Abstract
The heavy metal cadmium (Cd) is highly toxic to humans and can enter food chains from contaminated crop fields. Understanding the molecular mechanisms of Cd accumulation in crop species will aid production of safe Cd-free food. Here, we identified a single recessive gene that allowed higher Cd translocation in rice, and also determined the chromosomal location of the gene. The Cd hyperaccumulator rice variety Cho-Ko-Koku showed 3.5-fold greater Cd translocation than the no-accumulating variety Akita 63 under hydroponics. Analysis of an F2 population derived from these cultivars gave a 1:3 segregation ratio for high:low Cd translocation. This indicates that a single recessive gene controls the high Cd translocation phenotype. A QTL analysis identified a single QTL, qCdT7, located on chromosome 7. On a Cd-contaminated field, Cd accumulation in the F2 population showed continuous variation with considerable transgression. Three QTLs for Cd accumulation were identified and the peak of the most effective QTL mapped to the same region as qCdT7. Our data indicate that Cd translocation mediated by the gene on qCdT7 plays an important role in Cd accumulation on contaminated soil.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cadmium (Cd) is one of the most toxic heavy metals with respect to human health, especially in Cd-contaminated areas, where it can enter the food chain. Cd is often present in deposits of valuable heavy metals, such as copper, silver, and zinc, and mining activities can release Cd into the environment, where it may remain as a contaminant for a number of years. The contamination of crop fields with Cd is an important problem, particularly for paddy fields irrigated with water that passes through mining sites. Crops grown on Cd-polluted soils take up and accumulate Cd, with potentially devastating consequences for human health.
Phytoremediation is a promising and environmentally friendly approach for removing Cd pollution from soils. The plant species selected for use in phytoremediation accumulate Cd at a high level, especially in their aerial parts. Two such species are Thlaspi caerulescens and Arabidopsis halleri, which can accumulate more than 100 mg kg−1 shoot dry weight of Cd under field conditions (Baker et al. 2000; Bert et al. 2002). In addition to their ability to accumulate Cd, the selected species also need to show rapid growth and large biomass productivity. Some rice (Oryza sativa) cultivars are promising candidates for phytoremediators of Cd contaminated paddy fields (Ibaraki et al. 2009; Murakami et al. 2009): they obviously grow on the same habitat as crop cultivars, can accumulate Cd in their aerial parts at relatively high concentrations, and can produce a large biomass (Arao and Ae 2003; Liu et al. 2003; Ishikawa et al. 2005a). The cultivation of rice, including mechanical planting and harvesting, is obviously well established.
Understanding the mechanisms of accumulation of Cd in the aerial part of plants will be indispensable for creating more efficient hyperaccumulators. Plants are presumed to uptake Cd from the soils through their root systems, load it to the xylem and then transport it to the aerial parts (Clemens 2006). It is therefore likely that several genes involved in these transport processes control Cd accumulation in plants. A recent study on the hyperaccumulator A. halleri, in comparison with the model plant A. thaliana, revealed that xylem loading is controlled by HMA4, which enables the hyperaccumulation of Cd in shoots (Courbot et al. 2007; Hanikenne et al. 2008).
The genus Oryza shows considerable genetic diversity among varieties and also displays large differences in ability to concentrate Cd in shoots and brown rice (Morishita et al. 1987; Arao and Ae 2003; Liu et al. 2003; Uraguchi et al. 2009). Such genotypic and phenotypic variations provide a valuable resource for identifying genes that confer the ability to hyperaccumulate Cd. Using chromosome segment substitution lines of Koshihikari and Kasalath, which accumulate different Cd concentrations in their brown rice, three chromosome segments were identified that enable accumulation of high Cd concentrations (Ishikawa et al. 2005b). Recently, three putative QTLs controlling shoot Cd concentration were identified in an F2 population between the Badari Dhan and Shwe War cultivars that have different abilities to concentrate Cd in their shoots (Ueno et al. 2009). However, the specific genes involved were not identified and, therefore, the molecular mechanisms of Cd accumulation in rice plants remained uncertain.
