Abstract
Crouzon syndrome (CS), a syndromic craniosynostosis, is a craniofacial developmental deformity caused by mutations in fibroblast growth factor receptor 2 (FGFR2). Previous CS mouse models constructed using traditional gene editing techniques faced issues such as low targeting efficiency, extended lineage cycles, and inconsistent and unstable phenotypes. In this study, a CRISPR/Cas9-mediated strategy was employed to induce a functional augmentation of the Fgfr2 point mutation in mice. Various techniques, including bone staining, micro-CT, histological methods, and behavioral experiments, were employed to systematically examine and corroborate phenotypic disparities between mutant mice (Fgfr2C361Y/+) and their wild-type littermates. Confirmed via PCR-Sanger sequencing, we successfully induced the p.Cys361Tyr missense mutation in the Fgfr2 IIIc isoform of the extracellular domain (corresponding to the p.Cys342Tyr mutation in humans) based on Fgfr2-215 transcript (ENSMUST00000122054.8). Fgfr2C361Y/+ mice exhibited characteristics consistent with the phenotypic features associated with CS, including skull-vault craniosynostosis, skull deformity, shallow orbits accompanied by exophthalmos, midface hypoplasia with malocclusion, and shortened skull base, notably without any apparent limb defects. Furthermore, mutant mice displayed behavioral abnormalities encompassing deficits in learning and memory, social interaction, and motor dysfunction, without anxiety-related disorders. Histopathological examination of the hippocampal region revealed structural abnormalities, suggesting possible brain development impairment secondary to craniosynostosis. In conclusion, we constructed a novel gene-edited Fgfr2C361Y/+ mice strain based on CRISPR/Cas9, which displayed skull and behavioral abnormalities, serving as a new model for studying genetic molecular mechanisms and exploring treatments for CS.
Key messages
-
CRISPR/Cas9 crafted a Crouzon model by enhancing Fgfr2-C361Y in mice.
-
Fgfr2C361Y/+ mice replicate CS phenotypes—craniosynostosis and midface anomalies.
-
Mutant mice show diverse behavioral abnormalities, impacting learning and memory.
-
Fgfr2C361Y/+ mice offer a novel model for cranial suture studies and therapeutic exploration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Crouzon syndrome (CS, MIM 123500) is a very rare autosomal dominant craniosynostosis syndrome (incidence: 16.5 per 1,000,000), associated with gain-of-function mutations in the human fibroblast growth factor receptor 2 (FGFR2) gene [1]. Patients with CS exhibit distinctive craniofacial characteristics, including premature closure of cranial sutures, cranial bone deformities, proptosis, and midfacial hypoplasia. Severe CS may lead to intracranial hypertension [2], upper airway respiratory disorders [3], and associated neurocognitive deficits [4,5,6,7,8]. Due to ethical constraints and technical challenges, establishing genetically modified mouse models has become a viable approach for studying the genetic molecular mechanisms and therapies.
Previous literature reported CS mouse models included Fgfr2C342Y/+, Fgfr2W290R/+, and Fgfr3A385E/+ [9,10,11]. Traditional methods for model construction suffer from low targeting efficiency, prolonged lineage establishment, and complexity in operation. The inconsistent and unstable phenotypes among positive mice affected the result reliability. CRISPR/Cas9 gene editing technology, however, offers simplicity and high efficiency, demonstrating remarkable precision in gene targeting, enabling direct manipulation of fertilized eggs to induce heritable changes in offspring traits, and facilitating direct biological identification [12, 13].
Selective splicing of the extracellular IgIII domain of FGFR2 generates distinct IIIb and IIIc isoforms [14, 15]. Gain-of-function point mutations in the human FGFR2 IIIc isoform are associated with CS, leading to ligand-independent constitutive activation of FGFR2 [9, 16]. Known mutations associated with CS include FGFR2 p.C342Y, p.C342R, p.A344A, p.Y340H, etc., with p.C342Y being the most common mutation [17,18,19]. CS with acanthosis nigricans is characterized by the FGFR3 p.A391E mutation [20]. This study utilized CRISPR/Cas9 gene editing technology to generate a novel CS model, which was designed to express the p.Cys361Tyr missense mutation in the Fgfr2 IIIc isoform (corresponding to the p.Cys342Tyr mutation in humans) (Fig. 1a). Compared to previous CS models, we aimed to enhance gene editing efficiency and reproducibility, developing mutant mice with a more comprehensive and consistent craniofacial phenotype, thereby laying the groundwork for the subsequent study of cranial suture biology, model gene modification, and therapeutic intervention.
