolpyruvate:glycose phosphotransferase system. J Biol Chem 275:33084–33090
Kharat AS, Mahadevan S (2000) Analysis of the beta-glucoside utilization (bgl) genes of Shigella sonnei: evolutionary implications for their maintenance in a cryptic state. Microbiology 146(Pt 8):2039–2049
Kricker M, Hall BG (1984) Directed evolution of cellobiose utilization in Escherichia coli K12. Mol Biol Evol 1:171–182
Lan R, Reeves PR (2002) Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect 4:1125–1132
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Parker LL, Hall BG (1990a) Mechanisms of activation of the cryptic cel operon of Escherichia coli K12. Genetics 124:473–482
Parker LL, Hall BG (1990b) Characterization and nucleotide sequence of the cryptic cel operon of Escherichia coli K12. Genetics 124:455–471
Patrick WM, Quandt EM, Swartzlander DB, Matsumura I (2007) Multicopy suppression underpins metabolic evolvability. Mol Biol Evol 24:2716–2722
Peng J, Yang J, Jin Q (2009) The molecular evolutionary history of Shigella spp. and enteroinvasive Escherichia coli. Infect Genet Evol 9:147–152
Plumbridge J, Pellegrini O (2004) Expression of the chitobiose operon of Escherichia coli is regulated by three transcription factors: NagC, ChbR and CAP. Mol Microbiol 52:437–449
Pupo GM, Lan R, Reeves PR (2000) Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA 97:10567–10572
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Shepherd JG, Wang L, Reeves PR (2000) Comparison of O-antigen gene clusters of Escherichia coli (Shigella) sonnei and Plesiomonas shigelloides O17: sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides in a recent event. Infect Immun 68:6056–6061
Singh J, Mukerji M, Mahadevan S (1995) Transcriptional activation of the Escherichia coli bgl operon: negative regulation by DNA structural elements near the promoter. Mol Microbiol 17:1085–1092
Singh NS, Das G, Seshadri A, Sangeetha R, Varshney U (2005) Evidence for a role of initiation factor 3 in recycling of ribosomal complexes stalled on mRNAs in Escherichia coli. Nucl Acids Res 33:5591–5601
Thompson J, Ruvinov SB, Freedberg DI, Hall BG (1999) Cellobiose-6-phosphate hydrolase (CelF) of Escherichia coli: characterization and assignment to the unusual family 4 of glycosylhydrolases. J Bacteriol 181:7339–7345
Verma SC, Mahadevan S (2012) The chbG gene of the chitobiose (chb) operon of Escherichia coli encodes a chitooligosaccharide deacetylase. J Bacteriol 194:4959–4971
Woodcock DM et al (1989) Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucl Acids Res 17:3469–3478
Wright BE (2004) Stress-directed adaptive mutations and evolution. Mol Microbiol 52:643–650
Yang F et al (2005) Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucl Acids Res 33:6445–6458
Zangoui P, Vashishtha K, Mahadevan S (2015) Evolution of aromatic beta-glucoside utilization by successive mutational steps in Escherichia coli. J Bacteriol 197:710–716
Acknowledgments
This work was supported by an institutional grant from the Department of Biotechnology (DBT), Govt. of India, to the Indian Institute of Science and infrastructural support from the Department of Science and Technology (DST) under the FIST Program and the Universities Grants Commission (UGC), Govt. of India. AMJ thanks the Council for Scientific and Industrial Research (CSIR), Govt. of India, for a senior research fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Joseph, A.M., Sonowal, R. & Mahadevan, S. A comparative study of the evolution of cellobiose utilization in Escherichia coli and Shigella sonnei . Arch Microbiol 199, 247–257 (2017). https://doi.org/10.1007/s00203-016-1299-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-016-1299-0