Abstract
We have newly identified five Terminal-repeat retrotransposon in miniature (TRIM) families, four from Brassica and one from Arabidopsis. A total of 146 elements, including three Arabidopsis families reported before, are extracted from genomics data of Brassica and Arabidopsis, and these are grouped into eight distinct lineages, Br1 to Br4 derived from Brassica and At1 to At4 derived from Arabidopsis. Based on the occurrence of TRIM elements in 434 Mb of B. oleracea shotgun sequences and 96 Mb of B. rapa BAC end sequences, total number of TRIM members of Br1, Br2, Br3, and Br4 families are roughly estimated to be present in 660 and 530 copies in B. oleracea and B. rapa genomes, respectively. Studies on insertion site polymorphisms of four elements across taxa in the tribe Brassiceae infer the taxonomic lineage and dating of the insertion time. Active roles of the TRIM elements for evolution of the duplicated genes are inferred in the highly replicated Brassica genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Retrotransposons comprise a significant fraction of the genomes in most organisms, particularly in plants. They may play an important role in maintaining chromosomal structure at the heterochromatin region, especially in the centromere and telomere, and in restructuring the euchromatin region in the eukaryotic genome (Kumar and Bennetzen 1999; Fedoroff 2000; Kumekawa et al. 2001; Yang et al. 2005b, c; Lim et al. 2006). Retrotransposons are classified into two types: long terminal repeats (LTR) retrotransposons and non-LTR retrotransposons. LTR retrotransposons are subclassified into the Ty1-copia and Ty3-gypsy groups, based on distinct sequence features and the order of encoded gene products (Kumar and Bennetzen 1999).
Recently, two unique groups of non-autonomous LTR retrotransposons were reported. They have some typical features of LTR retrotransposons but lack the coding capacity for mobility-related proteins. One group is the large retrotransposon derivatives (LARD) in grasses (Kalendar et al. 2004), and the other is terminal-repeat retrotransposons in miniature (TRIM) (Witte et al. 2001; Yang et al. 2005b; Antonius-Klemola et al. 2006). TRIM elements have terminal repeats (TR) ranging from 100 to 250 bp in length that encompass an internal domain of ∼300 bp and create 5 bp target site duplications (TSD). The internal sequence begins with a complement of the primer binding site (PBS) of tRNA-methonine and ends with typical polypurine tract (PPT) motifs. Although TRIM elements have important roles in restructuring plant genomes by affecting the promoter, coding region, and intron-exon structure of genes (Witte et al. 2001), they have not been well studied.
Recent studies revealed that Brassicaceae family compose 25 tribes, 338 genera, and ∼3,700 species (Al-Shehbaz et al. 2006; Beilstein et al. 2006). In the tribe Brassiceae, three basic Brassica species, B. rapa (A, n = 10), B. nigra (B, n = 8), and B. oleracea (C, n = 9), diverged during approximately 7.9–3.7 million years ago (MYA); the first divergence of the rapa-oleracea (A–C genome) clade with B. nigra (B genome) 7.9 MYA; and later divergence between B. rapa and B. oleracea 3.7 MYA (Truco et al. 1996; Inaba and Nishio 2002; Rana et al. 2004; Lysak et al. 2005; Al-Shehbaz et al. 2006; Beilstein et al. 2006). Also, three amphidiploids (allotetraploids), B. juncea (AB, n = 18), B. napus (AC, n = 19), and B. carinata (BC, n = 17) were synthesized by the natural allopolyploidization of the three basic Brassica species fewer than 10,000 years ago (U 1935; Rana et al. 2004). Many studies suggested that the tribe Brassiceae has triplicated chromosome segments (Lagercrantz and Lydiate 1996; O’neill and Bancroft 2000; Rana et al. 2004; Lysak et al. 2005; Parkin et al. 2005; Yang et al. 2005a; Nelson and Lydiate 2006) or much more complex duplication of chromosome segments which is collinear with the Arbidopsis genome (Lan and Paterson 2000; Babula et al. 2003; Lukens et al. 2003) after Arabidopsis-Brassica divergence about 20 MYA (Blanc et al. 2003; Bowers et al. 2003).
Recently we reported that a triplicated FLOWERING LOCUS C regions of Brassica rapa maintain sequence-level collinearity with the homoeologous Arabidopsis chromosome region with a concerted deletions and a few insertions. About 50% of triplicated genes are deleted and about 9% of genes including transposons are newly inserted during diploidization process of the triplicated genome (Yang et al. 2006). During the analysis, we found that new TRIM elements inserted into Brassica counterparts. We postulated that transposable elements such as TRIM elements that are activated after divergence with Arabidopsis may do an important role for evolution of duplicated genes and genome restructuring in the triplicated Brassica genome.
