Abstract
Key message
BnA10.LMI1 positively regulates the development of leaf lobes in Brassica napus, and cis-regulatory divergences cause the different allele effects.
Abstract
Leaf shape is an important agronomic trait, and large variations in this trait exist within the Brassica germplasm. The lobed leaf is a unique morphological characteristic for Brassica improvement. Nevertheless, the molecular basis of leaf lobing in Brassica is poorly understood. Here, we show that an incompletely dominant locus, BnLLA10, is responsible for the lobed-leaf shape in rapeseed. A LATE MERISTEM IDENTITY1 (LMI1)-like gene (BnA10.LMI1) encoding an HD-Zip I transcription factor is the causal gene underlying the BnLLA10 locus. Sequence analysis of parental alleles revealed no sequence variations in the coding sequences, whereas abundant variations were identified in the regulatory region. Consistent with this finding, the expression levels of BnLMI1 were substantially elevated in the lobed-leaf parent compared with its near-isogenic line. The knockout mutations of BnA10.LMI1 gene were induced using the CRISPR/Cas9 system in both HY (the lobed-leaf parent) and J9707 (serrated leaf) genetic backgrounds. BnA10.LMI1 null mutations in the HY background were sufficient to produce unlobed leaves, whereas null mutations in the J9707 background showed no obvious changes in leaf shape compared with the control. Collectively, our results indicate that BnA10.LMI1 positively regulates the development of leaf lobes in B. napus, with cis-regulatory divergences causing the different allelic effects, providing new insights into the molecular mechanism of leaf lobe formation in Brassica crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaves are essential organs in crop plants, playing a major role in photosynthate accumulation, gas exchange, nutrient distribution and water transport (Tsukaya 2006). Leaf morphology can significantly affect canopy evapotranspiration, the penetration of sunlight and chemical pesticides, pest preference, and ultimately crop yield and quality (Zhu et al. 2016). An important variation in leaf shape is the outline of the leaf margin, which can be entire, serrated or lobed. Leaf shape diversity reflects natural selection operating on leaf function and is a valuable resource that can increase our understanding of the ecological and evolutionary drivers of diversification in leaf shape and the functional significance therein (Nicotra et al. 2011).
Rapeseed (Brassica napus) is one of the world’s most important sources of oilseed crop. Extensive leaf shape diversity, including lobed leaves, exists in Brassica species. The functional significance of lobed leaves in rapeseed has been identified with potential advantages for high-density planting and hybrid production (Pu et al. 2001; Tu et al. 2013). High planting density benefits mechanized harvesting in mechanical production, yet the deteriorative canopy microclimate would increase the incidence of pests and diseases (Ballare et al. 2012; Li et al. 2014). Lobed leaves with deep sinuses at the leaf margin can improve heat transfer and canopy architecture (Vogel 2009; Zhu et al. 2016); thus, lobed-leaf lines perform better than entire leaves to relieve this problem. Furthermore, leaf lobes are easily recognized even in the early stages, serving as a perfect indicator trait in hybrid production. Thus, a better understanding of the molecular regulatory mechanism of leaf lobing in rapeseed will contribute to the manipulation of leaf shape and its utilization in production.
Previously, lobed-leaf genes have been genetically analysed and mapped in different Brassica species. In B. rapa, the lobed-leaf phenotype is controlled by major genes or polygenic effects with the dominant trait being the presence of leaf lobes (Song et al. 1995; Kubo et al. 2010; Wang et al. 2015), and a quantitative trait locus (QTL) in the A10 linkage group was found to be one of the major loci involved in regulating leaf lobing. Similarly, leaf lobing was controlled by an incompletely dominant locus at the distal end of chromosome A10 in B. napus (Ni et al. 2015). Thus, these results provide a target for the molecular cloning and characterization of the lobed-leaf gene. Here, we report the isolation and functional characterization of a LATE MERISTEM IDENTITY1 (LMI1)-like gene, BnA10.LMI1, as the major determinant of leaf lobing underlying the BnLLA10 locus in rapeseed.
