Abstract
MiRNAs, a class of non-coding RNA molecules, have emerged as critical modulators of telomere length and telomerase activity by finely tuning the expression of target genes (and not gene targets) within signaling pathways involved in telomere homeostasis. The primary objective of this systematic review was to compile and synthesize the existing body of knowledge on the role, association, and involvement of miRNAs in telomere length. Additionally, the review explored the regulation, function, and activation mechanism of the human telomerase reverse transcriptase (hTERT) gene and telomerase activity in tumor cells. A comprehensive analysis of 47 selected articles revealed 40 distinct miRNAs involved in these processes. These miRNAs were shown to exert their function, in both clinical cases and cell line models, either directly or indirectly, regulating hTERT and telomerase activity through distinct molecular mechanisms. The regulatory roles of these miRNAs significantly affected major cancer phenotypes, with outcomes largely dependent on the tissue type and the cellular actions within the tumor cells, whereby they functioned as oncogenes or tumor suppressors. These findings strongly support the pivotal role of miRNAs in modulating telomere length and telomerase activity, thereby contributing to the intricate and complex regulation of telomere homeostasis in tumor cells. Moreover, they emphasize the potential of targeting miRNAs and key regulatory genes as therapeutic strategies to disrupt cancer cell growth and promote senescence, offering promising avenues for novel cancer treatments.
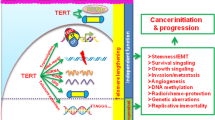
Graphical abstract
Mechanisms of action of miRNAs in regulating the hTERT complex and their impact on the tumor cells phenotypes.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
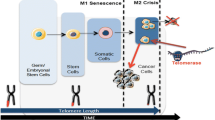
Telomeres, non-coding repeated sequences of TTA GGG nucleotides, are located at the final portions of the chromosomes and are responsible for maintaining the integrity and stability of the genome by protecting the ends of eukaryotic chromosomes in a sheltering protein complex [1,2,3].
In dividing somatic cells, telomeres shorten by about 60–150 bp within each cell division, inducing cell senescence and apoptosis [4,5,6]. This telomere shortening leads to loss of telomere capping, resulting in chromosomal instability, such as the formation of aberrant chromosomes and end to end fusions [7]. These events trigger DNA damage response (DDR) mechanisms, in which a cascade of cell signaling, mediated by p53, p21 and other cell cycle proteins, controls cell cycle arrest and the fate of cell survival [8, 9].
The synthesis of the TTAGGG repeats at the telomeric ends occurs by the telomerase enzyme, which controls the maintenance of telomere length. This enzyme, a ribonucleoprotein reverse transcriptase, is actively expressed in reproductive and stem cells but not expressed in somatic cells [10, 11]. In most cancer cells, however, telomerase expression is active, enabling the cells to bypass senescence and apoptosis, which results in continuous cell proliferation [12,13,14].
The human telomere reverse transcriptase (hTERT) gene, located at 5p15.33, codifies the catalytic subunit of the telomerase enzyme and is responsible for regulating telomerase activity [15]. The mechanisms that regulate hTERT expression and telomerase activity are extensively studied and can occur via multiple genetic and epigenetic mechanisms, including mutations in the hTERT promoter regions, mRNA splicing alterations, amplification, methylation, and/or disruption of telomere position effect (TPE) machinery [16, 17]. In a small fraction of tumors (5–15%), an alternative lengthening of telomeres (ALT) can also occur [18].
MicroRNAs (miRNAs/miRs) are highly conserved, non-coding single strand RNAs of approximately 20 to 22 nucleotides that regulate gene expression post-transcriptionally [19]. These molecules play a crucial role in tumor development and progression by regulating the expression of genes involved in several cancer-associated signaling pathways, including the ones that modulate cell proliferation, survival, and senescence [20,21,22,23]. MiRNAs also regulate telomere length and telomerase activity, mediating many aspects of telomere homeostasis, including the sheltering and the telomerase complex [24, 25].
Considering the essential role and function of the telomere length in cancer cells and the role of miRNAs in actively regulating driver oncogenes and tumor suppressors genes in this process, this systematic review main objective was to compile and synthesize the existing knowledge on the role, association and/or involvement of miRNAs in telomere length. Additionally, the review explored the regulation, function, and activation of the hTERT and telomerase in tumor cells.
Methods
The review protocol followed the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) guidelines [26, 27] and was registered at the International Prospective Register of Systematic Reviews (PROSPERO) database under the identifier CRD42021282906.
Data sources and search strategy
The selected terms “Telomerase” and synonyms (Telomerase OR Telomere OR hTERT), “Cancer” and synonyms (Cancer OR Tumor OR Carcinoma) and “microRNA” and synonyms (microRNA OR miR OR miRNA OR small non-coding RNA OR small ncRNA) in Title/Abstract), were searched in six distinct databases: EBSCO, EMBASE, Lilacs, Pubmed, Scielo, and Scopus. The searches were independently carried by two reviewers, using Rayyan Web App. Duplicate articles were excluded and the remaining articles were screened based on the title and abstract. The full text of the selected articles was then assessed for relevance and eligibility. The end of the search date was October 2nd, 2023.