Here we describe finding of eight TRIM lineages from Arabidopsis and Brassica through intensive search of genomics data. And we try to infer their utility as DNA markers to identify the phylogeneic clade in the Brassica relatives and their role for evolution of the duplicated genes in the highly replicated Brassica genome.
Materials and methods
Data analysis
Three TRIM elements, Br1, Br2, and Br3, were identified by sequence comparison of four paralogous B. rapa BAC clones, KBrH052O08 (52O08), KBrH117M18 (117M18), KBrH004D11 (4D11), and KBrH080A08 (80A08) (from the HindIII BAC library of B. rapa L. ssp pekinensis inbred line ‘Chiifu’) (Park et al. 2005). These four BAC clones, located in different chromosomal regions, show overall collinearity with one another and with an Arabidopsis sequence (3.0–3.3 Mb region of Arabidopsis pseudo chromosome five sequence, GenBank accession no. NC_003076 Version 4:). The presence of multiple instances of the homologous sequences in B. rapa has been inferred to the result of genome triplication (80A08 vs. 4D11 vs. 52O08–117M18 clade). Two BACs 52O08 and 117M18 that are highly homologous and located near at the same chromosome are thought to be derived from a recent segmental duplication (Yang et al. 2006).
Sequence comparison in the regions of InDel, such as the gaps indicated by arrows in Supplemental Fig. 1, revealed closely spaced direct repeats flanking short sequences that begin with the primer binding site (PBS) of tRNA methionine and end with a polypurine tract (PPT), that is a distinct feature of TRIM elements (Witte et al. 2001). Pairwise sequence comparison was conducted using the program PipMaker (http://bio.cse.psu.edu) and manual inspection. TRIM homologues were identified by subsequent BLAST-NR searches to GenBank (http://www.ncbi.nlm.nih.gov/BLAST/), the B. oleracea database (http://www.tigr.org/tdb/e2k1/bog1/release.shtml), and 11,204 B. rapa BAC end sequences of B. rapa (GenBank accession numbers CW978640–CW988843) using the terminal repeat (TR) sequences of each element. Only a few full length TRIM elements are detected from the relatively short sequences of BAC ends and shotguns. Therefore, hits containing at least one entire terminal repeat sequence, were pooled for further analysis.
For phylogenetic analysis, we used only one TR sequence of each element. The internal sequences were not compared because most elements extracted from BAC ends and shotgun sequences had truncated features. We included Katydid-At1, -At2, and -At3 (Witte et al. 2001) for the analysis. A total of 146 TRIM elements were included in the phylogenetic analysis, and their positions and accession numbers are listed in Supplemental Table 1.
Phylogenetic analyses were performed using distance and parsimony approaches in PAUP* 4.0b10 (Swofford 2002). Neighbor-joining (NJ) analysis was based on the Kimura-2-parameter model. Tree topologies were evaluated using bootstrap analysis with 1,000 replicates for neighbor-joining methods. The multiple sequence alignment of selected TRs was created using CLUSTALW and the program Boxshade (http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html).
Copy number estimation
We estimated the copy numbers of Br elements in B. rapa and B. oleracea genomes using hit numbers to 96.5 Mb of B. rapa BAC end sequences (133,644 BAC end sequences with an average length of 723 bp) and 434.3 Mb of B. oleracea shotgun sequences (538,418 whole genome shotgun sequences with an average length of 807 bp), respectively. All the B. rapa BAC end sequences derived from three BAC libraries using B. rapa L. ssp pekinensis inbred line ‘Chiifu’ are collected from GenBank (ftp://ftp.ncbi.nih.gov/genomes/BACENDS/brassica_rapa/) and B. oleracea shotgun sequences are collected from TIGR (http://www.tigr.org/tdb/e2k1/bog1/release.shtml) on January 2006. Only one hit is counted for each element based on BLASTN (E-value < E−2). The equation to estimate the copy numbers is followed to the previous report (Zhang and Wessler 2004); (1/genome coverage) × Hits/2[1 + (average read length-TR length × 2)/(average read length + TR length × 2)].
Identification and mapping of the At members on Arabidopsis genome are conducted by MegaBLAST of At1, At2, At3, and At4 into Arabidopsis psedu chromosome sequences (GenBank accession numbers NC_003070, NC_003073, NC_003074, NC_003075, and NC_003076 for chromosome number 1, 2, 3, 4, and 5, respectively).