On Cd-contaminated soil, the indica rice cultivar Cho-Ko-Koku was found to accumulate the metal at a much higher level than other tested cultivars (Matsumoto et al. 2005; Uraguchi et al. 2009; Murakami et al. 2009). We therefore selected this cultivar for use in our study on the mechanisms of Cd hyperaccumulation in rice grown under defined hydroponic conditions. In this study, we identified a single recessive gene (named qCdT7) located on chromosome 7 that conferred a high Cd translocation ability to Cho-Ko-Koku. We also found that the QTL with the greatest influence on Cd accumulation in rice grown in a Cd contaminated field also mapped to the same location as qCdT7. From these results, we conclude that the gene located at the qCdT7 locus will be of importance for phytoremediation.
Materials and methods
Plant materials
Two rice (Oryza sativa L) cultivars, Cho-Ko-Koku (indica cultivar) and Akita 63 (japonica cultivar), were used in this study. Cho-Ko-Koku and Akita 63 are high and low Cd accumulation varieties, respectively. An F2 population between Cho-Ko-Koku and Akita 63 was used for a QTL analysis of genes associated with Cd accumulation. One F2 plant carrying heterozygous allele for qCdT7 was selected from the F2 population. As much as 20 seedlings were selected from the self-pollinated F3 progeny of the F2 plant by analyzing the genotypes of RM6776 and RM5436 of their leaf tips and were used for analysis of Cd translocation rate.
Hydroponic culture and Cd treatment
Rice seeds were surface sterilized with 5% sodium hypochlorite, rinsed with water, and then incubated for 24 h at 32°C. Germinating seeds were transferred onto a nylon mesh floating on a nutrient solution (pH 5.4) containing 0.23 mM MgSO4, 0.18 mM NH4NO3, 0.18 mM CaCl2, 0.14 mM K2SO4, 0.09 mM Na2HPO4, 0.09 mM SiO2, 22.5 μM Fe(III)-EDTA, 9.2 μM H3BO3, 2.3 μM MnSO4, 0.78 μM CuSO4,0.77 μM ZnSO4, and 0.5 μM (NH4)6Mo7O24. The seedlings were cultured for 10 days in a growth cabinet (14 h light at 26°C, 10 h dark at 22°C). Each seedling was then transferred to a glass tube (φ35 mm × 20 cm) covered with aluminum foil and containing 200 mL of the nutrient solution supplemented with 1 μg of CdCl2 and was cultured in the growth cabinet for 20 days or 27 days (for the F3 seedlings). The nutrient solution was renewed every 2 days. At the end of the culture period, the roots of the seedlings were washed with deionized water 3 times. The seedlings were divided into shoot and root tissues and dried at 105°C for 24 h. The shoot and root tissues were weighed and then digested with 12 mL of HNO3–HClO4 (2:1 v/v) mix solution. Inductively coupled plasma-atomic emission spectrometry (ICP-AES) (Nippon-Jarrell-Ash, Tokyo, Japan) was used to determine the concentrations of Cd, Fe, Mg, Mn, and Zn in the digest solutions. The Cd translocation rate was estimated as the percentage of cadmium in the shoot compared to the whole plant.
Plants grown on a Cd-polluted paddy field
Rice seeds of the Cho-Ko-Koku and Akita 63 cultivars and of the F2 population were sown on 21st April 2008, and seedlings were transplanted to the Cd-polluted paddy field located in the Akita prefecture the northern part of Japan with spacing 15 cm × 15 cm on 28th May, and the soil in this paddy field classified as gray lowland soil (glysol). The irrigated water was drained on 29th July. The Cd concentration in the soil of the paddy field was 1.23 mg kg−1 (0.1 M HCl). Akita 63 and Cho-Kou-Koku headed on 12th and 20th August, respectively. Most of F2 individuals headed within August. We selected F2 plants which had headed between 9th and 29th August, and harvested on 1st October. Fertilizer was applied before planting: N, 70 kg ha−1; P, 100 kg ha−1; and K, 100 kg ha−1. The harvested plants were divided into shoots (leaves and stem) and ear parts. After weighing the dried materials, samples were ground to a powder, then 0.5 g was digested in 12 mL of HNO3–HClO4 (2:1 v/v) mixture. The Cd concentrations in the digest solutions were determined with the ICP-AES.