Study selection and eligibility criteria
Two reviewers evaluated the studies independently, according to the established inclusion and exclusion criteria. Inclusion criteria: (1) articles describing the interaction of miRNAs and the telomerase enzyme activation/function for telomere length maintenance in human tumor cells and cell lines; (2) articles that have been peer-reviewed and written in English. Exclusion criteria: (1) articles on telomerase enzyme activation/function that did not report on the role, association, and/or involvement of miRNAs; (2) articles on miRNAs analysis performed in animals; (3) articles on clinical intervention and/or other types of clinical trials; (4) non-original (review) articles, letter from reviewers, book chapters, unpublished studies; (5) articles for which the full-text was not available; (6) articles considered with low-quality risk of bias.
Data extraction
Two reviewers independently extracted the following information from the selected articles: name of the first author, year of publication, methodology (miRNA related and other relevant methods), sample source (biological material (patient’s samples and/or cell lines), miRNAs analyzed, miRNA target genes, and main results and conclusions.
Quality and bias evaluation
The Quality in Prognosis Studies (QUIPS) tool was used to assess the quality of the studies and the risk of bias. This tool evaluates studies based on the following categories: study participation and attrition, prognostic factor and outcome measurements, study confounding, and statistical analysis, and reporting [28]. Based on these categories, the article quality and risk of bias were classified into high quality ( +): with little or no risk of bias, acceptable ( ±): with moderate risk of bias, and low quality (-): with high risk of bias, and unsure (?). According to this classification, the articles received a general evaluation as low, moderate, or high risk of bias.
Results and discussion
Search results
The search of the selected terms in the six selected databases resulted in 741 articles. After removing duplicate entries, 380 articles were screened for relevance and compliance based on the established inclusion and exclusion criteria. Forty-seven full-text articles were identified and further proceeded for qualitative synthesis analysis. The entire search process, comprising four distinct strategy steps, along with the respective number of articles is presented in Fig. 1.
Risk of bias and quality of the studies
The assessment of the risk of bias in the six established categories by the QUIPS tools, resulted in 47 studies. From these, 22 presented a low risk of bias and 25 a moderate risk of bias (Supplementary Table 1). Following these analyses 47 studies were included for final qualitative synthesis.
Selected final articles
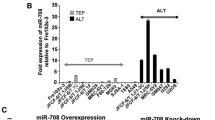
The 47 original articles were further subjected to qualitative synthesis based on the established inclusion and exclusion criteria. All the articles met the criteria and therefore, their findings were detailed described in this systematic review. The most frequent types of cancer studied were cervical (six studies), melanoma and colorectal (five studies each), followed by bladder, breast and gastric (three studies each), colon, glioblastoma, glioma, hepatocellular, ovarian, and thyroid cancer (two studies each), and osteosarcoma, lymphoma, lung, pancreatic, oropharyngeal, mesothelioma, head and neck squamous cells, laryngeal, and non-small-cell lung cancer (NSCLC) (one study each). In addition, one study was conducted in 56 different human cell lines, one in the Isogenic Telomerase Positive (TEP), and one in Alternative Length of Telomeres (ALT) cancer cells (Table 1).
Altogether 40 different miRNAs were described as potential regulators of gene and telomerase expression, and activity in these studies (Table 1). The most frequent miRNAs described in the 47 articles were: miR-138-5p (nine articles), miR-1182 and miR-34a-5p (three articles), and miR-195-5p, miR-21, miR-296-5p, miR-29a-3p, miR-346, miR-491-5p, and miR-512-5p (two articles). The other 30 miRNAs were described in one article each. Nine miRNAs were described regulating telomere length and function: miRNA-34 (two articles), miR-155, miR-182-3p, miR-185, miR-193b-3p, miR-29a, miR-376, miR-490, and miR-708 (one article each).
MiRNAs that directly regulate the hTERT gene
Among the 47 articles selected, 27 of them reported 24 miRNAs that directly regulate the hTERT gene (Supplementary Table 2). In most of the studies the miRNA-mRNA (hTERT) interactions were experimentally validated by luciferase assay [study references 30,31,32, 34,35,36,37,38,39,40, 45, 48, 51,52,53, 55, 57,58,59, 69, 70, 74], RT-qPCR [30,31,32,33,34,35,36,37,38,39,40, 44, 53, 55, 57,58,59,60, 69, 70], and Western Blot [29, 31, 32, 34,35,36,37, 39, 40, 44, 45, 51,52,53,54,55, 57,58,59, 69]. In five studies [39, 54, 55, 59, 74] the miRNA regulation of telomerase activity was described by the Telomerase Repeated Amplification Protocol (TRAP) assay.
Among the miRNAs identified directly targeting the hTERT gene, the miR-138 was described in a higher number of studies (nine studies). The downregulation of miR-138 in clinical cancer cases compared to normal tissue was reported in six studies, in the following types of cancer: cervical [30], colorectal [32, 33], anaplastic and papillary thyroid [37], bladder cancer [29] and melanoma [35]. In the cell line models of most of these cancers, the downregulation of miR-138 led to the up-regulation of the hTERT expression and induced cell growth. However, in another study in melanoma, overexpression of miR-138 was observed in the tumor tissue when compared to adjacent non-tumor tissue [34]. In the cell line models its overexpression was observed in five studies in the three different types of cancer: cervical [31], colorectal [32] and melanoma [34,35,36]. In these studies, the overexpression of miR-138 was shown to reduce the expression of hTERT and inhibit cell proliferation, migration, invasion, and metastasis. These last studies suggested that miR-138 acts in the described cancer types as a tumor suppressor, decreasing the expression of hTERT and reducing tumorigenicity.