PCR analysis to detect insertion polymorphism of TRIM element
To find insertional polymorphisms among taxa in the tribe Brassiceae, pairs of PCR primers were designed from the flanking sequence of the insertion sites of the selected TRIM elements. Primer sequences and their estimated PCR product sizes are represented in Table 1. PCR analysis followed the previous report (Yang and Park 1998). Plant materials, 15 species in the tribe Brassiceae and two Arabidopsis thaliana are listed in Table 2. To inspect their insertion polymorphism among F1 varieties, we randomly purchased 15 Chinese cabbage F1 varieties that are commercially available in Korea. Names of the 15 F1 varieties are listed in an order of lane numbers for PCR: (1) Guembit; (2) Noranja; (3) Norangkimjang; (4) Manjeom; (5) Maeryuk; (6) Bulam-2, 7) Ssatnorang; (8) Seoul; (9) Speed 60 days; (10) Shinchoon; (11) Ssammat; (12) Jangmi; (13) Jangwon; (14) Jeongsang; (15) Hanyeorum.
Results
Identification of TRIM elements Br1, Br2, and Br3
TRIM elements were identified during sequence comparison of four paralogous B. rapa BAC clones, 52O08, 117M18, 80A08, and 4D11, containing flowering locus C (FLC) gene homologs and the homoeologous Arabidopsis counterpart. These sequences are collinear with InDels. Two BACs 52O08 and 117M18 containing FLC-3, 80A08 containing FLC-1, and 4D11 containing FLC-2 gene are located on gene-rich regions of long arm terminals of chromosome 2 (linkage group R3), 10 (linkage group R10), and 6 (linkage group R2), respectively (Yang et al. 2006).
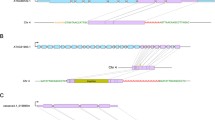
Three TRIM elements were identified by pairwise comparison of two highly homologous BACs 52O08 and 117M18 and their Arabidopsis counterpart. The collinear sequence of these two BACs showed over 98% sequence similarity, with the exception of the InDels (Supplemental Fig. 1). Cytogenetic inspection and sequence comparison revealed that these two regions were segmentally duplicated about 0.8 MYA (Yang et al. 2006). Two TRIM elements were identified at the InDel sites in 52O08 (Br1-52O08) and 117M18 (Br1-117M18), respectively. About 5 bp target site duplication (TSD) is detected by comparison with the related empty site (RESite) in each BAC sequence (Fig. 1a, b; Supplemental Fig. 1). A stowaway-type miniature inverted repeat transposable element (MITE), named BrMi1 (357 bp), was further identified as a nest insertion within a TA rich region of a TR of Br1-52O08.
Finding TRIM elements between homologous sequences and alignment of their RESites Inspection of the InDel region in the dotplots between two homologous BAC sequences, 52O08 and 117M18 (Left panel of Supplemental Fig. 1) and between 117M18 and the counterpart Arabidopsis sequence (Right panel of Supplemental Fig. 1) resulted in finding TRIM elements, Br1 in 52O08 (Br1-52O08) (a), Br1 in 117M18 (Br1-117M18) (b), and Br3 in both BAC clones (Br3-52O08) (c) based on their features, the related empty sites (RESites) and the 5 bp target site duplication (TSD). Another feature, a MITE (BrMi1, 357 bp), is identified as a nest form in the Br1-52O08 (a). Similarly, a Br2 is detected in a BAC clone 80A08 based on its RESites in another paralogous Brassica BAC clone 4D11 and a Arabidopsis sequence on chromosome 5 (3,189,981-3,190,167 bp) (d). BACs 52O08, 117M18, 4D11, and 80A08 containing flowering locus C (FLC) homologs are located on cytogentic chromosomes 2 (linkage group R3), 2 (linkage group R3), 6 (linkage group R2), and 10 (linkage group R10), respectively (Yang et al. 2006)
Another TRIM element, termed Br3, was found in both BAC clones 52O08 and 117M18 in comparison with the Arabidopsis counterpart sequence (Fig. 1c, Supplemental Fig. 1c) indicating that Br3 inserted into the region before the segmental duplication event. Another TRIM element, Br2-80A08, was identified by comparing BAC clones 80A08 and 4D11 and the counterpart sequence of Arabidopsis (NC_003076: 3, 189, 9813, 190, 157 bp) (Fig. 1d).