Construction of Crouzon mouse model based on CRISPR/Cas9 system. a Target gene expression strategy diagram based on mouse Fgfr2-215 (ENSMUST00000122054.8) transcript. b DNA template sequence of gRNA and sequences of the F1 forward and R1 reverse oligonucleotides required for synthesis. c The location of gRNA targeting sequence, PAM sequence, and donor DNA sequence in exon 8 of mouse FGFR2-215 transcript. TMS, target mutation site; SMS, synonymous mutation site, introduced near PAM in order to prevent re-recognition and cleavage by the CRISPR/Cas9 system. d Homologous sequence alignment of human FGFR2 gene and mouse Fgfr2 mutation model. The mutation position of bibliographic Fgfr2C342Y/+ mice was C342 (blue arrow), while the mutation position of Fgfr2C361Y/+ mice in this study was C361 (red arrow, Ensembl database). The discrepancy may be due to the previous bibliographic numbering being derived from the human C342Y or changes in annotation numbering caused by updates in the reference database transcript. e Representive result of Sanger sequencing of the bands from WT and Fgfr2C361Y/+ mice. The red box represents the missense mutation. The black box represents the synonymous mutation
Materials and methods
Animal models
Animal experiments were carried out with the approval of the Ethics Committee of the Chinese Academy of Medical Sciences and Peking Union Medical College (Approval No. 2023[45]), following the ARRIVE guidelines. In the Ensembl database, the mouse Fgfr2-215 transcript, labeled “Ensembl canonical” and “Genocode basic,” was selected for analysis (transcript length 3323 bp, encoding 840 amino acids, spanning 18 exons). Based on the mouse Fgfr2 gene sequence, the CRISPR design tool (http://crispr.mit.edu/) was used to design two highly target-specific, low off-target effect gRNA fragments (gRNA1-2). The gRNA DNA template sequence included the T7 promoter sequence and the crRNA/tracrRNA constant region (Fig. 1b). Using the Precision gRNA Synthesis Kit (A29377, Thermo Fisher Scientific, USA), the DNA template was assembled, followed by in vitro transcription and purification to generate full-length gRNA. The donor vector, containing the desired mutation and homology arms, was custom chemically synthesized by GenScript (GenScript Biotech, China). The location of gRNA target sequences, PAM, and the donor DNA are illustrated in Fig. 1c. Subsequently, TrueCut™ Cas9 Protein v2 (A36498, Thermo Fisher Scientific, USA), along with gRNA and the donor vector, were mixed at appropriate concentrations and microinjected into C57BL/6JGpt mouse zygotes. The zygotes were then implanted into the uteri of pseudopregnant C57BL/6JGpt female mice for gestation and offspring production. Genomic DNA was extracted from mouse tails 5–7 days post-birth, using the Mouse Genotyping Kit (P520, Vazyme, China). The target region was amplified by PCR with the following primers: F1: 5ʹ-TGCAGTTGGAATCTCCTGATGG-3ʹ; R1: 5ʹ-GCCCACTGAGTCAACAATTCAGC-3ʹ. Sanger sequencing of the amplified products was performed by Collective Pharmachem Biotechnology Co., Ltd (Nanjing, China).
Skeletal staining
Six-week-old mice skeletons were dissected, fixed for 72 h in 95% ethanol, soaked in 1% KOH until clear bone was evident, and then stained with Alizarin Red. After rinsing with distilled water, the skeleton was treated with mixtures of 20%, 50%, and 80% glycerol and 1% KOH until the soft tissue disappeared. Finally, it was stored in pure glycerol.
Skull micro-CT
Six-week-old mice skulls were scanned using the micro-CT system (INVEON MM GANTRY; Siemens, Germany) operating at 60 kV and 400 µA, with an effective pixel size of 20.93 µm. The DICOM data were then imported into Mimics 21.0 software (Materialise, Leuven, Belgium) to generate three-dimensional (3D) reconstruction. CT cephalometry was performed on mutant and wild-type (WT) mice (n = 5) based on the reported biological markers of mouse skull [21,22,23]. The skull cavity was manually segmented using the “Edit Masks” module in coronal, axial, and sagittal views. The “Cavity Fill” module filled the skull cavity, saved it as an object, and calculated intracranial volume (ICV).
Hematoxylin-eosin (HE) staining
Tissue samples were collected and fixed in 4% paraformaldehyde for 24 h. Bone tissues were decalcified with 10% EDTA for 7 days. All tissues were then dehydrated, cleared, embedded in paraffin, and sectioned. Sections were deparaffinized in xylene, followed by 100%, 95%, and 70% ethanol, then rehydrated with distilled water. After 10 min in hematoxylin, sections were rinsed, examined, and differentiated with 1% hydrochloric acid alcohol until the cytoplasm was clear. They were soaked in water for 5 min, air-dried, stained with eosin for 1 min, washed, dehydrated through ethanol, cleared with xylene, and mounted with PVP medium.
Femoral growth plate measurement
Sagittal longitudinal HE sections of the femur from 6-week-old mutant and WT mice (n = 3) were scanned and imported into ImageJ software (version 1.8.0). The dense, translucent, and light-colored cartilage region between the epiphysis and the diaphysis was identified as the location of the growth plate (hypertrophic zone and proliferating zone). The width of the cartilage zone was measured to roughly evaluate the growth plate length. For accuracy and reproducibility, at least three measurements were taken in different regions and averaged.
Behavioral assays
Prior to the experiment, 10-week-old mutant and WT female mice (n = 12) were conditioned for 3 days. At least an hour before testing, the mice were transferred to the experimental environment. Data analysis and behavioral testing were conducted in a genotype-blinded manner.
Novel object test
During habituation, mice were placed in an empty 30 × 40 × 30 cm3 area for 5 min. They then explored two identical objects for 10 min during familiarization. In the test phase, mice were presented with one familiar and one new object of different shape and size and given 5 min to explore. Interaction time percentages were calculated as Tn(To)/(Tn + To) × 100%. The preference index was calculated as (Tn–To)/(Tn+To) × 100% where Tn (To) represents the time spent exploring new (old) objects.