MiR-1182 was investigated in three studies [38,39,40]. In the only study performed in clinical cases (ovarian tumors), miR-1182 was observed down-regulated in the tumor tissues compared to the adjacent normal tissues [40]. In cancer cell lines, overexpression of miR-1182 was described in bladder [38], and ovarian [40] cells. In these cells, the ectopic expression of miR-1182 suppressed hTERT expression and inhibited cell proliferation, invasion, metastasis development, and induced chemosensitivity of the cells to cisplatin [38]. Conversely, in gastric cancer cell lines [39], downregulation of miR-1182 increased hTERT expression, promoting cell proliferation and conferring metastatic potential to the cancer cells. These studies pointed out to the tumor suppressor action of miR-1182.
The expression of miR-195-5p was observed downregulated in two studies in clinical cases [44, 45]. In the study of Liu et al. (2021) [45], miR-195-5p was observed downregulated in thyroid tumor tissues when compared to non-tumor tissues, and in the study of Chai et al. (2018) [44], the same was observed in melanoma samples. Conversely its upregulation in the cell line models of these tumors, showed reduced expression of hTERT, and the inhibition of cell proliferation and invasion, metastasis, and induction of apoptosis [44, 45]. The results of both studies suggested that this miRNA presents a tumor suppressor role in these cancer cells.
MiR-346 was observed down-regulated and up-regulated in gliomas [52] and cervical [30] tumor tissue compared to normal tissues, respectively. In the cell line models, overexpression of miR-346 increased the expression of hTERT and promoted cell growth [44, 45].
The regulation of hTERT expression by miR-491-5p was described in two studies in cervical cancer [53, 54]. In the Zhao et al. study [53], the overexpression of miR-491-5p decreased the expression of hTERT, inhibiting the PI3K/AKT signaling pathway and resulting in cell growth inhibition. Additionally, the authors demonstrated that the expression of miR-491-5p in cervical tumor clinical cases was lower when compared to adjacent normal tissue, supporting its tumor suppressive action [53]. In the other study, the overexpression of miR-491-5p reduced telomerase activity in Hela cells leading to the reduction of cell proliferation [54].
MiR-512-5p was observed directly regulating hTERT in one study in breast cancer [48] and one in head and neck squamous cell carcinoma [55]. In breast cancer, miR-512-5p was downregulated in the tumor tissue when compared to non-tumor tissues [48]. In the functional analysis both in breast cancer and head and neck cancer cell lines, the overexpression of miR-512-5p led to hTERT reduced expression and decreased tumorigenesis [48, 55].
Seventeen miRNAs (let-7g*, miR-1207-5p, miR-1255b-5p, miR-1266, miR-128, miR-133a, miR-135a, miR-296-5p, miR-299-3p, miR-29a, miR-3064, miR-342-5p, miR-455-3p, miR-497-5p, miR-532, miR-541-3p, and miR-615-3p) that also directly regulate the hTERT gene, were described (only once each) in eight different types of cancers: breast [48], cervical [54, 59], colorectal [58], gastric [51, 57], laryngeal [69], melanoma [44], non-small lung cancer [60], and ovarian [70] cancer.
Chen et al. (2014) [57] showed in gastric cancer cells that miR-1207-5p and miR-1266 interacted with the 3' UTR of hTERT, and their overexpression inhibited tumor growth. In these studies, both miRNAs were downregulated in the gastric tumor cases compared to non-tumor tissues. Zhang et al. (2020) [58] described the interaction between miR-1255b-5p and the hTERT and BRG1 genes in colorectal cancer. The downregulation of miR-1255b-5p increased the expression of hTERT and BRG1 promoting the epithelial-mesenchymal transition.
Guzman et al. (2018) [59] described that miR-128 significantly reduced the mRNA and protein levels of hTERT in HeLa cells. In another study, also in HeLa cell lines, [54] the overexpression of let-7g*, miR-133a, miR-342-5p, and miR-541-3p was shown to reduced telomerase activity, affecting cell proliferation. The miR-135a was associated with hTERT gene espression regulation in Choi et al. (2023) study [60]. The expression of mir-135a was observed downregulated in tumor tissues and cell lines of non-small-cells lung cancer, leading to the upregulation of the hTERT gene.
In gastric cancer, miR-29a was described directly regulating the hTERT gene in only one study [51]. In gastric tumor tissues, the lower expression of miR-29a was observed compared to precancerous gastric tissues. The same authors showed that in gastric cancer cell line models hTERT enhanced ITGB1 protein levels via the downregulation of miR-29a expression, leading to the invasion and metastasis [51].
Dinami et al. (2017) [48] described the downregulation of miR-296-5p expression in breast cancer compared to healthy controls. In the breast cancer cell line models, overexpression of miR-296-5p reduced hTERT expression, suppressing tumor progression, metastasis, and neovascularization. In addition to hTERT, this miRNA targets HMGA1, IKBKE, MAP2K3, MMP1, PUMA and SCRIB genes. Li et al. (2015) [69] described in the Hep-2 laryngeal cancer cell line that the overexpression of miR-299-3p was associated with the downregulation of hTERT mRNA and protein expression resulting in the inhibition of cell growth.
