Abstract
Phytophthora infestans, the causal agent of late blight, threatens potato production worldwide. An important tool in the management of the disease is the use of resistant varieties. Eleven major resistance genes have been identified and introgressed from Solanum demissum. However, new sources of resistance are continually sought. Here, we report the characterization and refined genetic localization of a resistance gene previously identified as Rber in a backcross progeny of Solanum tuberosum and Solanum berthaultii. In order to further characterize Rber, we developed a set of P. infestans isolates capable of identifying each of the 11 R-genes known to confer resistance to late blight in potato. Our results indicate that Rber is a new resistance gene, different from those recognized in S. demissum, and therefore, it has been named R Pi-ber according to the current system of nomenclature. In order to add new molecular markers around R Pi-ber , we used a PCR-based mapping technique, named MASP-map, which located R Pi-ber in a 3.9 cM interval between markers CT240 and TG63 on potato chromosome X. The location of R Pi-ber coincides with an area involved in resistance to different pathogens of potato and tomato.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato is the fourth most important crop in the world. In 2004, more than 328 million metric tons of potatoes were produced worldwide (FAOSTAT 2005). Potato production is threatened by diverse pathogens, the worst of which is the oomycete Phytophthora infestans, the causal agent of late blight. Late blight epidemics can be devastating, sometimes causing total crop losses (Fry and Goodwin 1997). It has been estimated that the management of the disease costs $3.5 billion annually in developing countries alone (GILB 2004).
Management strategies for late blight include the application of fungicides, the use of healthy seed tubers for planting and host resistance. However, the occurrence of isolates resistant to some modern fungicides emphasizes the need for host resistance (Deahl et al. 1993; Goodwin et al. 1996; Grunwald et al. 2001). Additionally, the high cost of fungicide applications, increasing awareness of health and environmental risks and world-wide pressures to minimize the use of chemical sprays (Fry and Goodwin 1997) also make the use of host resistance a priority.
Two types of resistance to late blight have been described in potato. First, general resistance slows the spread of the disease and it is understood to be often polygenic (Leonards-Schippers et al. 1994; Umaerus and Umaerus 1994), and strongly correlated with maturity type (Bormann et al. 2004; Simko 2002) which makes it difficult to breed into new varieties (Wastie 1991). Second, specific resistance confers immunity or near immunity to the plant through a hypersensitive response and is thought to be monogenic. Genes governing such resistance have been termed R-genes, and are thought to produce proteins involved in pathogen recognition and the initiation of defense responses. Eleven resistance genes have been introgressed into the cultivated potato (Solanum tuberosum) from its wild relative Solanum demissum (Van der Plank 1963; Wastie 1991), and they are “named” R1, R2,..., R11. Of these, only five have been located on the genetic map of potato: R1 on chromosome V (Leonards-Schippers et al. 1992); R2 on chromosome IV (Li et al. 1998); and R3a, R3b (Huang et al. 2004), R6, and R7 on chromosome XI (El-Kharbotly et al. 1996). However, R5–R11 have recently been identified as alleles of R3 (Huang 2005), which would also locate them in chromosome XI. Of the R-genes from S. demissum, only R1(Ballvora et al. 2002) and R3a (Huang et al. 2005) have been cloned and sequenced.
R-genes from other hosts of P. infestans have been reported. Three resistance genes against P. infestans have been mapped in tomato—a close relative to potato: Ph-1 on chromosome 7, Ph-2 on chromosome 10 (Moreau et al. 1998), and Ph-3 on chromosome 9 (Chunwongse et al. 2002). An R-gene from Solanum bulbocastanum on chromosome VIII (Helgeson et al. 1998; Naess et al. 2000), named RB, has now been cloned (Song et al. 2003). Rpi has been found in S. pinnatisectum and mapped to chromosome VII (Kuhl et al. 2001). We previously reported an R-gene located chromosome X of Solanum berthaultii (Ewing et al. 2000), named Rber, since it segregates from the S. berthaultii parent in a backcross with S. tuberosum.
Unfortunately, P. infestans has been shown to rapidly overcome the classic R-genes from S. demissum when they were deployed in potato cultivars. The result is that such specific resistance has had short durability (Wastie 1991), and the immunity hoped for in such R-genes has contributed little to practical late blight management. However recent reports (Bormann et al. 2004; Stewart et al. 2003) suggest that there may be some residual disease suppression effect of R-genes even when interacting with a “compatible” isolate. Knowledge of signaling pathways and downstream events may open new approaches to disease suppression, so it is important to learn as much as possible about such genes.
Rber was mapped to a 13 cM interval between the restriction fragment length polymorphism (RFLP) markers CT214b and TG63 in a diploid backcross progeny. However, it was uncertain if Rber was a new gene or an allele of a resistance gene previously introgressed from S. demissum. The isolate of P. infestans used for the identification of Rber was a complex race, compatible with (i.e., not recognized by) R-genes: R1, R2, R3, R4, R5, R6, R7, and R10. Therefore, we inferred that Rber might be a new R-gene (Ewing et al. 2000).
The goal of the research reported here was to determine if Rber was different from R8, R9 or R11, and to provide a finer map location of this R-gene. We determined the identity of Rber using a set of P. infestans isolates known as a “tester set”, since they allowed potato plants to be screened for the 11 known R-genes and therefore the identification of new R-genes. We also investigated a series of markers from diverse sources to provide a finer map location of this R-gene.
Materials and methods
Plant material
Host differential set
We used a differential set of S. tuberosum genotypes, each one containing one known resistance gene (Table 1). This differential set was obtained from the US Department of Agriculture—Potato Germplasm Introduction Station in Sturgeon Bay, WI, USA and had been used previously (Abu-El Samen et al. 2003; Spielman et al. 1989). Historical and collection data are available for each accession (USDA 1999). The cultivars Norchip or Katahdin were used as R-gene-free controls with no known R-genes, and served to indicate the success of inoculations (Mastenbroek 1952). For all experiments, plants were grown and maintained under greenhouse conditions at the Cornell University facilities (Ithaca, NY).
Backcross mapping population
The backcross population was developed by Bonierbale et al. (1994). Briefly, an interspecific progeny was developed by crossing a dihaploid S. tuberosum clone (USW-2230 Saco, GS 193 in the GRIN database) (USDA 1999) as female with an individual of the accession PI473331 of S. berthaultii from Cochabamba, Bolivia (USDA 1999). One individual from the F1 progeny (M200-30) was then backcrossed with the 2x S. tuberosum parent clone HH1-9 to generate the backcross to S. tuberosum (BCT) progeny (Bonierbale et al. 1994).