Materials and methods
Plant materials
For QTL analysis and gene mapping, two rapeseed cultivars, the lobed-leaved line HY and the serrated-leaved line Z9, were used as parents in a reciprocal cross (HY × Z9 and Z9 × HY) to produce F1 lines. Subsequently, the F1 line derived from HY × Z9 was self-pollinated and backcrossed successively with the recurrent parent, HY, to produce F2, F3, BC1F2, BC2F2 and BC3F2 populations. A BC3F2 individual with the homozygous Z9-allele at the BnLLA10 locus was selected by molecular markers developed in the study and used as a near-isogenic line (NIL) of HY (i.e. Z9-NIL). A BAC library of the serrated-leaved inbred line J7005 was used for comparative sequencing analysis. A semi-winter B. napus pure line, J9707 (serrated leaves), was used as the transformation receptor in this study.
In B. rapa, an F2 population was derived from a cross between the parental lines S1365 (female, lobed leaves) and P10 (male, serrated leaves).
Trait measurement
To quantify the extent of leaf lobing and complexity, the leaf dissection index was calculated as (perimeter-squared)/(4π × area) according to Bilsborough et al. (2011). For the preliminary linkage analysis, the leaf lobe phenotype was categorized by visual inspection using the following scale: 0–3 (0 = serrated leaf, 1–2 = intermediate and 3 = lobed leaf, as shown in Fig. S1).
To test the effects of leaf lobing on photosynthetic features, the chlorophyll content and net leaf photosynthetic rate (Pn) of HY and Z9-NIL were measured in the field. Seedlings were grown during the 2016–2017 season on the experimental farm of Huazhong Agriculture University, Wuhan, China. The field experiment followed a randomized block design with three replications. The chlorophyll content of the leaves from 80-day-old seedlings was measured using a chlorophyll meter (SPAD-502, Minolta Camera Co., Osaka, Japan). For each measurement, the ten uppermost fully expanded leaves were selected from each field block. Three SPAD readings were taken around the midpoint of each leaf margin beside the midrib. Ten SPAD readings were averaged to represent the mean value of each block. Pn was measured on the newest fully expanded leaves using a portable photosynthesis system (LI-6400XT; Li-COR Inc., Lincoln, NE, USA) between 09:00 am and 11:00 am during the bolting stage, as described previously (Sun et al. 2016).
DNA extraction and SNP analysis
For single-nucleotide polymorphism (SNP) genotyping (Illumina Brassica 60-K array), genomic DNA was extracted from fresh leaves of individual F2 plants, one F1 hybrid and the two parents using a plant genomic DNA extraction kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Two lobed-leaved bulk DNA samples were constructed with equivalent amounts of DNA from 12 lobed-leaved individuals. Each bulk included DNA from six individuals. The two parents (HY and Z9), their F1 hybrid and two lobed-leaved bulks were used for SNP genotyping with an Illumina Brassica 60-K array, according to the method described by Li et al. (2016). SNPs distributed in the target region are listed in Table S1.
Molecular marker development, linkage analysis and QTL mapping
Molecular markers, including simple sequence repeat (SSR), insertion/deletion (INDEL) and SNP markers in the BnLLA10 locus region, were newly developed (Table S2) based on genome assemblies of B. rapa (http://brassicadb.org/brad/) and B. napus (http://www.genoscope.cns.fr/brassicanapus/). SSR primers were designed using the web-based SSR finder tool (http://www.geboc.org/index), and INDEL primers were designed using Primer5.0 software. The SNAPER programme (Drenkard et al. 2000) was used to design the SNP primers. All primer pairs were synthesized by Sangon Biological and Engineering Co. (Shanghai, China).
Linkage analysis was performed using Mapmaker/Exp 3.0 (Lincoln et al. 1992), and the Kosambi function was used to calculate genetic distance. QTLs were detected using the composite interval mapping with the program QTL Cartographer V2.0 (Wang et al. 2004) at a threshold of LOD = 3.0.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was prepared using the EasyPure Plant RNA Kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. A 2-µg sample of RNA was converted into cDNA following the manufacturer’s instructions (TransScript RT Kit, TransGen Biotech). Real-time PCR was performed using the TransStart Top Green qPCR SuperMix Kit (TransGen Biotech) on a CFX96 Real-Time System (Bio-Rad). Relative quantification was performed using the comparative cycle threshold method, and the relative amount of PCR product that was amplified using the designed primer sets (listed in Table S3) was normalized to the reference gene ornithine transcarbamylase (OTC), as described by Cnops et al. (2004).