Three-chamber social interaction test
A 60 × 40 × 30 cm3 area with transparent partitions creating three chambers allowed free mouse movement. Stainless steel cylinders were placed in the center of the left and right chambers. During the familiarity stage (10 min), mice roamed freely in the middle chamber. In the social ability stage (10 min), an unfamiliar mouse (M1) matched in strain, age, and sex was introduced into the cylinder in the right chamber, while the left chamber’s cylinder contained an object. During the new social preference phase (10 min), another unfamiliar mouse (M2), matched similarly, replaced M1 in the right chamber’s cylinder, with M1 moved to the left chamber’s cylinder. The preference index was calculated as (T2 – T1)/(T2 + T1) × 100%, where T1 and T2 represented the time exploring old (M1) and new (M2) acquaintances.
Rotarod test
In the initial training phase, mice were trained to stay on a rotating rod at a constant 5 rpm speed for 60 s. Adaptation to the rod continued by gradually increasing the speed from 5 to 15 rpm within 5 min. During the test stage, the bar speed accelerated from 8 to 40 rpm over 5 min, and the time until the mice fell was recorded. This test was repeated three times.
Open field test
Mice were placed in a 40 × 40 cm2 area and allowed 30 min to explore. The total distance travelled was recorded. In three intervals, 0–10 min, 10–20 min, and 20–30 min, the percentage of time spent in the central area (24 × 24 cm2) was calculated.
Elevated plus maze test
A 50-cm-high elevated plus maze device consisted of four arms, with two open and two closed. For 10 min, the mice were allowed to roam freely in the maze, facing the open arms. The time of mice entering the open arm within a fixed time was recorded.
Immunohistochemistry (IHC)
Paraffin sections from coronal brain slices were baked, deparaffinized, and hydrated. After soaking in 3% H2O2 for 10 min, they were blocked with serum for 30 min. Anti-NeuN antibody (Abcam, UK, 1:3000) was incubated overnight at 4 °C, followed by Goat Anti-Rabbit IgG (Abcam, UK, 1:1000) incubation at 25 °C for 1 h. DAB chromogen was applied, and color development was monitored under a microscope. After counterstaining with hematoxylin for 3 min, sections were dehydrated and mounted. Positive DAB staining exhibited a brown-yellow color. The images were scanned and imported into ImageJ software. Total counts of positive neurons were conducted separately for the CA1-4 and DG regions of the hippocampal sections from mutant and WT mice (n = 3).
Statistical analysis
For comparisons between two groups, a two-tailed Student’s t test was used. The qualitative variables are presented as mean ± SD. Statistical significance was set to P < 0.05. All statistical analyses were performed using GraphPad Prism 9.4.0 software (San Diego, CA, USA).
Results
Mouse genotype identification
Based on the mouse Fgfr2-215 transcript (ENSMUST00000122054.8, CCDS52413, Uniprot match: E9QK53, NCBI match: NP_034337.2), we introduced the C361Y missense mutation in the Fgfr2 IIIc isoform encoded by exon 8 (corresponding to the human FGFR2 p.C342Y mutation) after human-mouse protein homology alignment (Fig. 1d). Sequence alignment showed that the gene editing location was consistent with the C342Y mutation in exon 9 of the mouse Fgfr2 gene introduced by Eswarakumar et al. [9] (Fig. 1d). The differences in amino acid and exon numbering may be related to the fact that previous bibliographic numbering was derived from human C342Y or updates in the selected mouse database transcript. This study used the Fgfr2 isoform IIIc precursor sequence (NP_034337.2, CCDS52413) for CRISPR/Cas9 system design to provide complete DNA sequence information. In the positive F1 generation mice, the corresponding c.1081-1083 TGC mutation to TAC was verified by PCR-Sanger sequencing (Fig. 1e).
Craniofacial phenotypes of Fgfr2 C361Y/+mice
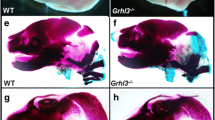
A photograph taken 6 weeks after birth revealed that the Fgfr2C361Y/+ mice were characterized by a round and short skull deformity, facial shortening, exophthalmos, and midface hypoplasia without apparent limb abnormalities (Fig. 2a). In addition, gross anatomy and alizarin red skull staining showed bilateral coronal suture closure and anterior skull base shortening (Fig. 2b, c); coronal suture HE sections further confirmed the closure of the coronal suture (Fig. 2d); micro-CT 3D reconstructions of craniofacial bones showed shallow eye orbits, maxillary retrusion, malocclusion, and fusion of the intersphenoidal synchondrosis (ISS) in mutant mice (Fig. 2e). Some positive heterozygotes (16.7%) exhibited nasal and jaw deviations (Fig. 2e).
Craniofacial phenotypes of Fgfr2C361Y/+ mice. a Photographs of 6-week-old live mice. b Gross anatomy of skull bones of 6-week-old mice. Red asterisks point to the bilateral closed coronal sutures. c Transparent whole calvaria staining of 6-week-old mice. The dotted box indicates that the anterior cranial base and midface appear shorter in mutant mice, while the posterior cranial base appears relatively unaffected. d HE sections of coronal suture from 6-week-old mice. Scale bar: 200 µm. e 3D reconstruction of 6-week-old mouse skull. Yellow arrowheads indicate nasal deviation in Fgfr2C361Y/+ mice. Red arrowheads exhibit fusion of the intersphenoidal synchondrosis suture (ISS), while the spheno-occipital synchondrosis suture (SOS) keeps patent (black arrowheads) in Fgfr2C361Y/+ mice. Blue arrowheads display maxillary retrusion and malocclusion in Fgfr2C361Y/+ mice
As shown in Fig. 3a, skull measurements based on craniofacial landmarks revealed that, compared to WT mice, Fgfr2C361Y/+ mice had reduced skull length (Nal-Opi) (P < 0.0001) and increased skull height (P < 0.0001), with no significant difference in skull width (P = 0.836). The interorbital distance (Ent-Ent) increased (P < 0.05), but nasal length (Nal-Brg) and upper and lower jaw length decreased (P < 0.05), indicating inhibited midfacial development. The frontal bone length (Nas-Brg) significantly decreased (P < 0.001), while parietal bone length (Brg-Lam) showed no significant difference (P = 0.051), suggesting limited compensatory growth near the coronal suture. A 2D sagittal plane constructed from Nas, Pbm, and Opi showed that the lengths of presphenoid bone (Con-Pbm) and basisphenoid bone (Pbm-Bom) in the anterior cranial base were shorter in mutant mice compared to WT mice (P < 0.05). There was no significant difference in the basioccipital length (Bom-Bas) in the posterior cranial base (P = 0.242). ICV reconstruction showed that mutant mice had a smaller ICV than WT mice (P < 0.05, Fig. 3b). These findings were consistent with previous skeletal staining results, indicating restricted cranial base and fossa development in mutant mice.