Originally the progeny consisted of 158 diploid individuals, of which 133 were still available for this study. A total of 665 genotypes of the BCT progeny were produced at Centro Internacional de la Papa (CIP–Lima, Peru) and shipped to Cornell University as a true seed to use in this study. They were tested for, and found to be free of, major potato viruses and other pathogens. For both the original and extended progenies, in vitro plants were obtained and maintained at 15°C until planting them in the greenhouse and field in July 1999. Plants were then stored as tubers for future use. For the characterization of Rber we used the pedigree clones of the BCT progeny.
P. infestans isolates
A set of 30 isolates of P. infestans was characterized for compatibility/incompatibility with each of the 11 known R-genes. The set of P. infestans isolates was obtained from the Cornell University P. infestans culture collection, and included strains from Ecuador, Kenya, Mexico, Uganda, United States, Peru, South Africa and Poland (Table 2). The strains were chosen on the expectation that phenotypic differences would occur among isolates from the geographically diverse regions. Specific information regarding the isolates is available upon request. For the assays, isolates were first grown on Rye B medium (Caten and Jinks 1968) in a growth chamber at 15°C to encourage sporulation and then transferred to new media every other week.
The isolates were tested on each host differential plant using a detached leaflet assay (Black et al. 1953; Dorrance and Inglis 1997). Each inoculation involved recently expanded detached leaflets from 5-, 6-week-old plants of each host differential. Fifty microliters of a suspension bearing at least 15,000 zoospores/ml were deposited on the abaxial side of the leaflets using Petri dishes with water agar (15 g/l) on the bottom as moist chambers (Tooley et al. 1986). Each inoculated leaflet was incubated for 7 days at 15°C and scored for presence/absence of infection (detected as sporulation from the leaflet) on the seventh day after inoculation (Dorrance and Inglis 1997; Rivera-Peña 1990).
Wastie (1991) and Dorrance et al. (1997) have suggested that small-scale tests, such as detached leaflets, should be interpreted with caution, because the conditions may not reliably reflect the real resistance or susceptibility interaction other than in extreme cases. Therefore, we scored as “compatible” only those interactions in which sporulation was evident. All cases without sporulation, and where hypersensitive response was present, were scored as incompatible. Each test was repeated at least five times and only those strains that showed consistent reactions were chosen to comprise a “tester set” (Table 2). A dichotomous key (Fig. 1) was then built to facilitate the identification of each of the 11 known R-genes from S. demissum.
Flow chart for identification of resistance genes to Phytophthora infestans in potato germplasm. The names in bold correspond to Table 2. C/I stand for compatible/incompatible reactions, respectively, and the numbers in parentheses are the resistance genes that show the corresponding reaction
Once the tester set of P. infestans isolates was developed, it was used to characterize the pedigree clones parents of the BCT progeny (USW22-30, HH1-9, and M200-30; the B11B clone was used as a representative of S. berthaultii), two of which were known to carry the Rber gene (M200-30 and B11B). Detached leaflet assays were again used to identify specific reactions against each isolate. The cultivar Norchip was used as a positive control in every inoculation due to its high susceptibility and absence of known R genes to P. infestans.
Phenotypic characterization of the BCT progeny
The presence or absence of Rber was determined in the extended progeny and reconfirmed in the original BCT progeny. We used isolate US940480 (ATCC# 208834, a member of the US-8 clonal lineage) obtained from the Cornell University P. infestans culture collection. We had previously shown that this isolate can distinguish Rber from 8 out of the 11 known R-genes (Ewing et al. 2000), and so it was used in both field inoculations and detached leaflet assays. Field inoculations are desirable because they reflect better resistance under agricultural conditions. The original BCT progeny had been previously exposed twice to the US8 isolate (US940480) (Ewing et al. 2000). The US8-compatible individuals from the original progeny and all individuals from the extended progeny were inoculated in the field at Freeville, NY, during the summer of 1999.
For field studies in 1999, plantlets from in vitro cultures were transplanted into Jiffy7-Peat Pellets (Jiffy Products) supports and grown under greenhouse conditions between May 15 and June 15; from June 15 to June 18 they were conditioned to the field environment in cold frames and transplanted to the field in single plant plots during the week of June 18. The spacing between plots was 0.86 m, and the space between rows was 0.9 m. The cultivar Atlantic was included as a positive control because of its high general susceptibility to late blight.
Potato production practices typical of northeastern USA were used to encourage plant growth. Fertilization was at the rate of 167 kg/ha of a 13-13-13 blend of nitrogen, phosphorous and potassium. Weeds were controlled as needed by applications of Lorox DF (lineron, Dupont, Wilmington, DE, USA) and Dual II (metalochlor, Syngenta, Greebsboro, NC, USA) at recommended label rates. Insects were controlled with the applications of Provado 3.75 oz/A (imidacloprid, Bayer CropScience, Durham, NC, USA), Sevin XLR 1 qt/A (carbaryl, Bayer CropScience), or Ambush 6.4 oz/A (permethrin, Amvac chemical corp, Newport Beach, CA, USA) when necessary.
Inoculation with P. infestans took place during the evening of August 13 by applying a suspension (10 ml) containing 150 sporangia/ml of isolate US940480 to each plant. Immediately prior to inoculation, the entire area had been sprinkler-irrigated to an equivalent of about 0.2 in. of rainfall. The inoculum was applied with a hand-held sprayer. Sporangia had been obtained from sporulating lesions on leaflets of potato cv. Atlantic.
The epidemic developed rapidly and rate of disease development on each progeny genotype was recorded. The amount of foliar disease (as a percentage of the total tissue affected) was evaluated every 3 days between September 5 and October 2 using the methods and guidelines previously described (Ewing et al. 2000). These data were used to calculate the area under the disease progress curve (AUDPC) as described by Shaner et al. (1977). Those plants that showed inconsistent results or that died in the field for reasons other than late blight were subsequently tested using detached leaflets from plants grown in the greenhouse as described before.
DNA extraction
DNA was obtained from each of the individuals in the original and extended BCT progeny. Expanding leaflets were collected from 826 plants growing under greenhouse conditions and were placed in liquid nitrogen. DNA was extracted using a CTAB extraction protocol (Doyle and Doyle 1990). The precipitated DNA was treated with 5 μl of a solution (10 mg/ml) of RNAse A for 30 min at 37°C. It was then visualized in a 0.8% agarose gel in TAE buffer to verify its integrity, and concentration was measured using a Hoefer DyNA QUANT 200 fluorometer (Amersham Biosciences, San Francisco, CA, USA).