DNA extraction and analysis with SSR markers
Total DNA was extracted from the leaf tips of F2 seedlings according to the method described previously (Kato et al. 2007). Leaves were dried at 70°C for 2 h and then ground to powder with a stainless ball (φ3 mm) on a Micro Smash (MS-100 TOMY) vibrator. An extraction buffer (Edwards et al. 1991) was added to the powder and the mixture was incubated at room temperature for 1 h. DNA was precipitated from the mixture by adding an equal volume of 2-propanol and used in the PCR analyses.
The PCR was carried out in a 20 μl of reaction volume consisting of 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 1 U of TAKARA Taq HS (TAKARA), 4 nmol dNTP, 10 ng of genomic DNA, and 10 pmol of each set of primers for SSR markers in a Thermal Cycler 9600 or 9700 (Perkin-Elmer, Foster City, Calif.). As much as 35 PCR cycles, each consisting of 10 s of denaturation at 94°C, 30 s of annealing at 55°C, and 1 min of polymerization at 72°C, were performed. Polymorphisms for SSR markers were identified using 2.5% MetaPhor Agarose gels (FMC, Rockland, Me).
Construction of linkage map and QTLs analysis
Genetic linkage maps for the F2 populations grown either under hydroponic conditions or in the paddy field were constructed with 114 SSR markers (Akagi et al. 1996; Temnykh et al 2001; McCouch et al. 2002; International Rice Genome Sequencing Project 2005) using Mapmaker ver.2 (Lander et al. 1987). QTL analyses for both F2 populations were performed using the composite interval mapping method with Windows QTL Cartographer ver. 2.5 (Wang et al. 2007). An LOD value of 2.5 was used as the threshold for detection of potential QTLs influencing Cd concentrations in the shoot or root, and for the translocation rate under hydroponic culture conditions.
Results
Characteristics of Cd accumulation in the Cd hyperaccumulator variety Cho-Ko-Koku
Analysis of Cd uptake in rice grown in a paddy field with moderate contamination by Cd (1.33 mg kg−1) showed that shoots of Cho-Ko-Koku averaged 0.88 mg plant−1 compared to 0.17 mg plant−1 for Akita 63. This difference in uptake was mirrored by a difference in Cd concentrations between the cultivars: Cho-Ko-Koku, 30.6 mg kg−1 DW; Akita 63, 4.03 mg kg−1 DW. Our results indicate that Cho-Ko-Koku has a higher rate of Cd translocation to the shoots and accumulates Cd at a higher concentration (more than 5-fold greater) than Akita 63.
No difference was observed in total amount of Cd uptake by the seedlings of Cho-Ko-Koku and Akita 63 after 20 days growth under hydroponic conditions (Fig. 1). However, the shoots of Cho-Ko-Koku seedlings accumulated 4.99 μg of Cd compared to 1.41 μg in Akita 63 (Fig. 1). In contrast, Cd accumulation in the roots of Akita 63 seedlings was 2.6-fold higher than those of Cho-Ko-Koku (Fig. 1). Thus, Cho-Ko-Koku seedlings were characterized by a high Cd translocation ability from the root to shoot not by a high Cd uptake ability when grown under hydroponic conditions.
Cadmium contents in shoots and roots of Cho-Ko-Koku and Akita 63 seedlings grown under hydroponic culture conditions. A relatively low level of CdCl2 (5 μg L−1) was applied for 20 days. Black and white bars indicate Cho-Ko-Koku and Akita 63, respectively. Bars represent standard deviations of 6 seedlings. ** shows the level of significance at P < 0.01
Next, we compared the rates of translocation of metals including Cd in the seedlings (Fig. 2). Akita 63 seedlings retained 80% of the Cd in the root but Cho-Ko-Koku seedlings translocated 70% of the Cd to the shoot (Fig. 2). In contrast, both cultivars translocated between 60 and 90% of Fe, Mg, Mn, and Zn to the shoot and there were no obvious differences between the two cultivars (Fig. 2). These findings suggest that Akita 63 seedlings have a specific mechanism that causes retention of Cd in the root.