Transgene construction and plant transformation
To construct the Cas9/sgRNA-expressing binary vectors, four sequence-specific sgRNAs in the target gene were selected using the web-based tool CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR). The binary pYLCRIPSR/Cas9 multiplex genome targeting vector system was provided by Prof. Yaoguang Liu (South China Agriculture University, Guangzhou, China) and used for construct assembly according to the method described by Ma et al. (2015). The oligos used in constructing the sgRNA vectors are listed in Table S3. The resulting construct contained a Cas9p expression cassette, sgRNA expression cassettes with target sequences and a hygromycin resistance cassette (Fig. 5a).
Following verification of the fused constructs via sequencing, the resulting constructs were transformed into B. napus via the Agrobacterium tumefaciens-mediated hypocotyl method (Zhou and Fowke 2002). The transgenic plants were screened and confirmed by antibiotic selection and PCR.
Identification of CRISPR/Cas9-induced mutations
PCR was performed to amplify the genomic region surrounding the CRISPR target sites using specific primers (Table S3). The PCR fragments were directly sequenced or cloned into the pEASY-T (TransGen Biotech) vector and then sequenced by the Sanger method to identify mutations.
Results
Leaf lobing in rapeseed is controlled by a nuclear locus BnLLA10 with major effect
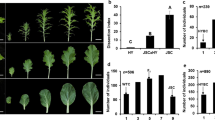
To investigate the inheritance of the lobed-leaf trait in B. napus, reciprocal crosses between the lobed-leaved parent Tongling huaye (HY) and the serrated-leaved parent Zhongshuang 9 (Z9) were conducted. The F1 plants from the reciprocal crosses exhibited intermediate leaf morphology (Fig. 1a, b). In each of the random F2 and BC1F2 subpopulations, the segregation ratio for the serrated, intermediate and lobed-leaf types was approximately 1:2:1 (χ2 = 2.33 and 2.99, p > 0.05; Table 1). These results indicated that the lobed-leaf trait is controlled by a nuclear locus with major effect, designated BnLLA10, without cytoplasmic genetic effects. A Z9-NIL was developed by introducing the homozygous Z9-allele at the BnLLA10 locus into the HY genetic background at the BC3F2 generation, which showed the same leaf morphology as that of Z9. Compared with Z9-NIL, the leaf chlorophyll content and photosynthetic rate in HY were significantly increased (Fig. 1c, d).
Morphological and physiological performance of the lobed-leaf trait in B. napus. a Leaves of Z9, F1 hybrid of Z9 × HY, F1 hybrid of HY × Z9 and HY (from left to right) of 50d-old plants. Bar, 1 cm. b Leaf dissection indices of parents and F1 hybrids (n = 10). Uppercase letters indicate a significant difference at the 0.01 probability level among parents and F1 hybrids based on a multiple comparison test. c Comparison of leaf chlorophyll contents (LCC) (n = 10) and d net photosynthetic rates between HY and Z9-NIL leaves (n = 3). Data represent mean ± SD. *** Significantly different at P < 0.001; * significantly different at P < 0.05
The tandemly duplicated LMI1-like genes BnA10.RCO and BnA10.LMI1 are candidate genes within BnLLA10 of B. napus
To map the BnLLA10 locus in B. napus, two lobed-leaved DNA bulks from the F2 population along with DNA from the two parents and one F1 hybrid were used for SNP detection with an Illumina Brassica 60-K array. After the removal of invalid markers, a total of 11,012 SNPs between the parents were used for further screening. Only 31 SNPs met the requirements that the genotype of both DNA bulks should be homozygous for the HY parental allele and that the F1 hybrid should be heterozygous. The subsequent BLAST analysis located 26 of these SNPs in a region corresponding to 15.2–17.4 Mb on chromosome A10 of the B. napus genome (Table S1).
A linkage map consisting of nine markers spanning the candidate region was subsequently constructed in a random BC1F2 subpopulation (Fig. 2a). The leaf lobe phenotype of each individual in the population was categorized by visual inspection using the following scale: 0 = serrated leaf, 1–2 = intermediate, 3 = lobed leaf (Fig. S1A). A major QTL with a LOD of 78.4, BnLLA10, was detected within the marker interval A10_87-A10_90 and found to account for 90.5% of the leaf lobe variation (Fig. 2a; Table 2).