Restriction fragment length polymorphism
Restriction fragment length polymorphism probes for chromosome 10 were chosen based on the tomato genetic map and obtained from Dr. Steve Tanksley’s lab (Cornell University). To obtain sufficient probe DNA for RFLP hybridization, each cloned probe was amplified by PCR using M13 universal primers in a 25 μl final volume. For the template, we used 1 μl of a liquid culture of Escherichia coli transformed with the probe which had been diluted to half concentration in distilled water and incubated in boiling water for 5 min. PCR conditions were: 2.5 mM MgCl2, 0.2 μM of each primer, 200 mM dNTPs, and 1 U of Taq Polymerase (Invitrogen Corporation, Carlsbad, CA, USA). Each probe was then amplified as follows: initial denaturation 5 min at 94°C, followed by 30 cycles of 94°C for 60 s, 55°C for 30 s, and 72°C for 90 s, and a final extension at 72°C for 5 min. The amplification products were visualized on 1% agarose gels and cleaned using the Wizard PCR preps DNA purification system (Promega, Madison, WI, USA).
To find polymorphisms in the progeny we performed Southern hybridizations with each tomato probe. DNA from each of the BCT pedigree clones was digested in separate reactions with: DraI, EcoRI, EcoRV, HaeIII (all from Invitrogen Corporation), and BstNI (New England Biolabs, Beverly, MA, USA). Digestion and Southern blot procedures were done as described by Sambrook et al. (1989) using Hybond N+ membrane (Amersham Pharmacia Biotech) for capillary transfer. For hybridization buffer we used Hyb-9 Hybridization solution (Gentra Systems, Plymouth, MN, USA).
Probes were labeled with 32Pα-dATP with the Random Primers DNA Labeling system (Invitrogen Corporation) and cleaned with Bio-Spin 30 Tris columns (Bio Rad, Hercules, CA, USA). Hybridization took place for at least 4 h at 65°C. Washes were performed with 2× SSC—0.1% SDS for 15 min, 1× SSC—0.1% SDS for 15 min, and 0.5× SSC—0.1% SDS for 15 min at 65°C. The membranes were then exposed to BioMax MS film (Eastman Kodak Company, Rochester, NY, USA) overnight at −80°C and developed with a Kodak X-Omat automatic developer.
Multiplex allele-specific polymorphism mapping
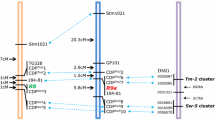
We used a mapping technique that we named Multiplex Allele-Specific Polymorphism (MASP-map), since it allows the simultaneous amplification of one allele and a control band. It is designed to follow a single allele through a mapping progeny, via a modification of the similar methods such as PASA (Okimoto and Dogson 1996) and tetra-primer ARMS-PCR (Ye et al. 2001). MASP-map allowed us to change RFLP probes into PCR markers. The first step is to design primers from known RFLP probe sequences and use them to amplify and sequence the parents of the cross (Fig. 2). These sequences are then used to find single nucleotide polymorphisms that allow the unique identification of one of the parental alleles. A new allele-specific primer is designed so that the 3′ end anneals to the polymorphic site. In addition a mismatch is added in the penultimate nucleotide on the 3′end of the specific primer to help improve allele specificity.
Diagram of the MASP-map method. See text for explanation of the methodology. FP and RP stand for forward and reverse primer, respectively. SFP stands for specific forward primer. The last box shows results of MASP-map: S.t. and S.b. stand for Solanum tuberosum and Solanum berthaultii, respectively. UHMB are U: S. t parent USW 22-30; H: S. t. Recurrent parent HH1-9; M: hybrid parent M200-30; B: S.b. representative B11B, respectively
To screen individuals of the progeny for the parental polymorphism of interest, PCR is carried out with three primers simultaneously; two that amplify both alleles and a third that is specific for the allele with the targeted polymorphism. Thus, individuals lacking the allele of interest amplify only the control band, while in individuals containing the allele of interest, both the specific allele and the control band are amplified. The amplification products are then separated and visualized in agarose gels.
In order to add markers to the interval between CT214 and TG63, the sequences of six tomato RFLP probes were obtained from the Solanaceae Genome Network (http://www.sgn.cornell.edu). From these, forward and reverse primers were designed using Primer Select from the DNASTAR DNA analysis software package (DNASTAR, Inc.), trying to localize them within the 100 initial or terminal base pairs on each probe.
These primers were used to amplify alleles in each individual parent from 50 ng of template DNA in a reaction containing 0.8 U of Taq DNA Polymerase (Invitrogen Corporation), 1× PCR buffer (10× stock: 200 mM Tris–HCl pH 8.4, 500 mM KCl), 2.5–3.0 mM MgCl2, 200 nM of forward and reverse primers, 200 nM of dNTPs, and double distilled sterile water. The reactions were carried out in a final volume of 50 μl set on ice to minimize nonspecific amplification. Prior to sequencing, 1 μl of the PCR product was visualized to verify amplification and size of the products on a 1% agarose gel, and the remaining 49 μl were purified using the Wizard PCR PrepDNA purification system (Promega). Amplification products were sequenced by the Cornell Biotechnology Sequencing Facility using only the forward primer for short products (around 500 bp) or both forward and reverse primers for longer products.
The two parental sequences obtained for each probe were compared with Sequencher (version 4.0.5, Gene Codes Corp. MI, USA) to look for single nucleotide polymorphisms that were specific to the S. berthaultii parent. Specific primers for S. berthaultii polymorphisms were designed from the parental sequences with a T m about 5°C lower than those of the initial primers (Table 3) and included a mismatch at the penultimate base at the 3′ end of the primer to improve specificity as described by Cha et al. (1992).
When the polymorphism was close to the 5′ end of the fragment a forward primer was designed (5/6 probes, Table 3); when polymorphisms were closer to the 3′ end, a reverse primer was designed (1/6 probes, Table 3). For the sake of clarity, the three primers used will henceforth be referred to as allele-specific (usually forward), general-forward and general-reverse.
To facilitate mapping the region of interest, we screened only individuals with a recombination at any point along chromosome X as first determined by Bonierbale et al. (1988a). Using this method, markers mCT11, mCD5, mCT238, mCT240, and mT1682 were added to the genetic map of chromosome X, the “m” refers to RFLP makers detected by MASP-map.
Tomato marker sequences have been published by Ganal et al. (1998). In order to assess percent similarity between potato and tomato sequences, they were aligned, visually inspected to identify sites with signal from more than one nucleotide (i.e., double peaks), and coded according to standard IUPAC degenerate code. Sequences were then trimmed to the same size using Sequencher version.4.2 (GeneCodes Corp.). Since tomato sequences were obtained from ESTs, for accurate comparison, introns were edited out of potato sequences. However, alignments among potato accessions were done using the whole length of the sequences. To find sequence similarity, the ClustalW utility of the software package MegAlign (Lasergene Corp, Clewley and Arnold 1997) was used. Multiple alignments were performed using the slow/accurate option weighted to account for diploidy.