Micro-CT quantitative analysis of 6-week-old mouse skull. a Craniofacial landmarks and CT cephalometry. Skull length: Nal-Opi; skull width: Eur-Eur; skull height: maximum height from the vertex of the skull to the lower edge of the auditory bulb; interorbital distance: Ent-Ent; nose length: Nal-Brg; zygomatic arch length: Apo-Lpo; upper jaw length: the horizontal distance from Nal to Zyt; lower jaw length: the horizontal distance from Inf to Goc; frontal bone length: Nas-Brg; parietal bone length: Brg-Lam; presphenoid bone length: Con-Pbm; basisphenoid bone length: Pbm-Bom; basioccipital bone length: Bom-Bas. b ICV reconstruction and measurement. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significant
Other systemic phenotypes of Fgfr2 C361Y/+mice
The growth curve showed that Fgfr2C361Y/+ mice had slower growth, with smaller height and weight than WT littermates starting from postnatal day 7 and a significant decrease in body weight (Fig. 4a). Due to feeding difficulties from malocclusion, the mutant mice suffered malnutrition-related deaths, with females too weak to conceive or experiencing miscarriages, resulting in lower lifespan and fertility compared to WT mice. Lower limb HE sections revealed that the femoral growth plate region length in mutant mice was shorter than that in WT mice (P < 0.05, Fig. 4b). Besides, heart HE sections showed disorganized myocardial fibers, with localized edema and scattered lymphocyte infiltration (Fig. 4c); the spleen tissue displayed extensive expansion of the germinal centers in the splenic nodules with congestion (Fig. 4d); the brain tissue exhibited a significant number of shrunken neurons in the CA1, CA4, and DG regions of the hippocampus, along with reduced cell layers, disorganized cell arrangement, hyperchromatic nuclei, blurred or absent nucleoli, and indistinct cytoplasmic and nuclear boundaries (Fig. 4e). IHC showed significantly fewer NeuN-labeled neurons in the hippocampus of mutant mice than WT (P < 0.05 for CA1, CA2, CA4, DG; P < 0.001 for CA3; Fig. 4f), indicating affected neuron maturation and development.
Other systemic phenotypes of Fgfr2C361Y/+ mice. a Photos and curves of mouse growth and development. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. b HE sections of femur tissue from 6-week-old mice. The black arrows indicate the growth plate region (hypertrophic zone and proliferating zone). Scale bar: 500 µm, 200 µm. *P < 0.05. c HE sections of heart tissue from 6-week-old mice. Red arrow indicates disordered muscle fiber arrangement. Blue arrow indicates a loose arrangement of connective tissue. Purple arrow indicates scattered lymphocyte infiltration. Scale bar: 1000 µm, 50 µm. d HE sections of spleen tissue from 6-week-old mice. Black arrow indicates extensive expansion of the germinal centers in the splenic nodules with congestion. Scale bar: 1000 µm, 50 µm. e HE sections of brain tissue from 6-week-old mice. Black arrows indicate shrunken neurons. Scale bar: 200 µm, 50 µm. f NeuN IHC total count in different zones of hippocampus (CA1-4 and DG) from 6-week-old mice. Scale bar: 200 µm. *P < 0.05, ***P < 0.001
Behavioral study in Fgfr2 C361Y/+mice
The novel object test primarily examines animals’ memory and cognition, generally revealing that rodents explore new objects more than familiar ones, indicating their recognition memory of previously encountered objects [24]. Compared to the WT group, Fgfr2C361Y/+ mice interacted for long duration with old objects (old objects Fgfr2C361Y/+, 59.47 ± 14.98% vs WT, 45.33 ± 6.26%, P < 0.01; new objects Fgfr2C361Y/+, 40.53 ± 14.98% vs WT, 54.67 ± 6.26%, P < 0.01), and the preference index was negative (Fgfr2C361Y/+, −18.95 ± 29.95% vs WT, 9.35 ± 12.52%, P < 0.01, Fig. 5a). The three-chamber social interaction test is primarily used to assess social interaction ability [25]. While the control mice showed the characteristic of rejoicing the new and loathing the old, the mutant mice exhibited social laziness, as reflected by a negative preference index (Fgfr2C361Y/+, −9.25 ± 24.56% vs WT, 18.30 ± 21.19%, P < 0.01; Fig. 5b). We employed the rotarod test to assess the motor skills [26]. While the control group successfully maintained on the rod for the entire testing period, mutant mice were unable to hold on until the end, with their rod-holding time (latency to fall) decreased (P < 0.05 for test1, P < 0.01 for test2, P < 0.001 for test3; Fig. 5c). The open field test can be used to evaluate anxiety and motor functions [27]. Since anxious mice tend to move at the edge of the field [28], we evaluated the central time percentage of mutant mice and controls, finding no significant difference (P = 0.1627 for test1, P = 0.0554 for test2, P = 0.3480 for test3; Fig. 5d). However, mutant mice showed less total movement distance than the control group (P < 0.05 for test1, P < 0.01 for test2, P < 0.05 for test3; Fig. 5d). The elevated plus maze test reflects anxiety and panic states by inducing conflicting behaviors, stemming from desire to explore new environments and fear of open elevated areas [29]. There was no statistical difference in the time taken for mutant and WT mice to enter the open arm within a fixed period (P = 0.1086, Fig. 5e).