Qin et al. (2015) [33] described in colorectal cancer cells that miR-422a expression was downregulated and inhibited hTERT expression. In melanoma cells, Chai et al. (2018) [44] showed the downregulation of miR-455-3p and miR-497-5p in tumor tissue compared to matched nevi tissues. The tumor suppressor action of these miRNAs by downregulating hTERT expression, led to the inhibition of cell proliferation, migration, invasion, and to the promotion of apoptosis. In ovarian cancer, Bai et al. (2017) [70] described the downregulation of miR-532 and miR-3064 in ovarian tumor tissue compared to normal tissues. In the cell lines, the ectopic expression of these miRNAs downregulated hTERT expression levels by directly targeting its 3’-UTR regions, inhibiting cell proliferation and invasion of the ovarian cells.
Finally, Yan et al. (2018) [74] in the analysis of diverse types of cell lines, demonstrated in rectal cancer cells (RKO cells), that the overexpression of miR-615-3p reduced the expression of hTERT, affecting cell proliferation.
Altogether, these studies demonstrated the miRNAs’ ability to directly regulate the expression of hTERT gene by the exogenous modulation of their own expression levels. The functional consequences of these expression modulations in the cancer phenotypes largely depend on the type of tissue and the cellular action of the miRNAs within the tumor cells, where they can function either as oncogenes or tumor suppressors. Among the miRNAs directly targeting the hTERT gene, miR-138 stands out exhibiting both tumor-suppressive and oncogenic effects across different cancer types. Similarly, miR-1182 has been shown to exhibit diverse roles, acting as tumor suppressors in some cancer types while promoting tumorigenesis in others.
MiRNAs that indirectly regulate the hTERT gene and telomerase expression
A total of 12 articles cited the association of miRNAs that indirectly regulate hTERT expression and/or telomerase function by modulating the expression of genes involved in the telomere/telomerase signaling pathways (Supplementary Table 3). In these articles, ten different miRNAs were described: miR-103 [56], miR-150 [61], miR-19b [66], miR-202 [67], miR-21 [46, 47], miR-22 [68], miR-296 [49], miR-34a [42, 43], miR-375 [71], and miR-380-5p [72]. The most frequent type of cancer studied (in two articles) was hepatocellular carcinoma [43, 56], followed by breast [49], cervical [68], colorectal [46], glioma [42], glioblastoma [47], HPV positive (including oropharyngeal primary tissue, cervical and tongue cell lines) [49], lymphoma [61], melanoma [66], malignant peritoneal mesothelioma cells [72], osteosarcoma [49], and pancreatic cancer [67].
MiR-34a was cited in two studies, one in gliomas [42] and one in hepatocellular carcinoma [43]. Li et al. (2019) [42] showed in glioma cell lines that the overexpression of miR-34a, by exogeneous expression from mesenchymal stem cells, regulated telomerase activity by SIRT1 downregulation (an anti-senescence factor) and caused the expression of senescence‑related genes TP53, CDKN1A, and CDKN2C resulting in cellular senescence. In the study of Xu et al. (2015) [43], overexpression of miR-34a in SMMC-7721 and HHCC cell lines, inhibited telomerase activity and telomere length in liver cancer cells by negatively regulating the hTERT activators, FOXM1 and c-MYC. These genes are transcription factors that regulate several biological processes with relevance to tumorigenesis, including cell cycle, cell proliferation, differentiation, renewal, survival, and senescence] [76,77,78]. In human HCC tissues, miR-34a was down expressed and was correlated with tumor malignant features and poor prognosis [43]. The authors of these two papers point out that miR-34a may offer a potential strategy for the treatment of the cancers studied [42, 43].
Other miRNAs which expression alterations were associated with the hTERT expression were miR-19b, miR-202, miR-21, and miR-22. The overexpression of miR-19b was shown to increase hTERT expression in melanoma cell lines, by the downregulation of PITX1, a homeobox gene that acts as a hTERT suppressor gene [79]. Lower expression of miR-202, mediated by the Adamantyl Retinoid-Related (ARR) molecule 3-Cl-AHPC, induced the increased expression of MXD1 (MAD1) gene, a transcription factor that mediates cellular proliferation, differentiation and apoptosis [80]. The MDX1 increased expression inhibited MYC and hTERT expression and promoted apoptosis in pancreatic carcinoma cell lines [67]. The up regulation of the MXD1 and its family members, directly bind to the hTERT promoter, inhibiting its expression. Yang et al. (2015) [46] described the regulation of hTERT expression by miR-21 via PTEN/PI3K/AKT signaling pathway, in colorectal tumors. This miRNA was also observed affecting hTERT expression in HCT166 cell line, inhibiting cell proliferation through inactivation of hTERT and ERK1/2 genes [46]. The PTEN/PI3K/AKT signaling pathway is one of the most affected signaling pathways in cancer cells, that regulates the signaling of multiple biological processes, such as apoptosis, metabolism, cell proliferation, and growth [81, 82]. AKT is a central protein of this pathway and was shown to be essential for telomerase activation, telomere protection, and maintaining telomere length [83]. MiR-21 was also described affecting hTERT in glioblastoma cell lines, where the repression inhibited hTERT mRNA expression by modulating the expression of STAT3, a signal transducer gene that targets critical cell signaling genes, such as CYCLIN D1 and c-MYC [84] and inhibited the cell proliferation and induced cell cycle G0/G1 phase arrest and cell apoptosis [47]. On the other hand, miR-22 was associated with the reduction of the hTERT expression in cervical cancer cells by inhibiting its direct target MYCBP, a MYC binding protein [85]. This reduction led to an increase in the radiosensitivity of the cells and indicated the potential use of miR-22 as a novel radiotherapy approach in cervical cancer cells [68].