Finally, sequences were compared to previously reported sequences at GenBank using the BLAST algorithm. ESTs were compared to the dbEST database, which contains GenBank, EMBL, and DDBJ sequences from ESTs. Sequences for the genomic marker TG63 were compared both with the nonredundant and the EST databases.
Cleaved amplified polymorphic sequences (CAPS)
The sequence of tomato probe TG63 was used to design primers to amplify homologous sequences in potato. However, due to multiple insertions and deletions in the sequence, it was not possible to design MASP-map primers. Therefore, we decided to use CAPS to transform this RFLP probe into a PCR marker. After screening the PCR products of all the pedigree accessions (USW22-30, HH1-9, M200-30, and B11B) with multiple restriction enzymes, a polymorphism unique to the S. berthaultii allele was discovered using EcoRV. This polymorphism could then be used as a cleaved amplified polymorphic sequence (CAPS, Konieczny and Ausubel 1993) marker for mapping. For the final screening, 50 ng of genomic DNA were amplified using primers TG63F1 and TG63R (Table 3), and a master mix identical to that described for amplification of RFLP probes, except that the MgCl2 concentration was 3 mM. The PCR conditions for this marker were: initial denaturation 95°C for 2 min; 35 cycles of 95°C for 30 s; 50°C for 30 s; 72°C for 90 s; and final extension at 72°C for 7 min.
The PCR product was digested with 2.5 U of EcoRV (Invitrogen Corporation) at 37°C for 3 h and visualized with ethidium bromide on 1.5% agarose gels in 1× TAE buffer. The gels were examined with a FotoAnalyst Investigation Column Mount System (Fotodyne Inc. Hartland, WI, USA).
Simple sequence repeats
Previous studies have shown that SSR marker STM1056 is located close to TG63 in potato (Milbourne et al. 1998), and hence was also screened for progeny that are recombinant on chromosome X. For amplification we followed the procedure and primers described by Milbourne et al. (1998). PCR products were run on a 5.5% Long Ranger Gel (Cambrex Corporation, East Rutherford, NJ, USA) in a SequiGen GT Nucleic Acid Electrophoresis Cell (Bio Rad Inc.). The gels were run in 0.6× TBE buffer at 2,000 V for about 2 h, dried and exposed to Kodak BioMaxMR film (Eastman Kodak Company) overnight at room temperature.
Search for new markers with bulk segregant analysis and resistance gene analogs (RGA)
To increase the chances of finding bands linked to Rber using RGA mapping (see subsequently), we used bulk segregant analysis (Michelmore et al. 1991). DNA of selected BCT progeny was bulked in two groups, one with 10 incompatible and the other with 10 compatible individuals. The respective bulks were made based on the genetic data (Bonierbale et al. 1994) and phenotypic (Ewing et al. 2000) information confirmed for the original BCT progeny in chromosome X. The resistant and the susceptible bulks each comprised individuals with recombination events similar in the area of interest between CT240 and TG63, but differing for the rest of the chromosome to minimize the area of interest. The bulks contained the same amount of DNA from each of 10 individuals in such a way that the incompatible group had the S. berthaultii allele (segregating from M200-30 and in common with B11B) for the area of interest, regardless of the rest of the chromosome; and the compatible bulk contained alleles from the S. tuberosum progenitors in the area between CT240 and TG63 but could differ in the rest of the chromosome.
We used resistance gene analog (RGA) amplification to search for markers linked to Rber because previously cloned R-genes to late blight are known to code for nucleotide binding sites (NBS) and leucine-rich repeats (LRR) (Ballvora et al. 2002; Song et al. 2003). Primers designed to amplify resistance gene analogs, including regions containing NBS, LRR, and protein kinase (Table 4) were obtained. PCR was carried out using 50 ng of DNA and 0.8 U of Taq DNA Polymerase (Invitrogen Corporation), 1x PCR buffer (10× stock, 200 mM Tris–HCl pH 8.4, 500 mM KCl), 2.5 mM MgCl2, 100 nm of 33P-γATP labeled forward primer and 200 nM of reverse primer, 200 nM of dNTPs, and bidistilled sterile water in a final volume of 15 μl. The thermocycler (Hybaid Touchdown TD7200, Thermo Electron Corporation, Waltham, MA, USA) was programmed as follows: initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s; annealing of 51–56°C depending on the primer pair for 30 s, extension at 72°C for 20 s, and a final extension step of 72°C for 2 min. Each of the reactions was mixed with 5 μl of formamide loading buffer (80% formamide, 10 mM EDTA, 1 mg/ml bromophenol blue, 1 mg/ml xylene cyanol), denatured for 3 min at 94°C and quickly chilled. RGA products were separated and visualized using the same procedure described for SSRs.
Data analysis
The BCT mapping data set was obtained directly from the Solgenes database (currently available at http://www.grain.jouy.inra.fr/gendatabasemirror.html). The RFLPs, MASP-map markers, RGA, and SSR gels were scored visually for presence or absence of the bands originating in the S. berthaultii parent. The genotype for each individual was coded as A (homozygous) or H (heterozygous) for each marker, and these data were combined with the previously published framework markers. The chromosome map was assembled with MapManager QTX, version 0.27 (Manly et al. 2001) using a backcross linkage evaluation with P=1×10−5 and the Kosambi mapping function. We added the information for the new markers to a different (new) chromosome and used the distribute command. Once the markers were located on chromosome X, we found the best localization for them using the ‘ripple’ command.
Results
Characterization of Rber
The 30 isolates of P. infestans we tested were diverse in their interaction (compatible/incompatible) with the differential plants (Table 2). Of these isolates, about half had unique phenotypes; 20% were compatible with R-genes 1, 2, 5, 6, 7, 10, and 11; 10% were compatible with 1, 2, 3, 4, 5, 6, 7, 10, and 11; 6.6% were compatible with 1, 3, 5, 7, 10, and 11; 6.6% with 1, 2, 5, 10, and 11; and 6.6% with 1, 2, 5, 6, and 7. In general, isolates with the same compatibility phenotype came from diverse geographical locations, except for some isolates from Mexico that showed the same compatibility pattern.
We designed a flow chart using the “tester set” of nine isolates (Table 2, Fig. 1) to identify known R-genes. The chart is used as a dichotomous key as follows. An unknown potato genotype is inoculated with each member of the tester set independently and the reactions recorded. The results are then read sequentially following the chart. For example, if a plant is incompatible with the strain UG980005, the next step is to follow the right branch. If it is compatible with the strain KE980002, the left branch should be followed, and so on, until a terminal point is reached, which will show the R-gene that is present. All interactions are recorded and analyzed to determine the number and identification of R-genes present in the potato genotype.