Mapping of the lobed-leaf gene locus in B. napus and B. rapa. a Locations of the BnLLA10 and BrLLA10 loci on molecular linkage maps. The left linkage map was constructed using a BC1F2 population. Numbers show the genetic distances between adjacent markers; the arrow denotes the QTL peak position. The middle is a physical map of B. napus A10 in the target region of the BnLLA10 locus. Numbers show the physical distances between adjacent markers. The right linkage map was constructed using an F2 population in B. rapa. The presence or absence of the lobed-leaf phenotype is used as a dominant marker of the BrLLA10 locus for linkage analysis; numbers show the genetic distances between adjacent markers. b Fine mapping of the BnLLA10 locus in B. napus. The numbers in parentheses behind the molecular marker names represent the number of recombinants at the corresponding markers. The corresponding genes annotated according to the reference genome are depicted in the 41.0-kb interval region
After a recombinant screening using 2981 plants with the parental phenotypes (extremely serrated or lobed leaves) from the F2, F3, BC1F2, BC2F2 and BC3F2 populations, the genomic region containing the BnLLA10 locus was further narrowed down to a 41.0-kb region bounded by A10_87 and A10_88 (Fig. 2b). Based on the B. napus reference genome database (http://www.genoscope.cns.fr/brassicanapus/), five genes, designated BnaA10g26310D through BnaA10g26350D, were identified in the mapping region (Table 3).
Among the mapped genes, BnaA10g26320D (hereafter BnA10.RCO) and BnaA10g26330D (hereafter BnA10.LMI1) are LMI1-like paralogues that encode class I HD-Zip transcription factors. Their homologues have been implicated in leaf shape diversity in Cardamine hirsuta (Sicard et al. 2014; Vlad et al. 2014) and Gossypium hirsutum (Andres et al. 2017). Of the remaining three putative genes, BnaA10g26310D is an exostosin family protein, BnaA10g26340D encodes a putative 3-deoxy-D-manno-octulosonate transferase, and BnaA10g26350D encodes a beta-mannan synthase that is required for Agrobacterium-mediated plant genetic transformation. Thus, BnA10.RCO and BnA10.LMI1 are the most likely candidate genes in BnLLA10.
BrLMI1 is the candidate gene for lobed leaves in B. rapa
To verify the results in B. napus, we conducted similar mapping of the lobed-leaf gene in B. rapa using a cross between S1365 (lobed leaves) and P10 (serrated leaves). F1 plants had leaves similar to S1365, suggesting that the lobed-leaf trait was dominant in this cross (Fig. S1B). The F2 individuals showed significant segregation of leaf shape phenotypes. A total of 123 F2 plants (43 individuals with extremely serrated leaves and 80 with lobed leaves) were selected for genotyping. Considering the dominant nature of the BrLLA10 locus, the presence or absence of the lobed-leaf phenotype was used as a dominant marker for the BrLLA10 locus in the linkage analysis. We localized the BrLLA10 locus to an interval delimited by markers A10_96 and A10_79, corresponding to an ~ 230-kb region on B. rapa chromosome A10 (Fig. 2a). Forty-three genes, designated Bra009506 to Bra009548, were predicted within this candidate region based on the B. rapa genome database (http://brassicadb.org/brad/index.php). Among these candidate genes, Bra009510 (hereafter BrLMI1) is an orthologue of B. napus BnA10.LMI1 and A. thaliana LMI1 (Saddic et al. 2006). Taken together, these results indicate a possibility that the conserved function of BnA10.LMI1/BrLMI1 homologues is important in the development of leaf lobes in both B. rapa and B. napus.
Promoter polymorphisms in BnA10.LMI1 underlie its allelic variations
To define the structure of the candidate gene, we located the exon–intron junctions by sequencing cDNA fragments amplified by RT-PCR using BnLMI1-specific primers. A comparison to genomic sequences revealed that BnA10.LMI1 (GenBank accession number: MF590053) consisted of three exons and two introns, and it encoded protein of 230 amino acids (Fig. 3a, b). The putative protein contains a homeobox domain and a leucine zipper domain (Fig. 3b).