The miRNAs shown to regulate the telomerase activity, included: miR-103 [56], miR-150 [61], miR-296 [49], miR-375 [71], and miR-380-5p [72] (one study each). In the study of Xia et al. (2016) [56], the overexpression of miR-103 increased telomerase activity by AKAP12 downregulation, a scaffold kinase protein [86] that functions as tumor suppressor in hepatocellular cells, and increasing PKCα activity in HCC cell lines, another kinase protein. The downregulation of miR-150 was shown in the study of Watanabe et al. (2011) [61] in lymphoma cell lines to activate telomerase expression via AKT2 and DKC1, critical components of telomerase complex [83, 87]. The authors pointed out miR-150 function as a tumor suppressor that could potentially be used as therapeutic target in NK/T-cell lymphoma. Yoon et al. (2011) [49] were unable to demonstrate a significant impact of miR-296 on telomerase activity, by the regulation of p-21, WAF1 and p53 protein levels. However, the overexpression of this miRNA was observed in immortalized cells that presented with telomerase activation. In the study of Jung et al. (2014) [71], the reduction of TERT transcription by the ectopic expression of miR-375 was observed. This regulation occurred via CIP2A, E6, and/or E6AP, HPV associated proteins, in SiHa, HEK293, and HeLa, increasing p53, p21, and RB activities resulting in cell cycle arrest and cell proliferation inhibition. Finally, the study of Cimino-Reale et al. (2017) [72] reported the repression of telomerase activity by the overexpression of miR-380-5p, that directly targets the telomerase associated protein 1 (TEP1) and the testis-specific protein, Y-encoded-like 5 (TSPYL5) genes, resulting in the induction of apoptosis. These studies are described in Supplementary Table 3.
The findings from these studies underscore the pivotal role of miRNAs in modulating hTERT gene expression by targeting key genes involved in biological processes and signaling pathways within cancer cells, particularly those associated with telomere maintenance. Moreover, the intricate interplay between several of these genes further contributes to the modulation of telomerase activity and can ultimately dictate the senescence or survival fate of the cancer cells. Among the genes implicated in these processes, MYC, AKT2, TP53, and p21 were frequently cited as pivotal regulators capable of modulating telomerase activity.
MiRNAs role in telomere length maintenance and protection
Nine studies described nine microRNAs that affected the telomere length and protection (Supplementary Table 4). MiR-155 [62], miR-182-3p [63], miR-185 [64], miR-193b-3p [65], miR-29a-3p [50], miR-34a (cited by two studies [41, 42], miR-376a-3p [50], miR-490 [73], and miR-708 [75]. The most frequent type of cancer studied was colon cancer (cited by two studies) [50, 65], followed by breast [62], colorectal [65] and gallbladder [41] cancer, glioma [42], and glioblastoma [73].
MiR-34a was shown to be associated with telomere length in gallbladder cancer [41] and glioma cell lines [42]. In gallbladder tumor tissues, the overexpression of miR-34a, decreased telomere length by downregulating the expression of PNUTS, a regulator of the Protein Phosphatase 1 [88], which resulted in tumor growth inhibition [41]. In glioma cell lines, miR-34a overexpression delivered by human mesenchymal cells (hMSCs) was associated with the downregulation of the autophagy and apoptosis regulator SIRT1 gene, which led to the higher expression of p53, decreased telomere length, and cellular senescence [42]. In both articles miR-34a was demonstrated to act as a tumor suppressor, opening new possibilities for its use as an effective therapy for these tumors.
MiR-155, miR-193b-3p, miR-29a-3p, miR-376a-3p, and miR-490 were also described affecting telomere length by the interaction mostly with shelterin complex proteins that directly regulate telomere length. The overexpression of miR-155 reduced the expression of the TRF1 gene, a member of the shelterin complex which protects chromosome ends, regulates telomere length, recombination, and DNA damage checkpoints [89], and decreased telomere length in breast cancer cell lines. In clinical cases, the overexpression of this miRNA was associated with poor clinical outcomes in patients with the luminal subtype of breast cancer [62]. In the Dinami et al. (2022) study [65] it was shown that the TRF2 gene, another member of the shelterin complex that protects telomere length [90], cooperated with CTCF, a chromatin organization factor [91], and led to the overexpression of miR-193b-3p. This upregulation resulted in the decrease of the expression of the SUV39H1 gene, a histone methyltransferase, and increased cell growth. Liu et al. (2021) [50] described in colon cancer, that miR-29a-3p and miR-376a caused dysregulation in telomere length by binding to the CTC1 gene, a member of the heterotrimeric CTC1-STN1-TEN1 (CST) complex which is involved in the repair of replication errors to facilitate telomeric DNA and genomic DNA replication [92, 93]. Re-expression of CTC1 in the colon cancer cells restored the telomere length. These two miRNAs however, acted in the colon cancer cells distinctly: miR-29a-3p expression alterations led to the increase in the frequency of telomere’s signals and induction of telomere replication stress, while alterations in miR-376a expression led to replicative telomere damage, resulting in cancer progression. In glioblastoma cell lines, the overexpression of miR-490 decreased telomere length by repressing TERF2, TNKS2 and SMG1 genes, causing activation of p53 pathway, suppression of tumorigenicity and DNA damage [73].