The tester set was applied to three of the pedigree genotypes of the BCT progeny and a representative individual of S. berthaultii PI 473331 (Table 5). The S. tuberosum ‘grand’parent USW-2230 was compatible with all isolates but US17, indicating the presence of R1. We also found that the recurrent S. tuberosum parent HH1-9 was compatible with all isolates, including US970001, which is only compatible with plants lacking all resistance genes; therefore, no resistance genes were detected in HH1-9. We do not have the original S. berthaultii donor (a genotype of PI473331), and therefore could not assess its phenotype. However, the hybrid parent M200-30 (product of a cross between PI473331 and USW22-30) carries Rber and is resistant to all isolates in the tester set, suggesting that Rber is a new resistance gene. Field tests performed in the Toluca Valley, Mexico allowed the isolation of several strains from plants carring R Pi-ber allele showing that R Pi-ber can be overcome by isolates MX990005, MX010003, MX010004, and Mx010008. Testing of these isolates with the differential set also demonstrated their compatibility with R1–R11 (unpublished results).
Genetic mapping of Rber
Phenotypic characterization
We confirmed the phenotype of all individuals in the original BCT progeny evaluated by Ewing et al. (2000) and determined the phenotype of the extended BCT progeny using either field or detached leaflet assays. For plants from the original progeny inoculated in the field with the US8 isolate US940480 during the summer of 1999, the AUDPC score ranged from 0 to 3,652 U. Only those individuals with AUDPC less than 1 were scored as incompatible and thus containing R Pi-ber . The average AUDPC for compatible individuals was 1,580. For the extended population, we tested 336 individuals, of which 180 were incompatible (53.57%). For the combined progeny (original and extended) 52% were resistant (428). For mapping purposes we coded incompatible individuals as carrying the S. berthaultii allele (R Pi-ber ) in heterozygous condition and the compatible ones as carrying only alleles from the tuberosum pedigree (for purposes of the mapping program these are considered being “homozygous” for the S. tuberosum allele, although the pedigree did permit heterozygosity).
MASP-map sequences
Given that MASP-map marker development required the characterization of nucleotide sequences in the pedigree clones of BCT (GenBank acc. AY874391 to AY874411), we thought it would be useful to find the level of similarity among them and with tomato. On average, potato and tomato sequences were over 90% similar. The biggest differences were found in noncoding sequences, e.g., for TG386, where tomato was only 70.1 and 69.2% similar to S. tuberosum and S. berthaultii, respectively. Regarding S. tuberosum and S. berthaultii, sequences were over 97% similar on an average. Interestingly the same average similarity was found between the S. tuberosum accessions (USW22-30 and HH1-9), most differences were caused by heterozygosity in one of the accessions, especially HH1-9.
Blast searches confirmed previous reports by Ganal et al. (1998) except for CT217, where we found the best homology with an Avr9/Cf9 elicited protein. Also, mCT11, mCT214, mCT238, and mCT240, each showed homology with sequences from EST libraries made from potato leaves challenged with P. infestans (P value > e−111). These results, the presence of an R-gene, and QTLs for resistance to other pathogens (Gebhardt and Valkonen 2001) may suggest that this area of chromosome X could be involved in resistance responses common to other pedigrees including tomato.
For the marker TG63, forward and reverse sequences from tomato, obtained from the Solanaceae Genomics Network, were compared to sequences in GenBank. No matches were found when the search used the nonredundant database, but the sequences share homology with one EST from tomato roots (EST303128, 1e−85) and one from the tomato callus (EST542946, 1e−55). However, since TG63 is a marker from a total genome library, these findings are inconclusive. We also sequenced the amplification products of all pedigree genotypes from primers TG63F1 and TG63R; however, heterozygosity for insertion/deletions precluded clean direct sequences that could be tested for homologies.
Genetic markers
Several new genetic markers were added to chromosome X in order to assess the precise location of R Pi-ber . First, we confirmed the identity of each individual from the original BCT progeny by testing with the flanking markers mCT214 and TG63. Doing this, we were able to assess previously missing genotypes for five individuals for CT214 and six individuals for TG63. There was disagreement between the genotyping results in this study and a previous one (Bonierbale et al. 1994) for three individuals that were originally scored for TG63 as double recombinants. In our study, these individuals were recorded as heterozygous (presence of berthaultii alleles), whereas in the previous study they were recorded as homozygous, which caused them to show as double recombinants. For a progeny of N=158 and given the original map distance between the markers TG63–Rber–CT214b of 4.8 and 8.2 cM, respectively (Ewing et al. 2000), the number of double recombinants should be about 1(0.62). The probability of the three double recombinants reported is 0.0012 suggesting that the original designation is highly unlikely. Using the revised designations determined in this study we found a new distance between TG63 and R Pi-ber of 0.6 cM, instead of the 4.8 cM originally reported (Ewing et al. 2000); the corrected distance between CT214 and R Pi-ber is 5.8 cM.
We added the RFLP markers CT217, CD32, CD72, and CT20, from the tomato genetic map to potato chromosome X. Because of the inversion between tomato and potato chromosomes, we found that only CD32 was located in the area of interest at 0.6 cM from CT214. Marker CT217 is located closer to TG52; CD72 and CT20 are located in a terminal position relative to TG63 (Fig. 3).
Localization of R Pi-ber on chromosome X in the potato map. Previously published framework markers are TG230, TG386, TG52, TG403, CT124, and TG420. RFLP markers are shown in bold; MASP-map markers are preceded by “m”; RGA markers have asterisks after the name. Approximate locations of RGA and SSR markers are shown in brackets, names of cosegregating markers are separated by commas. Resistance genes R Pi-ber and Ph2 (position inferred from tomato map) are surrounded by boxes. Inversion point between tomato and potato is indicated by an arrow
Using MASP-map, three new markers (mCD5, mCT238, and mCT240) were placed within the interval between mCT214 and TG63. Markers mCT240 and TG63 are the closest to R Pi-ber at 3.9 and 0.63 cM, respectively (Fig. 3). Two other markers, mCT11 and mT1682, were added outside the interval which confirmed the area where an inversion took place in potato chromosome X with respect to tomato chromosome 10.
When the entire progeny (original and extended; 826 individuals) was tested with the flanking markers mCT214 and TG63 to identify recombinant individuals in this interval, we found 54 recombinants, very close to the 52 expected for a 6.4 cM distance. There were six additional individuals for which results were inconsistent and from which we did not use data. By adding marker mCT240 to the 54 recombinants, 29 recombination events were found between CT240 and R Pi-ber (a genetic distance of 3.51 cM) and seven recombinants between R Pi-ber and TG63 (0.84 cM).
The microsatellite STM1056 resulted in a total of 12 markers, most of which are located on chromosome VIII. The band STM1056-7 showed linkage to markers on chromosome X, with LOD scores between 10.9 for AS1-13 and 26.3 for R Pi-ber . It is most likely located between R Pi-ber and TG63 but precise localization was not possible.