Metal translocation rate from roots to shoots. Rice seedlings were grown in a nutrient solution containing (5 μg L−1) of CdCl2 for 20 days. The metal translocation rate was estimated as the percentage of cadmium in the shoot compared to the whole plant. Black and white bars indicate Cho-Ko-Koku and Akita 63, respectively. Bars represent standard deviations of 6 seedlings. ** and * shows the levels of significance at P < 0.01 and 0.05, respectively
Segregation of Cd translocation rates in the F2 population
We investigated the genetic control of the different rates of accumulation of Cd in the two cultivars by an analysis of an F2 population between Cho-Ko-Koku and Akita 63. In the F2 population, the Cd accumulation rate in the shoot was highly correlated with the Cd translocation rate (r 2 = 0.96). Moreover, the F2 population could be divided into higher and lower accumulation rate groups. Of the 144 F2 individuals, 36 showed the higher (55–79%) Cd translocation rates similar to Cho-Ko-Koku, whereas 102 showed lower (12–48%) Cd translocation rates similar to Akita 63 (Fig. 3). Segregation rate was significantly fit to 1:3 ratio (high:low) (χ2 = 0.084 for 1:3, P = 0.768). A similar frequency distribution with two peaks was observed for Cd accumulation and concentration in the shoot (Figs. S1, S2). In contrast to shoots, the Cd concentration in the roots showed continuous variation in the F2 population (Fig. S2). The 1:3 Mendelian inheritance seen in the F2 population indicates that the higher Cd translocation present in Cho-Ko-Koku is controlled by a single recessive gene.
Segregation pattern of Cd translocation rates in the F2 population. The 144 F2 seedlings were grown in a nutrient solution with 5 μg L−1 of CdCl2 for 20 days. Means and standard deviations for the parental translocation rates are also included. CKK and A63 represent Cho-Ko-Koku and Akita 63, respectively
Analysis of the gene increasing Cd translocation
The chromosomal location of the recessive gene for Cd translocation was determined by a QTL analysis of the F2 population derived from Cho-Ko-Koku and Akita 63. A QTL with a LOD value of 68.6 was detected on chromosome 7 (Table 1). At the same position, QTLs were also detected for Cd accumulation and concentration both in shoot and in root (Table 1). The QTL controlling Cd translocation explained 88% of the phenotypic variation. The dominance effect of the QTL indicated that only the homozygous genotype for the Cho-Ko-Koku allele increased the rate of Cd translocation (Table 1). Since the QTL characteristics were consistent with the results of the segregation analysis, there is a gene responsible for Cd translocation between RM6776 and RM5436 on chromosome 7, and the locus of the gene was named as qCdT7 (Table 1, Fig. 7).
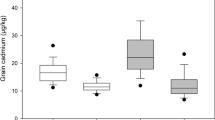
The F3 plants were classified into three types based on their genotypes of qCdT7 using markers, RM6776 and RM5436. The homozygotes for the Cho-Ko-Koku allele showed significantly higher Cd translocation rate (68.3 ± 7.97) than either the heterozygotes (40.4 ± 5.19) or homozygotes for the Akita 63 allele (33.2 ± 2.55; P = 0; Fig. 4). No significant difference for Cd translocation rate was observed between F3 plants heterozygous and homozygous for the Akita 63 allele, indicating that Cho-Ko-Koku allele for qCdT7 increase Cd translocation *** as a recessive manner (Fig. 4). This result confirmed the existence of a gene increasing Cd translocation from root shoot at the locus of qCdT7 on chromosome 7.
Frequency distribution for Cd translocation rate in F3 plants. The F3 plants were classified by the genotypes of qCdT7 region. Black, gray, and white bars indicate homozygous for Cho-Ko-Koku, heterozygous and homozygous for Akita 63, respectively. Means and SD for each genotype were indicated. Data marked with same letter are not significant level at P = 0.01 according to Tukey’s test
Another QTL affecting Cd concentration in the root was detected on chromosome 1; in this instance, the Cho-Ko-Koku allele decreased the concentration of Cd. Because the Cd concentration varied only in roots continuously under the hydroponic condition but not in shoots (Figs. S1, S2), this QTL promoted the root Cd concentration quantitatively in addition to environmental effects (Table 1, Fig. S2).