Finally, other three miRNAs, miR-182-3p, miR-185, and miR-708, were described associated with DNA damage. MiR-182-3p [63] caused DNA damage by regulating TRF2 expression and affecting the telomeric and pericentromeric regions in breast cancer; miR-185 overexpression led to downregulation of the POT1 (Protection of telomere 1) gene, and increased of the ATR (Ataxia Telanxiectasia and Rad3-related kinase) signaling pathway [94] causing telomere DNA damage and promoting senescence in lung cancer cells [64]; and miR-708 overexpression caused downregulation of the MRE11A- and BRCA1 genes (involved in DNA damage response) [95, 96], expression and resulted in DNA damage, suppression of cell migration, invasion, and angiogenesis and telomere dysfunction [75]. In TEP cells the overexpression of miR-708 caused DNA damage by affecting genes of the CARF-p53 pathway, a tumor suppression pathway that acts as a molecular sensor and regulator of cellular stress, senescence, and immortalization [75].
In this section, a notable trend observed across most studies (detailed in Supplementary Table 4) was the involvement of miRNAs in regulating genes associated with the telomere shelterin complex, particularly the TRF family of genes. This observation is not unexpected, given the fundamental role of the shelterin complex in protecting chromosome ends, regulating telomere length, facilitating telomere replication, and coordinating DNA damage responses. By targeting genes within the shelterin complex, miRNAs exert regulatory control over these critical aspects of telomere biology, thereby influencing telomere maintenance pathways and impacting cellular senescence. This emphasis on shelterin complex regulation also highlights the significance of miRNA-mediated mechanisms in fine-tuning telomere dynamics and underscores their importance in maintaining genomic stability and cellular homeostasis.
Conclusions
In this comprehensive systematic review, we have identified 47 articles that collectively described the involvement of 40 different miRNAs in modulating hTERT expression, telomerase activity, and telomere length and function. These miRNAs exert their influence through distinct molecular mechanisms, acting both directly and indirectly and functioning both as oncogenes and tumor suppressors across several types of cancer cells. The scope of the selected studies encompassed clinical samples and cell line models, including the isogenic telomerase positive (TEP) and alternative mechanism of lengthening of telomeres (ALT) cell lines. It is important to acknowledge that the diverse methodologies employed, and the wide variety of tumor cell models utilized across these studies introduce inherent variability and complexity. In addition, the intricate interplay between miRNAs and telomere biology is influenced by numerous biological factors, including cell type, genetic background, and tumor microenvironment, which can lead to distinct experimental outcomes. Therefore, reaching a definitive conclusion regarding the precise role of the cited miRNAs may be difficult. However, despite these challenges, the findings from these studies strongly support the role of miRNAs in telomere biology. The intricate regulatory mechanisms of miRNAs, involving the expression regulation of target genes within critical cancer associated signaling pathways, highlights the complex regulatory networks underlying telomere maintenance. Moreover, these findings emphasize the potential of targeting miRNAs and key regulatory genes as a strategy to disrupt cancer cell growth and promote senescence. By directly or indirectly blocking hTERT gene expression and telomerase activity, tumorigenesis can be inhibited, and cancer progression contained. Overall, understanding the intricate interactions between miRNAs, target genes, and cellular signaling pathways represents a crucial step toward the development of novel target therapies directed to the manipulation of telomerase activity and induction of cellular senescence in cancer.
Availability of data and material
All the data can be available upon request.