From all the RGA primers tested we found three bands linked to chromosome X. These were detected with primers S1 × AS1, which were designed to amplify the P-loop motif and the nucleotide binding site common to resistance genes (Leister et al. 1996). They are located at 4.1 cM distal to TG230 (AS1-13), and between markers CT20 and TG420 (AS1-11 and AS1-12). There were two other bands, AS2-15 and LM5, with linkage to the chromosome (LOD 18.3 and 18.6, respectively), but we were not able to locate them confidently between other markers on the chromosome. It is possible that the S. tuberosum and S. berthaultii alleles do not differ enough to show differential amplification with the RGA primers used; therefore, we were unable to find markers closely linked to R Pi-ber . Approximate locations are shown in Fig. 3.
Discussion
The objective of this study was to determine the novelty and precise location of the Rber resistance gene from S. berthaultii. The first step was to design a method that could rule out known R-genes in potato which are responsible for the resistance reaction conferred by the gene. Using a detached leaflet assay, a host differential set and P. infestans isolates from diverse geographic regions. We were able to select nine isolates that together differentiate each of the previously characterized resistance genes through their known incompatibility reactions. Using this tester set and the flow diagram illustrated (Fig. 1), we determined that Rber is a new previously uncharacterized R-gene against P. infestans. In order to avoid confusion with the recently published RB gene and maintain the currently used nomenclature, that describes R-genes as Rpathogen−donor of resistance (Kuhl et al. 2001; Solomon-Blackburn and Barker 2001) we renamed the gene R Pi-ber .
The rationale for characterizing R Pi-ber as a locus with a novel specificity, is as follows: it is incompatible with UG980005 suggesting that it could only be R3, R4, R8, R9, R10, R11, or new; it is incompatible with KE980002, ruling out R10 and R11; it is again incompatible with MX980100, eliminating R3 and R4, besides R3 is located on Chr. XI; incompatibility with US8 rules out R9. Finally, M200-30 is incompatible with the R8-compatible isolate Ke980004, allowing us to conclude that R Pi-ber is also not R8. Besides, new findings by Huang (2005) suggest that R5–R11 are allelic to R3, ruling them out from chromosome X. This leaves only the proposition that R Pi-ber is different from previously identified and “named” R-genes. Finally, as previously stated, R Pi-ber is overcome by several Mexican isolates including Mx990005 and Mx010003. In order to keep the current nomenclature of resistance genes to P. infestans in potato, we will refer to Rber (Ewing et al. 2000) as R Pi-ber .
While the development of the R-gene tester set proved to be a useful tool, its widespread application is unfortunately limited. Some avirulent characteristics in a few of the isolates have changed over time. Others have also observed this phenomenon (Abu-El Samen et al. 2003). Thus, we found it necessary to include the appropriate host differential plant as positive controls in all inoculation experiments.
The use of a new genotyping method for this study allowed us to add several specific RFLP markers to the map with relative ease. The MASP-map method provides an efficient alternative to RFLPs for mapping in large populations. It is based on the use of primers that specifically amplify one parental allele and that can be used to screen segregation in a progeny. This system is inexpensive and efficient, needing only one PCR step, three primers per set, the usual PCR reagents, and visualization on agarose gels. Another significant advantage of this method is the quantity of DNA required; only 50 ng of DNA per reaction was used in this study. In contrast, the same study using RFLP methods would have required at least 10 μg of DNA per probe. Possibly its most important advantage is that it provides amplification of a control band in addition to the allele-specific band. The lack of the target allele is visualized by amplification of just the control band; the absence of all amplification products in a sample indicates a failure of the PCR.
The original map placed R Pi-ber between markers CT214b and TG63, an area spanning 13 cM. Through confirmation of phenotypes and genotypes at these markers, we have demonstrated this area to be smaller than originally thought (6.4 cM). While the precise map position of R Pi-ber is still not known, we were able to add four new markers within the interval and locate R Pi-ber between CT240 and TG63, to an area spanning 3.8 cM. Map distances calculated for the original progeny increased slightly when the extended BCT progenies were added, from 3.2 between CT240 and R Pi-ber and 0.63 cM between R Pi-ber and TG63 to 3.51 and 0.84 cM in the same interval. A possible explanation for this difference is the dissimilar environments in which the crosses were made, as suggested by Paterson (1996). Since the size of the potato genome is close to 1,000 cM (Meksem et al. 1995), which corresponds to 1,000 Mb (haploid genome equivalent, Leister et al. 1997), 1 cM is roughly equivalent to 1 Mb. This means that (on average) there is predicted to be about 3.8 Mb of DNA in the area of interest, and the closest marker TG63 may be only 630 Kb away from R Pi-ber .
The molecular maps of potato and tomato show a strikingly high level of colinearity (Bonierbale et al. 1988a; Gebhardt et al. 1991; Tanksley et al. 1992). The tomato map, consisting of more than 1,030 markers (Tanksley et al. 1992), has been saturated to a higher degree than the potato map. For this reason, our effort to saturate the area with new markers began by using orthologous probes from tomato. Despite the similar genetic structure and colinearity between potato and tomato, their chromosome X homologues differ by a paracentric inversion (Bonierbale. et al. 1988b), close to R Pi-ber . We found the break point to be in a 2.7 cM area between markers TG280 and TG408, in agreement with Tanksley et al. (1992). In contrast to previous expectation, however, our results indicate that CT217 in potato is located closer to TG386 and is not within the inversion (Tanksley et al. 1992).
One question of interest is whether resistance genes found in one species are orthologous to those found in other species. This is particularly interesting because a recent study by Grube et al. (2000) showed that there are clusters of resistance genes that are conserved across the Solanaceae. Not all proposed homologies, however, turn out to be accurate. For example, R6 was originally thought to be present in both S. demissum and S. stoloniferum (Schick et al. 1958). Later work showed these to be two different loci (Malcomson and Black 1966). The same seems to be the case with R Pi-ber . A previous study (Ewing et al. 2000) considered the possibility that R Pi-ber was orthologous to Ph2 on tomato chromosome X (Moreau et al. 1998) due to the map position of both genes. Based on the position of the inversion point, we show here that they are not located at the same map position. Ph2 is located between markers CT240 and TG233 in tomato, which are upstream of R Pi-ber in the potato map, and which therefore occupy different loci in the long arm of Chromosome X (Moreau et al. 1998).