Contribution of qCdT7 to Cd accumulation in plants grown in a Cd-polluted paddy field
We performed a QTL analysis for Cd accumulation in an F2 population grown on a Cd-polluted paddy field to determine the effect of qCdT7 under normal culture conditions. In shoots of the F2 population, Cd accumulation showed a continuous and wide variation ranging from 0.04 to 2.14 mg plant−1 (Fig. 5). A similar frequency distribution was also observed for shoot Cd concentration and shoot dry weight (Fig. S3, Fig. 4). As much as 43 segregants showed greater accumulation of Cd than the parental cultivar Cho-Ko-Koku, one of these segregants accumulated Cd at a 2.5-fold greater rate than Cho-Ko-Koku (Fig. 5). The Cd accumulation in shoot was highly correlated with Cd concentration in shoot in the F2 population grown on Cd-polluted paddy field (r = 0.80) (Fig. 6). The low correlation coefficient (r = 0.26) between Cd accumulation and dry weight in shoot was observed. In this population, either Cd concentration or dry weight in shoots produced higher Cd accumulating plants than Cho-Ko-Koku (Fig. 6). The transgressive segregants in Cd accumulation should be feasible to breed rice varieties with a Cd accumulation performance that exceeds Cho-Ko-Koku which accumulated Cd at a much higher level in rice.
Frequency distribution of the Cd accumulation in shoots of the F2 population grown on the Cd-polluted paddy field. The cadmium concentration and dry weight in the shoots of 142 F2 plants were analyzed. CKK and A63 represent means and standard deviations of Cd accumulation in Cho-Ko-Koku and Akita 63, respectively
Three QTLs for Cd accumulation in shoots were detected on chromosomes 4, 7, and 12 (Table 2). The Cho-Ko-Koku alleles of the QTLs on chromosomes 7 and 12 increased shoot Cd accumulation, as did the Akita 63 allele on chromosome 4 (Table 2). Two Cho-Ko-Koku alleles of putative QTLs on chromosomes 7 and 12 increased Cd accumulation in shoots (Table 2). On chromosomes 7 and 12, QTLs for shoot Cd accumulation were mapped to the same positions as QTLs for shoot Cd concentration (Table 2). At these loci, there are probably genes responsible for Cd accumulation as a result of controlling the concentration of Cd in the shoot of plants grown on the Cd-polluted paddy field.
The QTL on chromosome 7 explained 39% of the Cd accumulation variation in plants grown on Cd-polluted soil (Table 2); it mapped to the identical position as qCdT7, which increased Cd translocation under hydroponic culture conditions (Fig. 7). Thus, the recessive gene at qCdT7 plays an important role in the high Cd uptake behavior of Cho-Ko-Koku on Cd-polluted soil.
Discussion
Cd is a harmful heavy metal and, in chronically exposed human populations, causes itai-itai disease. Cd contamination of crop fields increases the risk that the metal will enter the human food chain, and it is therefore important that it is removed from the environment. As a first step in developing a plant that can perform as an effective phytoremediator of Cd-polluted soils, we sought to identify genes in rice that influence Cd accumulation.
Unique characteristics of Cd accumulation in Cho-Ko-Koku
Rice plants have a wide distribution worldwide and display considerable genetic variation that is reflected by their diversity in Cd accumulation abilities (Arao and Ae 2003; Uraguchi et al. 2009). The Cho-Ko-Koku cultivar is known to be a Cd hyperaccumulator (Matsumoto et al. 2005; Murakami et al. 2009). In this study, we confirmed that Cho-Ko-Koku plants accumulate Cd in their shoots at approximately 5 times the level found in the non-Cd accumulating variety Akita 63. There are two key processes for high Cd accumulation in the shoots: a higher rate of Cd uptake from the soil through the root system; a higher rate of Cd translocation from the roots to the shoots through the xylem system (Clemens et al. 2002). Cho-Ko-Koku plants were assumed to have unique mechanisms for Cd accumulation, such as a higher xylem loading system of Cd in the Cd hyperaccumulator plants, A. halleri (Courbot et al. 2007; Hanikenne et al. 2008).