TRIM elements At4 and Br4
Based on subsequent BLAST and data mining, another TRIM lineage, named Katydid-At4 (At4), was newly identified from Arabidopsis. The At4 element contains 136–258 bp internal sequences with flanking 198 bp TRs. Seventeen At4 elements including one tri-TR and six solo-TR elements were identified (Supplemental Tables 1, 2). Their homologous elements, Br4, were identified from the B. rapa BAC end sequences (BES) and B. oleracea shotgun sequence databases (Supplemental Table 1), based on the similarities of their sequence and structure.
All members have distinct features of TRIM elements with a miniature terminal repeats and an internal sequence, even though there is variability in their sequences and sizes (Fig. 2). The Br2 element is slightly larger than that of Br1 (385 vs. 364 bp, respectively), and both showed significant sequence similarity with the Arabidopsis At1 and At2 families. In contrast, the Br3 elements show no sequence similarity with any Arabidopsis members and the size is much larger than the TRIM elements reported before (Supplemental Fig. 2) (Witte et al. 2001). The Br4 family shows significant sequence similarity with the At4 and their internal sequence is hyper variable in its length even though the sequence itself is conserved (Supplemental Fig. 3).
Schematic representation of TRIM elements Br1, Br2, Br3, and Br4 TRs are denoted as boxed triangles showing the same features based on sequence similarity. In all four families the internal sequences begin with primer binding sites (PBS) of tRNA methionine and end with a polypurine tract (PPT). Numerals indicate nucleotide length
Phylogenetic analysis of TRIM elements
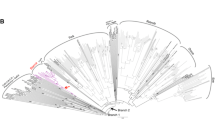
A total of 146 TRIM elements, included the At1, At2, and At3, which are identified in a previous report (Witte et al. 2001), are analyzed for phylogentic analysis. Tree topologies derived from neighbor-joining methods show eight independent lineages. Members derived from Arabidopsis are grouped as At1, At2, At3, and At4 and members derived from Brassica are grouped as Br1, Br2, Br3, and Br4 (Fig. 3). All the members in each lineage show almost same feature of the family represented in Fig. 2. Although TRIM elements derived from Arabidopsis and Brassica belonged to independent lineages, elements derived from B. rapa (red dots), B. oleracea (gray dots), and B. napus (green dots) are intermingled in each Br lineage (Fig. 3). This suggests that At and Br members were independently activated in each genus after the divergence of Brassica and Arabidopsis, respectively.
Phylogenetic tree of TRIM elements derived from the genus Brassica and Arabidopsis Bootstrap values are denoted as percentages in parenthesis. A total of 146 TRIM elements are listed in Supplemental Table 1 by their nucleotide positions from the GenBank accessions. Elements from Arabidopsis, B. rapa, B. oleracea, and B. napus are denoted as pink, red, gray, and green dots, respectively. Representative TRIM families are designated as numbers in parenthesis
PCR analysis to confirm the insertion polymorphisms
Activation time of Br members can be estimated by inspection of the insertion of each member among various taxa in Brassica relatives. We inspected the insertion sites of independent Br element by PCR analysis against 17 taxa in the tribe Brassiceae. Nine Br elements were inpected and five of them were not included in this paper because these five amplified the target bands in Brassica rapa but did not in other species or amplified unspecific several bands through species. Four primer sets produced bands that represent the insertion polymorphism against most of 17 taxa (Fig. 4; Tables 1, 2). Three of four cases of TRIM insertions are unique in B. rapa lineage (B, C, D) and one is unique in rapa-oleracea clade. All four cases of insertion are not detected in B. nigra as well as other sister genera such as Diplotaxis, Eruca, Erucastrum, Sinapis, Raphanus, and Arabidopsis indicating that these are inserted into B. rapa lineage after the rapa-oleracea clade diverged from the B. nigra and other genera.