In our study, resistance gene analogs (RGAs) were amplified with primers designed for nucleotide-binding sites, leucine-rich repeats and Pto-kinase. We identified five bands located in chromosome X, three of which could be added to the map (Fig. 2). The other two had high LOD scores for linkage to markers in chromosome X, but we were unable to map them confidently due to too many apparent double crossovers. Another RGA sequence was reported by Leister et al. (1996) on the long arm of chromosome X (St 1.2.4), which could be allelic to AS1-11 or AS1-12. These results may be suggestive of the presence of a resistance gene cluster on chromosome X.
Our results provide a more detailed characterization and improved understanding of R Pi-ber , including a more precise map position on potato chromosome X. Because we converted RFLP markers into MASP-map and CAPS markers, selection in the future for individuals containing R Pi-ber will be easier and faster, particularly if the gene is used in breeding programs. In addition, the improved localization of the inversion on chromosome X will make it easier to identify new markers from the always-growing tomato map to saturate the area between mCT240 and TG63. Further steps are needed to clone and characterize R Pi-ber . These future analyses will provide insights on resistance interactions between potato and P. infestans, and perhaps contribute to the management of the disease.
Abbreviations
- AUDPC:
-
Area under the disease progress curve
- BCT:
-
Backcross to S. tuberosum progeny
- BSA:
-
Bulk segregant analysis
- CAPS:
-
Cleaved amplified polymorphic sequences
- MASP-map:
-
Multiplex allele-specific PCR mapping
- R-gene:
-
Resistance gene
- chr:
-
Chromosome
- RFLP:
-
Restriction fragment length polymorphisms
References
Abu-El Samen FM, Secor GA, Gudmestad NC (2003) Variability in virulence among asexual progenies of Phytophthora infestans. Phytopathology 93:293–304
Ballvora A, Ercolano MR, WeiB J, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C (2002) The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J 30:361–371
Black W, Mastenbroek C, Mills WR, Peterson LC (1953) A proposal for an international nomenclature of races of Phytophthora infestans and genes controlling immunity in Solanum demissum derivatives. Euphytica 2:173–179
Bonierbale MW, Plaisted RL, Pineda O, Tanksley SD (1994) QTL analysis of trichome-mediated insect resistance in potato. Theor Appl Genet 87:973–987
Bonierbale MW, Plaisted RL, Tanksley SD (1988a) Construction of a genetic map of potato based on molecular markers from tomato. Am J Potato Res.65:471–472
Bonierbale MW, Plaisted RL, Tanksley SD (1988b) RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120:1095–1103
Bormann CA, Rickert AM, Castillo Ruiz RA, Paal J, Lübeck J, Strahwald J, Buhr K, Gebhardt C (2004) Tagging quantitative trait loci for maturity-corrected late blight resistance in tetraploid potato with PCR-based candidate gene markers. Mol Plant-Microbe Interact 17:1126–1139
Caten CE, Jinks. JL (1968) Spontaneous variability of single isolates of Phytophthora infestans. I. Cultural variation. Can J Bot 46:329–348
Cha RS, Zarbl H, Keohavong P, G.Thilly W (1992) Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl 2:14–20
Chen XM, Line RF, Leung H (1998) Genome scanning for resistance-gene analogs in rice, barley and wheat by high resolution electrophoresis. Theor Appl Genet 97:345–355
Chunwongse J, Chunwongse C, Black L, Hanson P (2002) Molecular mapping of the Ph-3 gene for late blight resistance in tomato. J Horticult Sci Biotechnol 77:281–286
Clewley JP, Arnold C (1997) MEGALIGN. The multiple alignment module of LASERGENE. Methods Mol Biol 70:119–129
Deahl KL, Inglis DA, Demuth SP (1993) Testing for resistance to metalaxyl in Phytophthora infestans isolates from northwestern Washington. Am J Potato Res 70:779–795
Dorrance AE, Inglis DA (1997) Assessment of greenhouse and laboratory screening methods for evaluating potato foliage for resistance to late blight. Plant Dis 81:1206–1213
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
El-Kharbotly A, Palomino-Sanchez C, Salamini F, Jacobsen E, Gebhardt C (1996) R6 and R7 alleles of potato conferring race-specific resistance to Phytophthora infestans (Mont.) de Bary identified genetic loci clustering with the R3 locus on chromosome XI. Theor Appl Genet 92:880–884
Ewing EE, Simko I, Smart CD, Bonierbale MW, Mizubuti ESG, May GD, Fry WE (2000) Genetic mapping of qualitative and quantitative field resistance to Phytophthora infestans in a population derived from Solanum tuberosum and Solanum berthaultii. Mol Breed 6:25–36
Fry WE, Goodwin SB (1997) Resurgence of the Irish potato famine fungus. BioScience 47:363–371
Ganal MW, Czihal R, Hannapel U, Kloos D, Polley A, Ling H-Q (1998) Sequencing cDNA clones from the genetic map of tomato (Lycopersicon esculentum). Genome Res 8:842–847
Gebhardt C, Ritter E, Barone A, Debener T, Walkemeier B, Schachtschabel U, Kaufmann H, Thompson RD, Bonierbale MW, Ganal MW, Tanksley SD, Salamini F (1991) RFLP maps of potato and their alignment with the homoeologus tomato genome. Theor Appl Genet 83:49–57
Gebhardt C, Valkonen JPT (2001) Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol 39:79–102
GILB (2004) Global initiative on late blight, http://www.cipotato.org/gilb/
Goodwin SB, Sujkowski LS, Fry WE (1996) Widespread distribution and probable origin of resistance to metalaxyl in clonal genotypes of Phytophthora infestans in the United States and Western Canada. Phytopathology 86:793–800
Grube RC, Radwanski ER, Jahn M (2000) Comparative genetics of disease resistance within the Solanaceae. Genetics 155:873–887
Grunwald NJ, Flier WG, Sturbaum AK, Garay-Serrano E, van den Bosch TBM, Smart CD, Matuszak JM, Lozoya-Saldana H, Turkensteen LJ, Fry WE (2001) Population structure of Phytophthora infestans in the Toluca valley region of central Mexico. Phytopathology 91:882–890
Helgeson JP, Pohlman JD, Austin S, Haberlach GT, Wielgus SM, Ronis D, Zambolim L, Tooley P, McGrath JM, James RV, Stevenson WR (1998) Somatic hybrids between Solanum bulbocastanum and potato: a new source of resistance to late blight. Theor Appl Genet 96:738–742
Huang S (2005) High allelic diversity of the potato R3 complex locus suggests a novel strategy for late blight control. Ph. D Thesis. Wageningen University, The Netherlands
Huang S, van der Vossen EAG, Kuang H, Vleeshouwers VGAA, Zhang N, Borm THA, van Eck H, Baker BJ, Jacobsen E, Visser RGF (2005) Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J (in press)