The ability of Cho-Ko-Koku plants to accumulate high levels of Cd was investigated in seedlings cultured in a hydroponic system. Overall, Cho-Ko-Koku plants absorbed similar amounts of Cd as Akita 63 under these conditions (Fig. 1). However, Cd accumulated in Cho-Ko-Koku shoots at 3 times the level present in Akita 63 (Fig. 1). This result showed that the higher rate of Cd accumulation in Cho-Ko-Koku plants was not due to higher uptake of the metal but rather was a consequence of a higher rate of Cd translocation. Since the xylem sap in Cho-Ko-Koku contains a higher concentration of Cd (Uraguchi et al. 2009), this cultivar is characterized by a high Cd translocation ability through xylem loading.
Analyses of other metals showed that more than 60% of Fe, Mn, Mg, and Zn were translocated from the roots to the shoots. Cho-Ko-Koku might have a specific transport system to Cd, not to other essential metals.
A single recessive gene in Cho-Ko-Koku controls Cd translocation
A 1:3 segregation ratio for high:low Cd translocation rates in the F2 population between Cho-Ko-Koku and Akita 63 indicated that a single recessive gene is responsible for the higher Cd translocation phenotype under hydroponic culture (Fig. 3). We detected a single QTL (qCdT7) with an extremely high value of LOD (68.6), which explained most of the variance (r 2 = 0.88) in the Cd translocation (Table 1). In addition, values of additive and dominance effects for qCdT7 indicate that the single recessive gene increasing Cd translocation locates on qCdT7 of Cho-Ko-Koku (Table 1). The existence of the recessive gene on qCdT7 locus was further confirmed by a significantly higher Cd translocation rate in the F3 plants homozygous for the Cho-Ko-Koku allele of qCdT7 (Fig. 4). Therefore, we concluded that the gene located in the qCdT7 region between RM6776 and RM5436 on chromosome 7 of Cho-Ko-Koku is responsible for the higher translocation rate of Cd from roots to shoots, and, consequently, produces the higher rate of Cd accumulation in shoots of this cultivar.
Previous studies detected several loci that were involved in Cd accumulation in shoots and unpolished seeds of rice (Ishikawa et al. 2005b; Ueno et al. 2009), suggesting that Cd accumulation is controlled by several genes. Three QTLs on chromosomes 3, 6, and 8 were identified as influencing Cd concentration in brown rice of the chromosome segment substitution lines of japonica variety Koshihikari and indica variety Kasalath grown on Cd-polluted soil (Ishikawa et al. 2005b). Additionally, three QTLs for Cd concentration in shoots were mapped to chromosomes 2, 5, and 11 in an F2 population between Badari Dhan (high Cd concentration variety) and Shwe War (low Cd concentration variety) under hydroponic conditions (Ueno et al 2009). Of these, the QTL located on chromosome 11 had the largest effect on shoot Cd concentration with a high LOD value (16.1) in a co-dominant manner (Ueno et al. 2009). In rice, Cd translocation via xylem loading is indicated to be important for Cd accumulation (Uraguchi et al. 2009). However, no gene for Cd translocation had been identified in rice. The gene, demonstrated here, located on qCdT7 determines Cd translocation phenotype as a major gene with recessive manner (Fig. 3, Table 1), demonstrated here, is unique.