PCR analysis for the detection of insertion polymorphism across 17 taxa in Brassicaceae The lane names follow the abbreviations of the plant materials based on genome compositions (Table 2). PCR analyses were conducted using flanking sequence of the four TRIM elements insertion sites, Br2-6P20 (a), Br2-80A08 (b), Br1-52O08 (c), and Br1-12I15 (d). Primers and estimated sizes are listed in Table 1. Dark and white arrowheads denote PCR bands with insertion and without insertion, respectively. e Phylogenetic lineages of three basic Brassica species and their allopolyploids are shown based on divergence dating (U 1935; Rana et al. 2004; Truco et al. 1996), and the estimated insertion points of each element are denoted as arrows based on appearance of the insertion polymorphism in the left panels A–D
The appearance of each TRIM element in lineage of Brassica species depends on the insetion time. A solo TR of Br2 found in the BAC end sequence (BES) of KBrH006P20 (Br2-6P20) occurred in B. rapa (A genome) and B. oleracea (C genome) and A-and C-subgenome of the three allopolyploids, B. juncea (AB), B. napus (AC), and B. carinata (BC) (Fig. 4a). The Br2-80A08 appeared in B. rapa (A genome) and the A-subgenome of B. juncea (AB genome) and B. napus (AC genome) (Fig. 4b). The Br1 element in BAC end KBrH012I15 (Br1-12I15) is unique in one of four inbred lines of B. rapa (Fig. 4d). Similarly, the Br1-52O08 insertion is detected in only one of four inbred lines of B. rapa, and it is additionally detected in the B. napus (Fig. 4c).
The insertions, Br2-80A08 and Br2-6P20 that are specifically appeared in A-and A and C-genome, respectively, were detected in all of 15 commercial F1 varieties of Chinese cabbage (Fig. 5a, b). The Br1-12I15 and Br1-52O08 that are recently activated appeared as heterozygous forms in four (Nos. 1, 8, 12, 14) and two F1 varieties (Nos. 7 and 8) (Fig. 5b).
PCR analysis for the detection of insertion polymorphism across 15 Chinese cabbage F1 varieties Primer information is the same as A–D in Fig. 4. The lineage with insertions are listed as numerals in panel C and D. Dark and white arrowheads denote PCR bands with insertion and without insertion, respectively
Insertion of TRIMs in genes
We found a TRIM insertion in one of two homologous gene predictions that existed in two sequenced BAC clones, KBrH004D11 and KBrH080A08, of B. rapa (Fig. 6). The gene 4D11_18 remains conserved with an Arabidopsis ortholog At5g10170, while the 80A08_12 is dramatically rearranged by a deletion and an insertion of the Br2. The Br2 provides novel first exon decoding 12 amino acids and 5′ UTR (Fig. 6b, c). PCR analysis revealed that the Br2 inserted into the 80A08_12 in the B. rapa lineage (Fig. 4b).
Gene rearrangement by insertion of Br2 in the BAC clone 80A08 Paralogous BACs 80A08 and 4D11 are located chromosome 10 (linkage group R2) and 6 (linkage group R2), respectively (Yang et al. 2006). a, b Predicted genes in the 80A08 and 4D11, 80A08_12 (52,621–54,970) and 4D11_18 (57,532–57,659) are orthologous with a Arbidopsis gene At5g10170. Exons and introns are denoted as black (gray in dotplots) and white boxes, respectively. Percent identity plot (PIP) indicates high, low, and no similarity as red, green, and white boxes, respectively. Deletion is denoted as red dotted lines. Br2 insertion is represented as blue dotted lines (c) Exon arrays of At5g10170 (NP_196579, putative IPS), 4D11_18, and 80A08_12 are denoted as the black boxes with numerals. Br2 insertion in the 80A08_12 is represented as blue lines compared to the 4D11_18 and the novel First exon is denoted as blue dotted lines compared to the Br2 element
Nucleotide sequences of Br elements occur as chimeric features in many expressed sequence tags (ESTs): the TR sequence of Br1 and Br2 in EST sequences of Lycoris longituba (GenBank ID gi46452940) and of B. napus (such as gi32499949, gi32500904, gi32505705); the internal sequence of Br1 in Arabidopsis ESTs (such as gi5841198, gi1053954); the internal sequence of Br3 in many full-length potato callus cDNAs (such as gi39799021); and the TR sequence of Br4 in Arabidopsis cDNA (such as gi42534613). Furthermore, we have identified that one expressed gene is chimeric between a Br1 and a known gene. One chimeric B. napus EST (gi56841787) begins with an inverted form of an almost intact Br1 element and ends with unique sequence, which is identical to Arabidopsis aldose 1-epimerase gene (Fig. 7). Additional intact form of the aldose 1-epimerase gene ortholog should be present in the B. napus genome because it is an allopolyploid of the B. rapa and B. oleracea genome (U 1935).