Huang SW, Vleeshouwers V, Werij JS, Hutten RCB, van Eck HJ, Visser RGF, Jacobsen E (2004) The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. Mol Plant-Microbe Interact 17:428–435
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J 4:403–410
Kuhl JC, Hanneman RE, Havey MJ (2001) Characterization and mapping of Rpi1, a late blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol Genet Genom 265:977–985
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Leister D, Berger A, Thelen H, Lehmann W, Salamini F, Gebhardt C (1997) Construction of potato YAC library and identification of clones linked to the disease resistance loci R1 and Gro1. Theor Appl Genet 95:954–960
Leonards-Schippers C, Gieffers W, Salamini F, Gebhardt C (1992) The R1 gene conferring race-specific resistance to Phytophthora infestans in potato is located on potato chromosome V. Mol Gen Genet 233:278–283
Leonards-Schippers C, Gieffers W, Schafer-Pregl R, Ritter E, Knapp S, Salamini F, Gebhardt C (1994) Quantitative resistance to Phytophthora infestans in potato: a case study for QTL mapping in an allogamous plant species. Genetics 137:67–77
Li X, van Eck HJ, Rouppe van der Voort JNAM, Huigen DJ, Stam P, Jacobsen E (1998) Autotetraploids and genetic mapping using common AFLP markers: the R2 allele conferring resistance to Phytophthora infestans mapped on potato chromosome 4. Theor Appl Genet 96:1121–1128
Malcomson JF, Black W (1966) New R-genes in Solanum demissum Lindl. and their complementary races of Phytophthora infestans (Mont) de Bary. Euphytica 15:199–203
Manly KF, Cudmore JRH, Meer JM (2001) Map manager QTX, cross platform software for genetic mapping. Mamm Genome 12:930–932
Mastenbroek C (1952) Investigations into the inheritance of the immunity from Phytophthora infestans De B. of Solanum demissum Lindl. Euphytica 1:187–198
Meksem K, Leister D, Peleman J, Zabeau M, Salamini F, Gebhardt C (1995) A high resolution map of the vicinity of the R1 locus on chromosome V of potato based on RFLP and AFLP markers. Mol Gen Genet 249:74–81
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulk segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Milbourne D, Meyer RC, Collins AJ, Ramsay LD, Gebhardt C, Waugh R (1998) Isolation, characterization and mapping of simple sequence repeat loci in potato. Mol Gen Genet 259:233–245
Moreau P, Thoquet P, Olivier J, Laterrot H, Grimsley N (1998) Genetic mapping of Ph-2, a single locus controlling partial resistance to Phytophthora infestans in tomato. Mol Plant-Microbe Interact 11:259–269
Naess SK, Bardeen JM, Wielgus SM, Haberlach GT, McGrath JM, Hegelson JP (2000) Resistance to late blight in Solanum bulbocastanum is mapped to chromosome 8. Theor Appl Genet 295:1–8
Okimoto R, Dogson JB (1996) Improved PCR amplification of multiple specific alleles (PAMSA) using internally mismatched primers. Biotechniques 21:20–26
Paterson AH (1996) Making genetic maps. In: Paterson AH (eds) Genome mapping in plants. Academic, Austin pp 22–39
Rivera-Peña A (1990) Wild tuber-bearing species of Solanum and incidence of Phythophthora infestans (Mont.) de Bary on the western slopes of the volcano Nevado de Toluca. 5. Type of resistance to P. infestans. Potato Res 33:479–486
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Plainview
Schick R, Schick E, Haussdorfer M (1958) Ein beitrag zur physiologischen spezialisierung von Phytophthora infestans. Phytopath Z 31:225–236
Shaner G, Finney RE (1977) The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knowx wheat. Phytopathology 67:1051–1056
Simko I (2002) Comparative analysis of quantitative trait loci for foliage resistance to Phytophthora infestans in tuber-bearing Solanum species. Am J Potato Res 79:125–132
Solomon-Blackburn RM, Barker H (2001) A review of host major-gene resistance to potato viruses X, Y, A and V in potato: genes, genetics and mapped locations. Heredity 86:8–16
Song J, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, Haberlach GT, Liu J, Kuang H, Austin-Phillips S, Buell CR, Helgeson JP, Jiang J (2003) Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci USA 100:9128–9133
Spielman LJ, McMaster BJ, Fry WE (1989) Dominance and recessiveness at loci for virulence against potato and tomato in Phytophthora infestans. Theor Appl Genet 77:832–838
Stewart HE, Bradshaw JE, Pande B (2003) The effect of the presence of R-genes for resistance to late blight (Phytophthora infestans) of potato (Solanum tuberosum) on the underlying level of field resistance. Plant Pathol 52:193–198
Tanksley SD, Ganal MW, Prince JP, Vicente MCd, Bonierbale MW, Broun P, Fulton TM, Giovanoni JJ, Grandillo S, Martin GB, Messenger R, Miller JC, Miller L, Paterson AH, Pineda O, Roder MS, Wing RA, Wu W, Young ND (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160
Tooley PW, Sweigard JA, Fry WE (1986) Fitness and virulence of Phytophthora infestans isolates from sexual and asexual populations. Phytopathology 76:1209–1212
Umaerus V, Umaerus M (1994) Inheritance of resistance to late blight. In: Bradshaw JE, Mackay GR (eds) Potato genetics. CAB International, Oxon pp 365–401
USDA and ARS (1999) National genetic resources program. Germplasm Resources information network (GRIN). On-line database National Germplasm Resources Laboratory, Beltsville, Maryland. Available http://www.ars-grin.gov/npgs/searchgrin.html
Van der Plank JE (1963) Plant diseases: epidemics and control. New York
Wastie RL (1991) Breeding for resistance. Adv Plant Pathol. 7:193–223
Ye S, Dhillon S, Ke X, Collins AR, Day INM (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucl Acids Res 29:e88
Yu YG, Buss GR, Maroof MAS (1996) Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide binding site. Proc Natl Acad Sci USA 93:11751–11756
Zhang LP, Khan AA, Niño-Liu D, Foolad MR (2002) A molecular linkage map of tomato displaying chromosomal locations of resistance gene analogs based on a Lycopersicon esculentum × Lycopersicon hirsutum cross. Genome 45:133–146
Acknowledgements
The present study was supported by Syngenta. We are grateful to Jason Rauscher, for critical reading and helpful suggestions on the manuscript, and to Kevin Myers and Barbara Esposito for their skilled technical assistance. RFLP markers were kindly provided by S. Tanksley.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Wenzel
Rights and permissions
About this article
Cite this article
Rauscher, G.M., Smart, C.D., Simko, I. et al. Characterization and mapping of R Pi-ber , a novel potato late blight resistance gene from Solanum berthaultii . Theor Appl Genet 112, 674–687 (2006). https://doi.org/10.1007/s00122-005-0171-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0171-4