The behavioral study related to brain function in Fgfr2C361Y/+ mice. a Novel object test. The percentage of interaction time was calculated as Tn(To)/(Tn + To) × 100%. The preference index was calculated as (Tn − To)/(Tn + To) × 100%, where Tn (To) represents the time spent exploring new (old) objects. b Three-chamber social interaction test. The preference index was calculated as (T2 − T1)/(T2 + T1) × 100%, where T1 and T2 represent the time spent exploring old (M1) and new (M2) friends, respectively. c Rotarod test. Performance was recorded as time on the rotarod. d Open field test. The proportion of time spent in the center area and total distances were measured. e High plus maze test. The time entering the open arm within a fixed time was recorded. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant
Discussion
Through craniofacial phenotype verification, the Fgfr2C361Y/+ mice exhibited brachycephaly, shallow orbits with exophthalmos, and midface hypoplasia with malocclusion, which were consistent with the phenotypic characteristics associated with CS [30]. Furthermore, the shortening of the skull base and the reduction of the subcranial space were in line with the measurement trends of skull base morphology and cranial fossa growth commonly seen in patients with CS [31, 32]. This affirmed the comparability of this mouse model to human CS.
Different from previous CS mouse models (Fgfr2C342Y/+, Fgfr2W290R/+), where significant variability existed among individuals in the types and degree of suture fusion [10, 22], Fgfr2C361Y/+ mice primarily exhibited fusion of the bilateral coronal sutures. Besides, midfacial development and malocclusion were more pronounced compared to Fgfr2W290R/+ mice [10]. With the high specificity and precision of CRISPR/Cas9 technology, mutant mice exhibited more comprehensive and consistent craniofacial phenotypes, aiding in the precise delineation of mutation-affected craniofacial regions. Additionally, the strain background of mice could influence phenotypes [33]. Previous models often involved crosses between different strains, but this study used C57BL/6JGpt mice from a single strain to clearly delineate the phenotypic effects of the Fgfr2 p.C361Y mutation in a uniform genetic background. Despite some limitations in generalizability, this provided crucial insights for subsequent mechanistic investigations.
The Fgfr2C361Y/+ mice did not exhibit finger or toe deformities, unlike the syndactyly observed in Apert Fgfr2P253R/+ mice [34]. Histopathological examination revealed shortened long bone growth plates, distinct from the pronounced limb shortening observed in Apert Fgfr2S252W/+ mice [35], with the possibility of feeding difficulties leading to malnutrition not being ruled out. Additionally, exploration of other systemic phenotypes suggested potential abnormalities in heart and spleen tissues in Fgfr2C361Y/+ mice, which are not commonly observed in human CS phenotypes. Considering the growth curves of the mutant mice, we speculated that the feeding difficulties and malnutrition were also closely related.
CS is generally less prone to mental retardation, but severe intracranial hypertension can still impact brain and neurocognitive development in patients [5, 36,37,38]. Fernandes et al. reported that patients scored at the lower end of the average range for oral, performance, and full-scale IQ [37]; Maximino et al. reported that 50% of ten patients had poor academic performance, 30% had learning disabilities, and 30% had CNS abnormalities [38]; Cohn et al. observed negative effects on social adaptation, occupation, and relationships in two non-surgical patients with CS [5]. Mouse behavioral assays mimicking human symptoms have been developed to assess mutation consequences and evaluate potential preclinical treatments [39,40,41]. Currently, aside from a few other syndromic craniosynostosis mouse models reporting related brain dysfunctions (i.e., Twist1+/− [42], Fgfr3A385E/+ [11], Fgfr2P252W/+, and Fgfr2P253R/+ [43]), there is a significant lack of neurocognitive studies on Fgfr2 mutant CS mouse models.
Studies have revealed the hippocampus as a key hub for cognitive functions, encompassing episodic memory and spatial navigation [44]. Fgfr2C361Y/+ mice displayed learning and memory deficits, impaired social interaction, as well as motor dysfunction, without anxiety-related disorders. Brain tissue sections confirmed that the morphology and maturation of partial neurons in the hippocampus region were impaired. Yu et al. found similar hippocampus-dependent cognitive deficits in Twist1+/− mice, but they displayed normal motor strength in open field test [42]. Cornille et al. reported that Fgfr3A385E/+ mice showed more severe impairments in episodic memory without locomotor or anxiety-related issues [11]. These differences could be related to the varying severity of craniofacial phenotypes. Fgfr2C361Y/+ mice exhibited more pronounced midface and occlusal abnormalities, and feeding difficulties could lead to malnutrition, affecting their motor abilities. Species-specific differences in brain composition, as well as the lower adaptability and plasticity of the mouse brain compared to humans, could render it more vulnerable to the impacts of craniosynostosis. Combined with the observed reduction in ICV in Fgfr2C361Y/+ mice, we hypothesize that the observed behavioral and brain abnormalities are primarily secondary to craniofacial deformities. This aligns with Yu et al.’s hypothesis that neurocognitive deficits in Twist1+/− mice are mediated by increased ICP, differing from Apert mice where the brain phenotype was primarily due to initial brain malformations [43].