The Cd hyperaccumulator species A. halleri transports Cd from the roots to shoots via a high level of xylem loading of Cd in the roots. This characteristic is controlled through increased transcription of the HMA4 gene that loads Cd and Zn to the xylem sap through companion cells (Courbot et al. 2007; Hanikenne et al. 2008). As only homozygotes of qCdT7 increased Cd translocation, a positive xylem loading system, such as HMA4 in A. halleri is not present in Cho-Ko-Koku. On the other hands, AtHMA3 belonging to the P1B-ATPase showed to transport Cd into vacuole, allowing storage toxic Cd in vacuole in A. thaliana (Morel et al. 2009), indicating proteins, such as AtHMA3 can store Cd in root. We hypothesized that Cho-Ko-Koku had lost a gene function involved in Cd storage in root cells, such as AtHMA3, that confers an increase in the xylem loading of Cd resulting in a higher level of Cd accumulation in the shoots. An increase in Cd translocation in recessive manner supports this hypothesis. Furthermore, as several genes for the metal transporter, including OsHMA3, located on qCdT7, OsHMA3 is a candidate for increasing Cd translocation in Cho-Kou-Koku.
The importance of qCdT7 for phytoremediation of Cd-polluted soil
We investigated the contribution of the gene located at qCdT7 for Cd accumulation in plants grown on Cd-polluted paddy field based on comparisons of both position and effect of QTLs on paddy field with that of qCdT7. In contrast to the hydroponic culture experiment, the shoot Cd accumulation segregated continuously with transgression in F2 population under field condition (Fig. 5). Since plants absorb Cd from the soil through multiple steps, various environmental factors can affect multiple genes under field conditions. However, we succeed in disclosing QTLs on chromosome 7 because of their high LOD value (Table 2). Since peaks of these QTL mapped to the same region as qCdT7 (Fig. 7) and Cho-Ko-Koku allele increased both Cd concentration and accumulation in shoot (Table 2), the gene at qCdT7 that confers higher Cd translocation also plays an important role in shoot Cd accumulation under Cd-polluted field.
We identified a recessive gene located at qCdT7 that controls the rate of Cd translocation in rice plants derived from the Cd hyperaccumulator variety Cho-Ko-Koku. The gene is also important for increased Cd concentration and accumulation in shoots of Cho-Ko-Koku plants grown on Cd-polluted soil. Overall, this gene will be of value in generating rice varieties for phytoremediation of Cd-polluted soils. In the future, we will clone the gene and clarify the molecular mechanisms of Cd accumulation in rice plants. Further analyses of the other QTLs identified here for Cd accumulation will also be important for breeding improved hyperaccumulating rice varieties for phytoremediation of Cd-polluted paddy fields.
References
Akagi H, Yokozeki Y, Inagaki A, Fujimura T (1996) Microsatellite DNA markers for rice chromosomes. Theor Appl Genet 93:1071–1077
Arao T, Ae N (2003) Genotypic variations in cadmium levels of rice grain. Soil Sci Plant Nutr 49:473–479
Baker AJM, McGrath SP, Reeves RD, Smith JAC (2000) Metal hyperaccumulator plants: a review of the ecology and physiology of a biochemical resource for phytoremediation of metal-polluted soils. In: Terry N, Banuelos G (eds) Phytoremediation of contaminated soil and water. Lewis Publishers, Boca Raton, Florida, pp 85–107
Bert V, Bonnin I, Saumitou-Laprade P, de Laguérie P, Petit D (2002) Do Arabidopsis halleri from nonmetallicolous populations accumulate zinc and cadmium more effectively than those from metallicolous populations? New Phytol 155:47–57
Clemens S (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88:1707–1719
Clemens S, Palmgren MG, Kramer U (2002) A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci 7:309–315
Courbot M, Willems G, Motte P, Arvidsson S, Roosens N, Saumitou-Laprade P, Verbruggen N (2007) A major quantitative trait locus for cadmium tolerance in Arabidopsis halleri colocalizes with HMA4, a gene encoding a heavy metal ATPase. Plant Physiol 144:1052–1065
Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19:1349
Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U (2008) Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453:391–395
Ibaraki T, Kuroyanagi N, Murakami M (2009) Practical phytoextraction in cadmium-polluted paddy fields using a high cadmium accumulating rice plant cultured by early drainage of irrigation water. Soil Sci Plant Nutr 55:421–427