Gene structure and protein alignment of BnA10.LMI1. a The BnA10.LMI1 gene structure, as well as natural variations between the alleles from HY and Z9, are shown; four sgRNA (S1-S4) positions are indicated as red vertical lines, and horizontal black arrows represent PCR genotyping primers. b Alignment of BnA10.LMI1 protein sequences from three inbred lines of B. napus (HY, Z9 and J7005). Full lines indicate the homeobox and dashed lines indicate the homeobox-associated leucine domains (color figure online)
To establish the causal relationship between the candidate genes and leaf shape variations, a comparative sequence analysis of the BnA10.LMI1 genomic DNA fragment was performed. A multiple sequence alignment of ~ 3.0 kb upstream of the start codon, the entire gene, and a 0.75 kb 3′ flanking sequence was conducted using the sequences from both parents and a bacterial artificial chromosome (BAC) library of inbred line J7005 (serrated leaves) (Fig. 3a). No sequence variations were detected in the open reading frame of BnA10.LMI1 (Fig. 3a), while a larger insertion (317 bp upstream of the transcription start site) and three SNPs in the BnA10.LMI1 promoter sequence were unique to the two serrated-leaved inbred lines Z9 and J7005, along with 12 SNPs in the 3′ flanking sequence among these lines (Fig. 3a; Fig. S2–S3), which were concluded to be the cause of the functional variation.
The lobed-leaf shape is positively correlated with enhanced transcript level of BnLMI1 during early leaf formation in rapeseed
Since the promoter polymorphism was the most critical variations between parental alleles of BnA10.LMI1, we speculated that expression levels of the gene would be affected by this regulatory diversification. To verify this hypothesis, the relative expression of BnLMI1 in HY and Z9-NIL was analysed during leaf development. BnLMI1 was dramatically up-regulated in HY compared with Z9-NIL in the shoot apex of 9d-old seedlings (Fig. 4a). In addition, expression of this gene was very low and comparable in the shoot apex of 9d-old seedlings, and the leaves of 10d-old and 15d-old seedlings in Z9-NIL; by contrast, it was down-regulated significantly in the leaves of 10d-old and 15d-old seedlings compared with the shoot apex of 9d-old seedlings in HY (Fig. 4a), suggesting a potentially important role during early leaf formation. The expression profiles of BnLMI1 in different tissues of seedlings revealed higher expression in the shoot apex than in other tissues (Fig. 4b). These findings also indicated an important function during early leaf development. Taken together, the gene expression differences between HY and its NIL strongly suggest that the lobed-leaf shape is positively correlated with enhanced BnLMI1 transcript levels.
Expression pattern of BnLMI1 in rapeseed. Expression of BnLMI1 in the shoot apex of 9d-old plants, and the leaves of 10d-old and 15d-old seedlings of HY and Z9-NIL (a), as well as the expression patterns of BnLMI1 in various tissues of HY seedlings (b), as determined by qRT-PCR with normalization to BnOTC. Values are the mean ± SD of three biological replicates. Z9-NIL is a BC3F2 individual with a homozygous Z9 genotype at the BnLLA10 locus in the HY background; **, significantly different at P < 0.01; *, significantly different at P < 0.05
CRISPR/Cas9-induced null mutations confirmed that BnA10.LMI1 positively regulates leaf lobe formation in B. napus
We tested whether BnA10.LMI1 functions in rapeseed leaf lobe development by engineering null mutations in BnA10.LMI1 using CRISPR/Cas9 gene-editing technology. A CRISPR/Cas9 construct containing four sgRNAs (Fig. 3a) within BnA10.LMI1 with Cas9 driven by P35S, i.e. Cas9P35s-BnLMI1 (Fig. 5a), was generated, and the resulting construct was transformed into HY using Agrobacterium-mediated transformation. Of the resulting 139 T0 positive transgenic plants, 19 showed a strongly reduced lobed-leaf phenotype. Three targeted mutants (SLMI1-9, SLMI1-21 and SLMI1-39) were verified with Sanger sequencing. In addition, various frameshift mutations were produced in proximity to all target sites except S2 in these lines (Fig. 5b, c).