Due to the shortcomings and complications of surgical treatment, prospective treatments based on drugs targeting FGFR2 or its downstream signaling molecules, bioactive molecules, and stem cell tissue engineering have received extensive attention [8, 42, 45]. The synchondroses formed by endochondral ossification at the skull base are important growth centers for the neurocranium, and their abnormalities can affect the development of adjacent craniofacial bones [46]. We observed ossification fusion of the skull base ISS in Fgfr2C361Y/+ mice, shortening of the presphenoid and basisphenoid bone length, and significant midface maxillary retrusion and malocclusion deformities, suggesting that ISS might be an important potential target for early intervention. Behavioral tests on Fgfr2C361Y/+ mice could assess the impact of craniosynostosis and its correction on brain development, offering a comparative therapeutic index for future research. This model could also serve as a long-term pharmacokinetic tool to identify effective drugs. Further utilization of CRISPR/Cas9 gene editing tools to repair mutations lays the groundwork for gene therapy. Simulating surgical intervention timing and outcomes in Fgfr2C361Y/+ mice allows for evaluating long-term results and complications, providing insights for human surgical practices.
Limitations of this study include the biological and genetic diversity differences between mice and humans, which present certain constraints and obstacles in model translation. There are significant differences in brain development between humans and mice, so caution should be exercised when extrapolating behavioral research results to humans, despite the value of early research findings. Secondly, the initial CS model mouse group was small, and there was a lack of phenotypic data for homozygous offspring. Adult female Fgfr2C361Y/+ mice struggled to conceive due to their weakened state. We have been actively working to improve the lifespan and fertility ability of deformed mice. Lastly, animal disease models using CRISPR-Cas9 have unique pros and cons compared to previous models. This study provides one possibility to increase the diversity of research designs.
Conclusions
In this study, we established a novel, stable, and efficient mouse strain, Fgfr2C361Y/+, using the CRISPR/Cas9 system. Fgfr2C361Y/+ mice showed CS-associated phenotypic features: skull-vault craniosynostosis, brachycephaly, shallow orbits with exophthalmos, midface hypoplasia causing malocclusion, and a shortened skull base, without apparent limb defects. Mutant mice also exhibited behavioral abnormalities, including learning and memory deficits, impaired social interaction, and motor dysfunction. Structural anomalies in the hippocampus suggested impaired brain development. In future research endeavors, there is an aspiration to conduct thorough investigations into the cranial suture pathophysiology and molecular mechanisms of CS through the development of Fgfr2C361Y/+ mouse models. This will involve discovering therapeutic targets to advance treatment methods and expedite drug development.
Data availability
All data generated during this study have been reported.
References
Cohen MM Jr, Kreiborg S (1992) Birth prevalence studies of the Crouzon syndrome: comparison of direct and indirect methods. Clin Genet 41:12–15. https://doi.org/10.1111/j.1399-0004.1992.tb03620.x
Lee ES, Lee SH, Han SW, Kim YO, Lim SY (2024) Association of cranial base suture/synchondrosis fusion with severity of increased intracranial pressure in Crouzon syndrome. Journal of cranio-maxillo-facial surgery : official publication of the European Association for Cranio-Maxillo-Facial Surgery 52:385–392. https://doi.org/10.1016/j.jcms.2024.02.011
Forte AJ, Lu X, Hashim PW, Steinbacher DM, Alperovich M, Persing JA, Alonso N (2019) Analysis of airway and midface in Crouzon syndromes. Ann Plast Surg 82:686–691. https://doi.org/10.1097/sap.0000000000001740
Maliepaard M, Mathijssen IM, Oosterlaan J, Okkerse JM (2014) Intellectual, behavioral, and emotional functioning in children with syndromic craniosynostosis. Pediatrics 133:e1608-1615. https://doi.org/10.1542/peds.2013-3077
Cohn ER, Hesky EM, Bradley WF Jr, McWilliams BJ, Hurwitz DJ, Wallace SB (1985) Life response to Crouzon’s disease. Cleft Palate J 22:123–131
Barden RC, Ford ME, Wilhelm WM, Rogers-Salyer M, Salyer KE (1988) Emotional and behavioral reactions to facially deformed patients before and after craniofacial surgery. Plast Reconstr Surg 82:409–418. https://doi.org/10.1097/00006534-198809000-00006
Pertschuk MJ, Whitaker LA (1985) Psychosocial adjustment and craniofacial malformations in childhood. Plast Reconstr Surg 75:177–184. https://doi.org/10.1097/00006534-198502000-00005