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Ishikawa S, Ae N, Sugiyama M, Murakami M, Arao T (2005a) Genotypic variation in shoot cadmium concentration in rice and soybean in soils with different levels of cadmium contamination. Soil Sci Plant Nutr 51:101–108
Ishikawa S, Ae N, Yano M (2005b) Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa). New Phytol 168:345–350
Kato H, Tezuka K, Feng YY, Kawamoto T, Takahashi H, Mori K, Akagi H (2007) Structural diversity and evolution of the Rf-1 locus in the genus Oryza. Heredity 99:516–524
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Liu JG, Li KQ, Xu JK, Liang JS, Lu XL, Yang JC, Xhu QS (2003) Interaction of Cd and five mineral nutrients for uptake and accumulation in different rice cultivars and genotypes. Field Crops Res 83:271–281
Matsumoto S, Ito M, Masaki S, Kodama I, Kawamoto T, Nakagawa S, Itani T (2005) Selection of rice varieties with high cadmium contents (in Japanese). Jpn J Crop Sci 74(2):150
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P (2009) AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol 149:894–904
Morishita T, Fumoto N, Yoshizawa T, Kagawa K (1987) Varietal differences in cadmium levels of rice grains of Japonica, Indica, Javanica and hybrid varieties produced in the same plot of a field. Soil Sci Plant Nutr 33:629–637
Murakami M, Nakagawa F, Ae N, Ito M, Arao T (2009) Phytoextraction by rice capable of accumulating Cd at high levels: reduction of Cd content of rice grain. Environ Sci Technol 43:5878–5883
Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (2001) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet 100:697–712
Ueno D, Kono I, Yokosho K, Ando T, Yano M, Ma JF (2009) A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa). New Phytol 182:644–653
Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S (2009) Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot. doi:10.1093/jxb/erp119
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL cartographer 2.5. North Carolina State University, Raleigh
Acknowledgments
The authors thank Dr Hiroyuki Hattori, Dr Shinichi Nakamura, Dr Yoshihiro Kaneta (Akita Prefectural University) and Mr Masashi Ito (Akita Agricultural Experimental Station) for analysis of metals. This research was supported by a grant from the Ministry of Agriculture and Forestry of Japan (Genomics for Agricultural Innovation, RGB-2404).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Xu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2009_1244_MOESM1_ESM.tif
Fig. S1. Frequency distribution of Cd accumulation in shoots and roots of F2 seedlings grown under hydroponic culture conditions. The 144 F2 seedlings were grown in a nutrient solution with 5 μg L−1 of CdCl2 for 20 days. Means and standard deviations of the parental translocation rates are also included. CKK and A63 represent Cho-Ko-Koku and Akita 63, respectively (TIFF 333 kb)
122_2009_1244_MOESM2_ESM.tif
Fig. S2. Frequency distributions of Cd concentrations in shoots and roots of F2 seedlings grown under hydroponic culture conditions. The 144 F2 seedlings were grown in a nutrient solution with 5 μg L−1 of CdCl2 for 20 days. Means and standard deviations of the parental translocation rates are also included. CKK and A63 represent Cho-Ko-Koku and Akita 63, respectively (TIFF 324 kb)
122_2009_1244_MOESM3_ESM.tif
Fig. S3. Frequency distribution of the Cd concentration in shoots of the F2 population grown on the Cd-polluted paddy field. The cadmium concentration and dry weight in the shoots of 142 F2 plants were analyzed. CKK and A63 represent means and standard deviations of Cd accumulation in Cho-Ko-Koku and Akita 63, respectively (TIFF 148 kb)
122_2009_1244_MOESM4_ESM.tif
Fig. S4. Frequency distribution of the shoots dry weights of the F2 population grown on the Cd-polluted paddy field. The cadmium concentrations and dry weights in the shoots of 142 F2 plants were analyzed. CKK and A63 represent means and standard deviations of Cd accumulation in Cho-Ko-Koku and Akita 63, respectively (TIFF 138 kb)
Rights and permissions
About this article
Cite this article
Tezuka, K., Miyadate, H., Katou, K. et al. A single recessive gene controls cadmium translocation in the cadmium hyperaccumulating rice cultivar Cho-Ko-Koku. Theor Appl Genet 120, 1175–1182 (2010). https://doi.org/10.1007/s00122-009-1244-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1244-6