CRISPR/Cas9-induced null mutants of BnA10.LMI1 in the lobed-leaf parent HY. a Binary constructs of Cas9P35 s-BnLMI1 (SLMI1). b Mutant genotypes and phenotypes of BnA10.LMI1-edited T0 and T1 plants. c Sequences at the sgRNA target sites of BnA10.LMI1-edited plants. The protospacer adjacent motif (PAM) is underlined, and nucleotide INDELs are marked in red, with details labelled at right; “A” represents the WT allele, and “a” represents the mutated allele. d Leaf dissection index of BnA10.LMI1-edited plants. e Leaf shape of BnA10.LMI1-edited plants (the upper, 15d-old; the lower, 50d-old). AA represents HY; Aa represents heterozygous mutant; aa represents homozygous or biallelic mutants; bar, 1 cm
We confirmed reduced leaf lobing in the T1 progeny of the CRISPR/Cas9-induced mutants (Fig. 5b, e). All tested plants with frameshift mutations at both BnA10.LMI1 alleles (homozygous or biallelic mutations, aa genotype) had unlobed-leaf shapes, and plants carrying frameshift mutations at only one allele of BnA10.LMI1 (heterozygous mutations, Aa genotype) had intermediate leaf shapes (Fig. 5b, e). Thus, the phenotypes of the different types of induced mutants indicated the incompletely dominant nature of the lobed-leaf trait.
Similarly, BnA10.LMI1-edited lines in J9707 (serrated leaf) genetic backgrounds were created using the Cas9P35s-BnLMI1 construct. The targeted mutations of four T1 progeny from two independent T0 lines (JSLMI1-22 and JSLMI1-35) were verified with Sanger sequencing (Fig. S4A-B). All tested plants with frameshift mutations at one or both BnA10.LMI1 alleles exhibited no obvious changes in leaf shape compared with the control (Fig. S4C), which indicated the recessive nature of the serrated leaf phenotype and was clearly in agreement with the incompletely dominant nature of the lobed-leaf trait. Taken together, these findings indeed provide strong evidence that BnA10.LMI1 positively regulates leaf lobe formation in B. napus.
Discussion
In this study, we identified an LMI1-like gene, BnA10.LMI1, which encodes an HD-Zip I transcription factor, as the causal gene underlying the lobed-leaf trait in B. napus. Sequence analysis of parental alleles revealed no sequence variations in the open reading frame of BnA10.LMI1 gene, whereas abundant variations, particularly a large insertion, were identified in the regulatory regions (Fig. 3a, Fig. S2). Consistent with this finding, the BnLMI1 expression level was substantially elevated in the lobed-leaf parent compared with its NIL (Fig. 4a), suggesting a positive connection between the expression level of this gene and leaf complexity. The knockout mutations of BnA10.LMI1 gene were induced using the CRISPR/Cas9 system in both HY (the lobed-leaf parent) and J9707 (serrated leaf) genetic backgrounds, and BnA10.LMI1 null mutations in the HY background were sufficient to produce unlobed leaves (Fig. 5b–e), whereas the null mutations in the J9707 genetic background showed no obvious changes in the leaf shape compared with its background (J9707). Collectively, these findings provide strong evidence that BnA10.LMI1 positively determines leaf shape complexity in B. napus and that cis-regulatory divergences cause the different allele effects. Interestingly, this finding is consistent with previous reports that promoter modifications to a LMI1 gene altered expression and leaf shape in upland cotton, which further strengthens the evidence that LMI1-mediated leaf shape diversification is an evolutionary hot spot in plants (Sicard et al. 2014; Vlad et al. 2014; Andres et al. 2017).
The amphidiploid species B. napus (AACC), an important oil seed crop, was formed by a recent allopolyploidy event between two diploid ancestors of B. rapa (AA) and B. oleracea (CC). Several studies have consistently detected a major locus with an effect on leaf lobing on chromosome A10 in B. rapa (Song et al. 1995; Kubo et al. 2010; Wang et al. 2015). In the present study, we localized the major locus BrLLA10 to an interval containing an LMI1-like gene (Fig. 2a). The evidence presented here indicates that the lobed-leaf trait may be controlled by the same LMI1 gene in B. rapa.