References
Blackburn EH (2001) Switching and signaling at the telomere. Cell 106(6):661–673. https://doi.org/10.1016/s0092-8674(01)00492-5
Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318(5851):798–801. https://doi.org/10.1126/science.1147182
Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42:301–334. https://doi.org/10.1146/annurev.genet.41.110306.130350
Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345(6274):458–460. https://doi.org/10.1038/345458a0
Crompton NE (1997) Telomeres, senescence and cellular radiation response. Cell Mol Life Sci 53(7):568–575. https://doi.org/10.1007/s000180050073
Karlseder J, Smogorzewska A, de Lange T (2002) Senescence induced by altered telomere state, not telomere loss. Science 295(5564):2446–2449. https://doi.org/10.1126/science.1069523
de Lange T (2009) How telomeres solve the end-protection problem. Science 326(5955):948–952. https://doi.org/10.1126/science.1170633
Jacobs JJ, de Lange T (2004) Significant role for p16INK4a in p53-independent telomere-directed senescence. Curr Biol 14(24):2302–2308. https://doi.org/10.1016/j.cub.2004.12.025
Zhang Y, Sturgis EM, Huang Z, Zafereo ME, Wei Q, Li G (2012) Genetic variants of the p53 and p73 genes jointly increase risk of second primary malignancies in patients after index squamous cell carcinoma of the head and neck. Cancer 118(2):485–492. https://doi.org/10.1002/cncr.26222
Uziel O, Yosef N, Sharan R, Ruppin E, Kupiec M, Kushnir M, Beery E, Cohen-Diker T, Nordenberg J, Lahav M (2015) The effects of telomere shortening on cancer cells: a network model of proteomic and microRNA analysis. Genomics 105(1):5–16. https://doi.org/10.1016/j.ygeno.2014.10.013
Hiyama E, Hiyama K (2007) Telomere and telomerase in stem cells. Br J Cancer 96(7):1020–1024. https://doi.org/10.1038/sj.bjc.6603671
Chen CH, Chen RJ (2011) Prevalence of telomerase activity in human cancer. J Formos Med Assoc 110(5):275–289. https://doi.org/10.1016/S0929-6646(11)60043-0
Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266(5193):2011–2015. https://doi.org/10.1126/science.7605428
Jafri MA, Ansari SA, Alqahtani MH, Shay JW (2016) Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med 8(1):69. https://doi.org/10.1186/s13073-016-0324-x
Leão R, Apolónio JD, Lee D, Figueiredo A, Tabori U, Castelo-Branco P (2018) Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer. J Biomed Sci 25(1):22. https://doi.org/10.1186/s12929-018-0422-8
Yuan X, Larsson C, Xu D (2019) Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene 38(34):6172–6183. https://doi.org/10.1038/s41388-019-0872-9
Roake CM, Artandi SE (2020) Regulation of human telomerase in homeostasis and disease. Nat Rev Mol Cell Biol 21(7):384–397. https://doi.org/10.1038/s41580-020-0234-z
MacKenzie D Jr, Watters AK, To JT, Young MW, Muratori J, Wilkoff MH, Abraham RG, Plummer MM, Zhang D (2021) ALT Positivity in Human Cancers: Prevalence and Clinical Insights. Cancers 13(10):2384. https://doi.org/10.3390/cancers13102384
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
Peng Y, Croce CM (2016) The role of MicroRNAs in human cancer. Signal Transduct Target Ther 1:15004. https://doi.org/10.1038/sigtrans.2015.4
Vannini I, Fanini F, Fabbri M (2018) Emerging roles of microRNAs in cancer. Curr Opin Genet Dev 48:128–133. https://doi.org/10.1016/j.gde.2018.01.001
Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ (2020) Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci 21(5):1723. https://doi.org/10.3390/ijms21051723
Dragomir MP, Knutsen E, Calin GA (2022) Classical and noncanonical functions of miRNAs in cancers. Trends Genet 38(4):379–394. https://doi.org/10.1016/j.tig.2021.10.002
Slattery ML, Herrick JS, Pellatt AJ, Wolff RK, Mullany LE (2016) Telomere Length, TERT, and miRNA Expression. PLoS ONE 11(9):e0162077. https://doi.org/10.1371/journal.pone.0162077
Rossi M, Gorospe M (2020) Noncoding RNAs controlling telomere homeostasis in senescence and aging. Trends Mol Med 26(4):422–433. https://doi.org/10.1016/j.molmed.2020.01.010
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6(7):e1000097. https://doi.org/10.1371/journal.pmed.1000097
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE et al (2021) The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372:n71. https://doi.org/10.1136/bmj.n71
Hayden JA, Côté P, Bombardier C (2006) Evaluation of the quality of prognosis studies in systematic reviews. Ann Intern Med 144(6):427–437. https://doi.org/10.7326/0003-4819-144-6-200603210-00010
El Ahanidi H, El Azzouzi M, HafidiAlaoui C, Tetou M, Bensaid M, Chaoui I, Benbacer L, Hassan I, Oukabli M, Michaud K et al (2022) Immune checkpoint and telomerase crosstalk is mediated by miRNA-138 in bladder cancer. Front Oncol 11:795242. https://doi.org/10.3389/fonc.2021.795242
Song G, Wang R, Guo J, Liu X, Wang F, Qi Y, Wan H, Liu M, Li X, Tang H (2015) miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively. Sci Rep 5:15793. https://doi.org/10.1038/srep15793
Zhou N, Fei D, Zong S, Zhang M, Yue Y (2016) MicroRNA-138 inhibits proliferation, migration and invasion through targeting hTERT in cervical cancer. Oncol Lett 12(5):3633–3639. https://doi.org/10.3892/ol.2016.5038