Stanton E, Urata M, Chen JF, Chai Y (2022) The clinical manifestations, molecular mechanisms and treatment of craniosynostosis. Dis Model Mech 15:dmm049390. https://doi.org/10.1242/dmm.049390
Eswarakumar VP, Horowitz MC, Locklin R, Morriss-Kay GM, Lonai P (2004) A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc Natl Acad Sci U S A 101:12555–12560. https://doi.org/10.1073/pnas.0405031101
Mai S, Wei K, Flenniken A, Adamson SL, Rossant J, Aubin JE, Gong SG (2010) The missense mutation W290R in Fgfr2 causes developmental defects from aberrant IIIb and IIIc signaling. Devel Dynam Off Pub Am Assoc Anatom 239:1888–1900. https://doi.org/10.1002/dvdy.22314
Cornille M, Moriceau S, Khonsari RH, Heuzé Y, Loisay L, Boitez V, Morice A, Arnaud E, Collet C, Bensidhoum M et al (2022) FGFR3 overactivation in the brain is responsible for memory impairments in Crouzon syndrome mouse model. J Exp Med 219:e20201879. https://doi.org/10.1084/jem.20201879
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi.org/10.1126/science.1231143
Ul Ain Q, Chung JY, Kim YH (2015) Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN. J Control Release 205:120–127. https://doi.org/10.1016/j.jconrel.2014.12.036
De Moerlooze L, Spencer-Dene B, Revest JM, Hajihosseini M, Rosewell I, Dickson C (2000) An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development (Cambridge, England) 127:483–492. https://doi.org/10.1242/dev.127.3.483
Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P (2002) The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development (Cambridge, England) 129:3783–3793. https://doi.org/10.1242/dev.129.16.3783
Beenken A, Mohammadi M (2009) The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov 8:235–253. https://doi.org/10.1038/nrd2792
Steinberger D, Mulliken JB, Müller U (1995) Predisposition for cysteine substitutions in the immunoglobulin-like chain of FGFR2 in Crouzon syndrome. Hum Genet 96:113–115. https://doi.org/10.1007/bf00214198
Wilkie AO (1997) Craniosynostosis: genes and mechanisms. Hum Mol Genet 6:1647–1656. https://doi.org/10.1093/hmg/6.10.1647
Wilkie AO, Byren JC, Hurst JA, Jayamohan J, Johnson D, Knight SJ, Lester T, Richards PG, Twigg SR, Wall SA (2010) Prevalence and complications of single-gene and chromosomal disorders in craniosynostosis. Pediatrics 126:e391-400. https://doi.org/10.1542/peds.2009-3491
Johnson D, Wilkie AO (2011) Craniosynostosis. Eur J Hum Genet 19:369–376. https://doi.org/10.1038/ejhg.2010.235
Richtsmeier JT, Baxter LL, Reeves RH (2000) Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Devel DynamOff Public Am Assoc Anatom 217:137–145. https://doi.org/10.1002/(sici)1097-0177(200002)217:2%3c137::Aid-dvdy1%3e3.0.Co;2-n
Perlyn CA, DeLeon VB, Babbs C, Govier D, Burell L, Darvann T, Kreiborg S, Morriss-Kay G (2006) The craniofacial phenotype of the Crouzon mouse: analysis of a model for syndromic craniosynostosis using three-dimensional microCT. Cleft Palate-craniofac J Off Public Am Cleft Palate-Craniof Assoc 43:740–748. https://doi.org/10.1597/05-212
Liu J, Nam HK, Wang E, Hatch NE (2013) Further analysis of the Crouzon mouse: effects of the FGFR2(C342Y) mutation are cranial bone-dependent. Calcif Tiss Int 92:451–466. https://doi.org/10.1007/s00223-013-9701-2
Chao OY, Nikolaus S, Yang YM, Huston JP (2022) Neuronal circuitry for recognition memory of object and place in rodent models. Neurosci Biobehav Rev 141:104855. https://doi.org/10.1016/j.neubiorev.2022.104855
Kaidanovich-Beilin O, Lipina T, Vukobradovic I, Roder J, Woodgett JR (2011) Assessment of social interaction behaviors. J Visual Exp JoVE. https://doi.org/10.3791/2473
Lubrich C, Giesler P, Kipp M (2022) Motor behavioral deficits in the cuprizone model: validity of the rotarod test paradigm. Int J Mol Sci 23:11342. https://doi.org/10.3390/ijms231911342
Pentkowski NS, Rogge-Obando KK, Donaldson TN, Bouquin SJ, Clark BJ (2021) Anxiety and Alzheimer’s disease: behavioral analysis and neural basis in rodent models of Alzheimer’s-related neuropathology. Neurosci Biobehav Rev 127:647–658. https://doi.org/10.1016/j.neubiorev.2021.05.005
Walz N, Mühlberger A, Pauli P (2016) A human open field test reveals thigmotaxis related to agoraphobic fear. Biol Psychiatry 80:390–397. https://doi.org/10.1016/j.biopsych.2015.12.016
Rodgers RJ, Dalvi A (1997) Anxiety, defence and the elevated plus-maze. Neurosci Biobehav Rev 21:801–810. https://doi.org/10.1016/s0149-7634(96)00058-9
Taylor JA, Bartlett SP (2017) What’s new in syndromic craniosynostosis surgery? Plast Reconst Surg 140:82e–93e. https://doi.org/10.1097/prs.0000000000003524
Lu X, Forte AJ, Wilson AT, Park KE, Allam O, Alperovich M, Steinbacher DM, Alonso N, Persing JA (2021) Respective roles of craniosynostosis and syndromic influences on cranial fossa development. Plast Reconst Surg 148:145–156. https://doi.org/10.1097/prs.0000000000008101