Schematic representation of a chimeric EST sequence A B.napus EST sequence (gi56841787) and it’s homologous gene in Arabidopsis (gi26452397) are represented as boxes (middle) and lines (top), respectively. The inverted form of Br1 element (bottom) shows similarity with the beginning 321 bp of the EST. The remaining sequence of the EST is similar to 3′ region of the Arabidopsis gene, a putative aldose 1-epimerase (At5g15140: gi26452397). The homologous regions are denoted as shade boxes with sequence similarity and nucleotide positions (numerals) in each sequence
Copy number estimation of the TRIM elements of B. rapa and B. oleracea
TRIM elements, including degenerate members, were identified at 92 positions in the 120 Mb Arabidopsis genome sequence (Supplemental Table 2). These distributed relatively evenly through five Arabidopsis chromosomes (Fig. 8).
In silico Mapping of TRIM elements on Arabidopsis chromosomes. The Katydid-At1, -At2, -At3 and the newly identified At4 are positioned as different colors on the chromosomal region based on the Arabidopsis pseudo chromosome sequence Version 4. Their exact positions are listed in Supplemental Table 2
The copy numbers of the TRIM elements in B. oleracea and B. rapa genome are estimated based on the occurrence of their homologs in 434 Mb of B. oleracea shotgun sequences and 96 Mb of B. rapa BAC end sequences, respectively. The numbers are roughly estimated to be present in 660 and 530 copies in B. oleracea and B. rapa genomes, respectively (Table 3).
Distribution of these elements are unknown yet in B. rapa. However, five TRIM elements were identified from BACs 52O08, 117M18, and 80A08 (Fig. 1 and Supplemental Fig. 1), that were mapped on gene-rich regions of B. rapa chromosomes based on BAC sequences (Yang et al. 2006).
Discussion
Dating of the insertion time based on the phylogenetic lineage
The distribution patterns of Br elements across taxa show their evolutionary lineages and permit inference of the time periods in which the insertion events occurred (Kumar and Bennetzen 1999). Inspection of various insertion sites of Br elements can be used as clade markers for the species in Brassicaceae such as the previous study using insertion sites of SINE element (Lenoir et al. 1997; Tatout et al. 1999).
The genus Brassica includes many important vegetable crops including three basic Brassica species, B. rapa (A genome), B. nigra (B genome), and B. oleracea (C genome), and also three allotetraploids, B. juncea (AB genome), B. napus (AC genome), and B. carinata (BC genome). Three basic Brassica species diverged during approximately 7.9–3.7 MYA (Truco et al. 1996; Inaba and Nishio 2002; Rana et al. 2004; Lysak et al. 2005; Al-Shehbaz et al. 2006; Beilstein et al. 2006) and three allotetraploids are resynthesized by the natural allopolyploidization fewer than 0.01 MYA (U 1935; Rana et al. 2004).
To identify insertion time of various TRIM elements and to know their utility for clade marker, we have searched the insertion polymorphism across the tribe Brassiceae. We have targeted the intact Br1 and Br2 element because Br1 and Br2 are relatively small (less than 350 bp) compare to Br3 and Br4 elements. The Br2-6P20, a solo-TR, appeared in the A and C genomes but not in the B genome, indicating that it inserted into the rapa-oleracea (A–C genome) clade after divergence from B. nigra (B genome) during 7.9–3.7 MYA (Inaba and Nishio 2002; Rana et al. 2004; Lysak et al. 2005). The Br2-80A08, B. rapa (A genome)-unique, is likely to have been inserted into the B. rapa lineage within 3.7–0.01 MYA, after the rapa-oleracea split, but before the allopolyploidization of the AB and AC genomes (Inaba and Nishio 2002; Rana et al. 2004). The Br1-52O08 is detected in only one of two highly homologous BACs 52O08 and 117M18 that are segmentally duplicated about 0.8 MYA (Yang et al. 2006). It is identified in only one of four inbred lines of B. rapa and one of allopolyploids (AC), indicating it was activated in a certain B. rapa lineage during 0.8–0.01 MYA. The Br1-12I15 is unique in only one of four inbred lines of B. rapa, indicating it was activated after allopolyploidization events fewer than 10,000 years ago (Fig. 4e).
The assumption of the insertion time is supported by inspection of the 15 F1 hybrid cultivars. Two Br elements, Br2-80A08 and Br2-6P20, that assumed to be inserted in an ancient B. rapa lineage remained in all of 15 commercial F1 varieties of Chinese cabbage (Fig. 5a, b). However, two Br elements, Br1-12I15 and Br1-52O08, that are recently activated in B. rapa appeared in four and two F1 varieties, respectively (Fig. 5b). All the six insertions are heterozygous in the F1 varieties indicate that they are derived from one of their parental lines. If an insertion of Br element is identified as unique in one breeding line, the insertion polymorphism can be used as DNA marker for the breeding line that protects the breeder’s right for the specific breeding line.