Previous studies indicated that the RCO gene, but not LMI1, plays a key role in leaf shape diversification in C. hirsuta, Arabidopsis lyrata and B. napus (Sicard et al. 2014; Vlad et al. 2014; Ni et al. 2017). In the present study, we provide strong evidence that BnA10.LMI1 positively regulates leaf lobe formation in B. napus, providing new insights into the molecular mechanism of leaf lobe formation in Brassica crops. In addition, whether BnA10.RCO plays a vital role in determining lobed leaves remains an intriguing question.
Leaves are the main photosynthetic organs of crop plants. Accumulating evidence indicates that lobed leaves are associated with production advantages such as reducing humidity, shade, and improving water use efficiency and thermoregulation, ultimately resulting in a favourable microclimate for photosynthesis and agronomic profitability (Vogel 2009; Zhu et al. 2016). For example, cotton with the okra leaf phenotype showed increased photosynthetic efficiency and leaf complexity together with enhanced LMI1 expression (Pettigrew et al. 1993; Chang et al. 2016). Recently, Vuolo et al. (2016) also reported that an LMI1-like gene positively influenced photosynthesis and leaf complexity in both C. hirsuta and A. thaliana. The effects of the LMI1-like gene on photosynthesis may ultimately influence resource allocation to seeds because it positively influenced seed yield (Vuolo et al. 2016). These findings are in line with transcriptomic analyses from tomato and cotton, showing that photosynthetic gene expression was positively correlated with increased leaf dissection, implying a broad connection between leaf morphology and photosynthetic capacity (Chitwood et al. 2013; Andres et al. 2017). In the present study, the leaf chlorophyll content and photosynthetic rate in HY were significantly increased compared with those in Z9-NIL (Fig. 1c, d). Thus, our findings further strengthen the potential to improve photosynthesis via manipulation of the leaf shape, and the positive effects of BnA10.LMI1 on photosynthesis may ultimately influence resource allocation to seeds, which has the potential to improve productivity through the modulation of BnA10.LMI1 activity in crops.
Author Contribution statement
HL, FC, ZY conceived and designed the experiments; HL, MQ, ZH performed the experiments; FC, HL, CC, MH wrote the manuscript; YQ, HS involved in bioinformatics analysis.
References
Andres RJ, Coneva V, Frank MH, Tuttle JR, Samayoa LF, Han SW, Kaur B, Zhu L, Fang H, Bowman DT, Rojas-Pierce M, Haigler CH, Jones DC, Holland JB, Chitwood DH, Kuraparthy V (2017) Modifications to a LATE MERISTEM IDENTITY1 gene are responsible for the major leaf shapes of Upland cotton (Gossypium hirsutum L.). Proc Natl Acad Sci USA 114:E57–E66
Ballare CL, Mazza CA, Austin AT, Pierik R (2012) Canopy light and plant health. Plant Physiol 160:145–155
Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M (2011) Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA 108:3424–3429
Chang L, Fang L, Zhu Y, Wu H, Zhang Z, Liu C, Li X, Zhang T (2016) Insights into interspecific hybridization events in allotetraploid cotton formation from characterization of a gene regulating leaf shape. Genetics 204:799–806
Chitwood DH, Kumar R, Headland LR, Ranjan A, Covington MF, Ichihashi Y, Fulop D, Jiménezgómez JM, Peng J, Maloof JN (2013) A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell 25:2379
Cnops G, Jover-Gil S, Peters JL, Neyt P, De Block S, Robles P, Ponce MR, Gerats T, Micol JL, Van Lijsebettens M (2004) The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. J Exp Bot 55:1529–1539
Drenkard E, Richter BG, Rozen S, Stutius LM, Angell NA, Mindrinos M, Cho RJ, Oefner PJ, Davis RW, Ausubel FM (2000) A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol 124:1483
Kubo N, Saito M, Tsukazaki H, Kondo T, Matsumoto S, Hirai M (2010) Detection of quantitative trait loci controlling morphological traits in Brassica rapa L. Breed Sci 60:164–171