Lu X, Forte AJ, Wilson AT, Park KE, Allam O, Mozaffari MA, Alperovich M, Steinbacher DM, Alonso N, Persing JA (2020) What is the difference in cranial base morphology in isolated and syndromic bicoronal synostosis? Plast Reconst Surg 146:599–610. https://doi.org/10.1097/prs.0000000000007068
Percival CJ, Marangoni P, Tapaltsyan V, Klein O, Hallgrímsson B (2017) The interaction of genetic background and mutational effects in regulation of mouse craniofacial shape. G3 Bethesda Md 7:1439–1450. https://doi.org/10.1534/g3.117.040659
Yin L, Du X, Li C, Xu X, Chen Z, Su N, Zhao L, Qi H, Li F, Xue J et al (2008) A Pro253Arg mutation in fibroblast growth factor receptor 2 (Fgfr2) causes skeleton malformation mimicking human Apert syndrome by affecting both chondrogenesis and osteogenesis. Bone 42:631–643. https://doi.org/10.1016/j.bone.2007.11.019
Chen L, Li D, Li C, Engel A, Deng CX (2003) A Ser252Trp [corrected] substitution in mouse fibroblast growth factor receptor 2 (Fgfr2) results in craniosynostosis. Bone 33:169–178. https://doi.org/10.1016/s8756-3282(03)00222-9
Yacubian-Fernandes A, Ducati LG, Silva MV, Abramides DV, Perosa GB, Palhares A, Gabarra RC, Giglio A, Portela L, Marinello JL et al (2007) Crouzon syndrome: factors related to the neuropsychological development and to the quality of life. Arq De Neuro-psiquiatria 65:467–471. https://doi.org/10.1590/s0004-282x2007000300020
Fernandes MB, Maximino LP, Perosa GB, Abramides DV, Passos-Bueno MR, Yacubian-Fernandes A (2016) Apert and Crouzon syndromes-cognitive development, brain abnormalities, and molecular aspects. Am J Med Gen Part A 170:1532–1537. https://doi.org/10.1002/ajmg.a.37640
Maximino LP, Ducati LG, Abramides DVM, Corrêa CC, Garcia PF, Fernandes AY (2017) Syndromic craniosynostosis: neuropsycholinguistic abilities and imaging analysis of the central nervous system. Arq De neuro-psiquiatria 75:862–868. https://doi.org/10.1590/0004-282x20170171
Tecott LH (2003) The genes and brains of mice and men. Am J Psychiatry 160:646–656. https://doi.org/10.1176/appi.ajp.160.4.646
Crawley JN (2008) Behavioral phenotyping strategies for mutant mice. Neuron 57:809–818. https://doi.org/10.1016/j.neuron.2008.03.001
Sukoff Rizzo SJ, Crawley JN (2017) Behavioral phenotyping assays for genetic mouse models of neurodevelopmental, neurodegenerative, and psychiatric disorders. Annu Rev Anim Biosci 5:371–389. https://doi.org/10.1146/annurev-animal-022516-022754
Yu M, Ma L, Yuan Y, Ye X, Montagne A, He J, Ho TV, Wu Y, Zhao Z, Sta Maria N et al (2021) Cranial suture regeneration mitigates skull and neurocognitive defects in craniosynostosis. Cell 184:243-256.e218. https://doi.org/10.1016/j.cell.2020.11.037
Aldridge K, Hill CA, Austin JR, Percival C, Martinez-Abadias N, Neuberger T, Wang Y, Jabs EW, Richtsmeier JT (2010) Brain phenotypes in two FGFR2 mouse models for Apert syndrome. Devel Dynam Off Public Am Assoc Anatom 239:987–997. https://doi.org/10.1002/dvdy.22218
Dalton MA, Zeidman P, McCormick C, Maguire EA (2018) Differentiable processing of objects, associations, and scenes within the hippocampus. J Neurosci 38:8146–8159. https://doi.org/10.1523/jneurosci.0263-18.2018
Perlyn CA, Morriss-Kay G, Darvann T, Tenenbaum M, Ornitz DM (2006) A model for the pharmacological treatment of Crouzon syndrome. Neurosurgery 59(210–215):210–215. https://doi.org/10.1227/01.Neu.0000224323.53866.1e
McBratney-Owen B, Iseki S, Bamforth SD, Olsen BR, Morriss-Kay GM (2008) Development and tissue origins of the mammalian cranial base. Devel Biol 322:121–132. https://doi.org/10.1016/j.ydbio.2008.07.016
Acknowledgements
We would like to thank the Institute of Laboratory Animals Science, CAMS&PUMC for assistance with the micro-CT scanning work. Schematic diagrams were done with the assistance of Figdraw platform.
Funding
This work was supported by the Research Fund for the National Major Disease Multidisciplinary Diagnosis and Treatment Cooperation Project, number 1112321004.
Author information
Authors and Affiliations
Contributions
Y.-Y.Y., C.-Z.L., G.-D.S., and X.-L.J. designed and supervised the study. Y.-Y.Y. and C.-Z.L. performed experiments related to Fgfr2C361Y/+ and WT mice. Y.-Y.Y., C.-S.Y., and Y.W. planned and conducted laboratory experiments. Y.-Y.Y. and C.-S.Y. performed statistical analyses. Y.-Y.Y., C.-Z.L., X.-S.G., and X.-L.J. drafted and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Animal experiments were carried out with the approval of the Ethics Committee of the Chinese Academy of Medical Sciences and Peking Union Medical College (Approval No. 2023[45]), following the ARRIVE guidelines.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yue, Y.Y., Lai, CZ., Guo, XS. et al. New CRISPR/Cas9-based Fgfr2C361Y/+ mouse model of Crouzon syndrome exhibits skull and behavioral abnormalities. J Mol Med (2024). https://doi.org/10.1007/s00109-024-02476-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00109-024-02476-y