TRIMs play an active role for rearrangement of duplicated genes
Transposons are genetic elements that can spread within genomes, and that constitute an important fraction of eukaryote genomes. Most transposons are activated by stress and have reshaped eukaryote genomes in many ways (Casacuberta and Santiago 2003). Plant centromeric and pericentromeric regions often contain a large fraction of retrotransposons that could be important for the functionality of these regions (Zhong et al. 2002; Yang et al. 2005b). Inter-specific hybridization or allopolyploidization might activates the transposition of a centromere specific retrotransposon to facilitate rapid karyotypic evolution (O’neill et al. 1998; Lim et al. 2006).
In spite of their mutagenic capacity, both LTR-retrotransposons and MITEs can be found associated to the evolution of plant genes. MITEs are distributed at gene-rich regions of the rice genome sequences (IRGSP 2005). The insertion of MITEs within genes can modify the promoter and terminator sequences, as well as the translational start and coding sequences. LTR-retrotransposon sequences are also frequently found associated to genes, suggesting that the modification of the regulation of target gene expression due to the presence of promoter and terminator sequences within the LTRs (Casacuberta and Santiago 2003).
In silico mapping of the insertion sites in Arabidopsis showed that the Katydid-At elements were evenly distributed throughout the genome (Fig. 8, Supplemental Table 2) and some were directly related to the structure of the expressed genes in Arabidopsis (Witte et al. 2001). The haploid genome size of B. rapa (529 Mb) and B. oleracea (696 Mb) is 3.5–4.5 times greater than that of Arabidopsis (157 Mb) (Johnston et al. 2005). However, the estimated copy numbers of Br elements are more than six times greater in Brassica species than in Arabidopsis. Chromosomal locations of these are unknown yet in B. rapa genome except five TRIM elements identified from BAC clones 52O08, 117M18, and 80A08. These three BACs are derived from gene-rich euchromatin of chromosome 2 and 10 (Yang et al. 2006). Based on evidence of distribution of TRIM elements in the Arabidopsis euchromatin regions and occurrence of chimeric feature of TRIM elements in lots of ESTs, the abundant TRIM elements seem to play an important role for restructuring the host genome in B. rapa as do MITE elements in the rice genome (Jiang and Wessler 2001; Jiang et al. 2003; IRGSP 2005). Moreover TRIM elements have promoter and terminator sequence in their small terminal repeat sequences that can provide active modification of gene feature by internal insertion.
We have identified many of expressed sequence tags (ESTs) that include a part of Br elements as chimeric forms. Sequence-level analysis of the triplicated B. rapa genome sequence revealed that about 12 and 44% of triplicate genes remained as triplet and doublet paralogues, respectively. Furthermore some of these genes are duplicated once more by a recent segmental duplicateion (Yang et al. 2006). These duplicated genes can attain a selective advantage by gaining new function or silencing in each duplicate in a complementary manner (reviewed in Hurles 2004). We found that one of the triplicated genes is dramatically rearranged at its 5′ coding and promoter region (Fig. 6). Collective data suggest that TRIM elements play an active role for gaining new function by rearrangement of the duplicate genes in the highly replicated Brassica genome.