Li YS, Yu CB, Zhu S, Xie L, Hu XJ, Liao X, Liao XS, Che Z (2014) High planting density benefits to mechanized harvest and nitrogen application rates of oilseed rape (Brassica napus L.). Soil Sci Plant Nutr 60:384–392
Li X, Wang A, Zu F, Hu Z, Lin J, Zhou G, Tu J (2016) Identification of a nuclear-recessive gene locus for male sterility on A2 chromosome using the Brassica 60 K SNP array in non-heading Chinese cabbage. Genes Genom 38:1–7
Lincoln S, Daly M, Lander E (1992) Constructing genetic maps with MAPMAKER/EXP 3.0
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Ni X, Huang J, Ali B, Zhou W, Zhao J (2015) Genetic analysis and fine mapping of the LOBED-LEAF 1 (BnLL1) gene in rapeseed (Brassica napus L.). Euphytica 204:29–38
Ni X, Liu H, Huang J, Zhao J (2017) LMI1-like genes involved in leaf margin development of Brassica napus. Genetica 145:269–274
Nicotra AB, Leigh A, Boyce CK, Jones CS, Niklas KJ, Royer DL, Tsukaya H (2011) The evolution and functional significance of leaf shape in the angiosperms. Funct Plant Biol 38:535–552
Pettigrew WT, Heitholt JJ, Vaughn KC (1993) Gas-exchange differences and comparative anatomy among cotton leaf-type isolines. Crop Sci 33:1295–1299
Pu HM, Fu SZ, Qi CK, Zhang JF, Wu YM, Gao JQ, Chen XJ (2001) Inheritance of divided leaf trait of rapeseed (Brassica napus) and application in hybrid breeding. Chin J oil Crop Sci 23:60–62
Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D (2006) The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development 133:1673–1682
Sicard A, Thamm A, Marona C, Lee YW, Wahl V, Stinchcombe JR, Wright SI, Kappel C, Lenhard M (2014) Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr Biol 24:1880–1886
Song K, Slocum MK, Osborn TC (1995) Molecular marker analysis of genes controlling morphological variation in Brassica rapa (syn. campestris). Theor Appl Genet 90:1–10
Sun J, Ye M, Peng S, Li Y (2016) Nitrogen can improve the rapid response of photosynthesis to changing irradiance in rice (Oryza sativa L.) plants. Sci Rep 6:31305
Tsukaya H (2006) Mechanism of leaf-shape development. Annu Rev Plant Biol 57:477–496
Tu YQ, Sun J, Dai XL, Tang J, Tu WF, Shao HQ (2013) Character and genetic analysis of lobed-leaf traits in Brassica napus. Chin J oil Crop Sci 35:93–96
Vlad D, Kierzkowski D, Rast MI, Vuolo F, Ioio RD, Galinha C, Gan X, Hajheidari M, Hay A, Smith RS, Huijser P, Bailey CD, Tsiantis M (2014) Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343:780–783
Vogel S (2009) Leaves in the lowest and highest winds: temperature, force and shape. New Phytol 183:13
Wang S, Basten CJ, Zeng ZB (2004) Windows QTL Cartographer 2.0. Department of Statistics, North Carolina State University, Raleigh
Wang Y, Liu X, Ji X, Zhang L, Liu Y, Lv X, Feng H (2015) Identification and validation of a major QTL controlling the presence/absence of leaf lobes in Brassica rapa L. Euphytica 205:761–771
Zhou Y, Fowke LC (2002) Control of petal and pollen development by the plant cyclin-dependent kinase inhibitor ICK1 in transgenic Brassica plants. Planta 215:248
Zhu QH, Zhang J, Liu D, Stiller W, Liu D, Zhang Z, Llewellyn D, Wilson I (2016) Integrated mapping and characterization of the gene underlying the okra leaf trait in Gossypium hirsutum L. J Exp Bot 67:763–774
Acknowledgements
We thank Prof. Yaoguang Liu (South China Agriculture University) for kindly providing the sgRNA-Cas9 plant expression vectors. This study was supported by Grants 2015CB150202 from the National Key Basic Research Program of China and the Fundamental Research Funds for the Central Universities (programme No. 2662015PY172).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Communicated by Maria Laura Federico.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, L., Zhang, H., Yang, Q. et al. Promoter variations in a homeobox gene, BnA10.LMI1, determine lobed leaves in rapeseed (Brassica napus L.). Theor Appl Genet 131, 2699–2708 (2018). https://doi.org/10.1007/s00122-018-3184-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3184-5