References
Al-Shehbaz IA, Beilstein MA, Kellogg EA (2006) Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Syst Evol 259:89–120
Antonius-Klemola K, Kalendar R, Schulman A (2006) TRIM retrotransposons occur in apple and are polymorphic between varieties but not sports. Theor Appl Genet 112:999–1008
Babula D, Kaczmarek M, Barakat A, Delseny M, Quiros CF, Sadowski J (2003) Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol Genet Genomics 268:656–665
Beilstein MA, Al-Shehbaz IA, Kellogg EA (2006) Brassicaceae phylogeny and trichome evolution. Am J Bot 93:607–619
Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13:137–144
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438
Casacuberta JM, Santiago N (2003) Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene 311:1–11
Fedoroff N (2000) Transposons and genome evolution in plants. Proc Natl Acad Sci USA 97:7002–7007
Hurles M (2004) Gene duplication: the genomic trade in spare parts. PLoS Biol 2:900–904
Inaba R, Nishio T (2002) Phylogenetic analysis of Brassiceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theor Appl Genet 105:1159–1165
IRGSP (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421:163–167
Jiang N, Wessler SR (2001) Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13:2553–2564
Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, Lopez R, Price HJ (2005) Evolution of genome size in Brassicaceae. Ann Bot (Lond) 95:229–235
Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166:1437–1450
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2001) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res. 8:285–290
Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144:1903–1910
Lan TH, Paterson AH (2000) Comparative mapping of quantitative trait loci sculpting the curd of Brassica oleracea. Genetics 155:1927–1954
Lenoir A, Cournoyer B, Warwick S, Picard G, Deragon JM (1997) Evolution of SINE S1 retroposons in Cruciferae plant species. Mol Biol Evol 14:934–941
Lukens L, Zou F, Lydiate D, Parkin I, Osborn T (2003) Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164:359–372
Lim KB, Yang TJ, Hwang YJ, Kwon SJ, Kim JA, Choi BS, Lim MH, Kim JS, Jin M, Park JY, et al (2006) Characterization of the major centromere and pericentromere retrotransposons in Brassica rapa and distribution in the related Brassica species. Plant J. In press
Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res. 15:516–525
Nelson MN, Lydiate DJ (2006) New evidence from Sinapis alba L. for ancestral triplication in a crucifer genome. Genome 49:230–238
O’Neill CM, Bancroft I (2000) Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 23:233–243
O’Neill RJW, O’Neill MJ, Graves JAM (1998) Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393:68–72
Park JY, Koo DH, Hong CP, Lee SJ, Jeon JW, Lee SH, Yun PY, Park BS, Kim HR, Bang JW, et al (2005) Physical mapping and microsynteny of Brassica rapa ssp. pekinensis genome corresponding to a 222 kb gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol Genet Genomics 274:579–588
Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781
Rana D, van den Boogaart T, O’Neill CM, Hynes L, Bent E, Macpherson L, Park JY, Lim YP, Bancroft I (2004) Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40:725–733
Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version. 4.0b10. Sinauer Associates, Sunderland, MA
Tatout C, Warwick S, Lenoir A, Deragon JM (1999) SINE insertions as clade markers for wild Crucifer species. Mol Biol Evol 16:1614–1621
Truco MJ, Hu J, Sadowski J, Quiros CF (1996) Inter-and intra-genomic homology of the Brassica genomes: implications for their origin and evolution. Theor Appl Genet 93:1225–1233
Tsunoda S, Hinata K, Gomez-Campo C (1980) Preservation of genetic resources. Japan Scientific Societies Press, Tokyo
U N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Witte CP, Le QH, Bureau T, Kumar A (2001) Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci USA 98:13778–13783
Yang TJ, Kim JS, Kwon SJ, Choi BS, Lim KB, Kim JA, Jin M, Park JY, Lim MH, Kim HI, et al (2006) Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C regions of Brassica rapa. Plant Cell 18:1339–1347
Yang TJ, Kim JS, Lim KB, Kwon SJ, Kim JI, Jin M, Park JY, Lim MH, Kim HI, Kim SH, et al (2005a) The Korea Brassica Genome Project: a glimpse of the Brassica genome based on comparative genome analysis with Arabidopsis. Comp Funct Genomics 6:138–146
Yang TJ, Lee S, Chang SB, Yu Y, de Jong H, Wing RA (2005b) In-depth sequence analysis of the tomato chromosome 12 centromeric region: identification of a large CAA block and characterization of pericentromere retrotranposons. Chromosoma 114:103–117
Yang TJ, Park HG (1998) Optimization of RAPD condition in Capsicum annuum. Kor. J. Breeding 30:16–23
Yang TJ, Yu Y, Lee S, Chang SB, Oh CS, Ahn SN, de Jong H, Wing RA (2005c) Toward closing rice telomere gap: mapping and sequence characterization of rice subtelomere regions. Theor Appl Genet 111:467–478
Zhang X, Wessler SR (2004) Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc Natl Acad Sci USA 101:5589–5594
Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK (2002) Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14:2825–2836
Acknowledgments
This work was supported by grants from the BioGreen 21 Program, RDA, and NIAB, Korea. The authors thank Macrogen (http://www.macrogen.com/english/index.html) for BAC end sequencing. We are grateful for Maryana Bhak and Chang-Bae Kim for valuable comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. S. Heslop-Harrison.
Tae-Jin Yang and Soo-Jin Kwon have equally contributed to this work.
Electronic supplementary material
Below are the electronic supplementary materials.
Rights and permissions
About this article
Cite this article
Yang, TJ., Kwon, SJ., Choi, BS. et al. Characterization of terminal-repeat retrotransposon in miniature (TRIM) in Brassica relatives. Theor Appl Genet 114, 627–636 (2007). https://doi.org/10.1007/s00122-006-0463-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0463-3