Wang X, Zhao Y, Cao W, Wang C, Sun B, Chen J, Li S, Chen J, Cui M, Zhang B et al (2017) miR-138-5p acts as a tumor suppressor by targeting hTERT in human colorectal cancer. Int J Clin Exp Pathol 10(12):11516–11525
Qin YZ, Xie XC, Liu HZ, Lai H, Qiu H, Ge LY (2015) Screening and preliminary validation of miRNAs with the regulation of hTERT in colorectal cancer. Oncol Rep 33(6):2728–2736. https://doi.org/10.3892/or.2015.3892
Ye T, Li Y, Ye J, Zhang C (2019) miR-138-5p promotes proliferation of human melanoma cells by inhibiting hTERT expression. Anal Quant Cytopathol Histpathol 41:39–46
Li C, Zang Z, Gao T, Du M (2017) MicroRNA-138 suppresses cell proliferation of human malignant melanoma cells by targeting hTERT. Int J Clin Exp Med 10:6517–6526
Tarazón E, de Unamuno BB, MurriaEstal R, Pérez Simó G, SahuquilloTorralba A, Simarro J, PalancaSuela S, Botella Estrada R (2021) MiR-138-5p suppresses cell growth and migration in melanoma by targeting telomerase reverse transcriptase. Genes 12(12):1931. https://doi.org/10.3390/genes12121931
Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu N et al (2008) Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci 99(2):280–286. https://doi.org/10.1111/j.1349-7006.2007.00666.x
Zhou J, Dai W, Song J (2016) miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem Biophys Res Commun 470(2):445–452. https://doi.org/10.1016/j.bbrc.2016.01.014
Zhang D, Xiao YF, Zhang JW, Xie R, Hu CJ, Tang B, Wang SM, Wu YY, Hao NB, Yang SM (2015) miR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett 360(2):151–159. https://doi.org/10.1016/j.canlet.2015.01.044
Hou XS, Han CQ, Zhang W (2018) MiR-1182 inhibited metastasis and proliferation of ovarian cancer by targeting hTERT. Eur Rev Med Pharmacol Sci 22(6):1622–1628. https://doi.org/10.26355/eurrev_201803_14569
Jin K, Xiang Y, Tang J, Wu G, Li J, Xiao H, Li C, Chen Y, Zhao J (2014) miR-34 is associated with poor prognosis of patients with gallbladder cancer through regulating telomere length in tumor stem cells. Tumour Biol 35(2):1503–1510. https://doi.org/10.1007/s13277-013-1207-z
Li Q, Wang C, Cai L, Lu J, Zhu Z, Wang C, Su Z, Lu X (2019) miR-34a derived from mesenchymal stem cells stimulates senescence in glioma cells by inducing DNA damage. Mol Med Rep 19(3):1849–1857. https://doi.org/10.3892/mmr.2018.9800
Xu X, Chen W, Miao R, Zhou Y, Wang Z, Zhang L, Wan Y, Dong Y, Qu K, Liu C (2015) miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget 6(6):3988–4004. https://doi.org/10.18632/oncotarget.2905
Chai L, Kang XJ, Sun ZZ, Zeng MF, Yu SR, Ding Y, Liang JQ, Li TT, Zhao J (2018) MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag Res 10:989–1003. https://doi.org/10.2147/CMAR.S163335
Liu Z, Zhang L, Chen W, Yuan F, Yang Z, Liu S, Le F (2021) miR-195-5p regulates cell proliferation, apoptosis, and invasion of thyroid cancer by targeting telomerase reverse transcriptase. Bioengineered 12(1):6201–6209. https://doi.org/10.1080/21655979.2021.1963908
Yang Y, Yang JJ, Tao H, Jin WS (2015) MicroRNA-21 controls hTERT via PTEN in human colorectal cancer cell proliferation. J Physiol Biochem 71(1):59–68. https://doi.org/10.1007/s13105-015-0380-5
Wang YY, Sun G, Luo H, Wang XF, Lan FM, Yue X, Fu LS, Pu PY, Kang CS, Liu N et al (2012) MiR-21 modulates hTERT through a STAT3-dependent manner on glioblastoma cell growth. CNS Neurosci Ther 18(9):722–728. https://doi.org/10.1111/j.1755-5949.2012.00349.x
Dinami R, Buemi V, Sestito R, Zappone A, Ciani Y, Mano M, Petti E, Sacconi A, Blandino G, Giacca M et al (2017) Epigenetic silencing of miR-296 and miR-512 ensures hTERT dependent apoptosis protection and telomere maintenance in basal-type breast cancer cells. Oncotarget 8(56):95674–95691. https://doi.org/10.18632/oncotarget.21180
Yoon AR, Gao R, Kaul Z, Choi IK, Ryu J, Noble JR, Kato Y, Saito S, Hirano T, Ishii T et al (2011) MicroRNA-296 is enriched in cancer cells and downregulates p21WAF1 mRNA expression via interaction with its 3’ untranslated region. Nucleic Acids Res 39(18):8078–8091. https://doi.org/10.1093/nar/gkr492
Liu Y, Zhao X, Wang B, Liu Z, Zhang M, Wang J, Xu C, Wang Y, Du L, Wang F et al (2021) miR-376a Provokes Rectum Adenocarcinoma Via CTC1 Depletion-Induced Telomere Dysfunction. Front Cell Dev Biol 9:649328. https://doi.org/10.3389/fcell.2021.649328
He B, Xiao YF, Tang B, Wu YY, Hu CJ, Xie R, Yang X, Yu ST, Dong H, Zhao XY et al (2016) hTERT mediates gastric cancer metastasis partially through the indirect targeting of ITGB1 by microRNA-29a. Sci Rep 6:21955. https://doi.org/10.1038/srep21955
Wolter M, Werner T, Malzkorn B, Reifenberger G (2016) Role of microRNAs Located on Chromosome Arm 10q in Malignant Gliomas. Brain Pathol 26(3):344–358. https://doi.org/10.1111/bpa.12294
Zhao Q, Zhai YX, Liu HQ, Shi YA, Li XB (2021) [Retracted] MicroRNA-491-5p suppresses cervical cancer cell growth by targeting hTERT. Oncol Rep 46(6):248. https://doi.org/10.3892/or.2021.8199
Hrdličková R, Nehyba J, Bargmann W, Bose HR Jr (2014) Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS ONE 9(2):e86990. https://doi.org/10.1371/journal.pone.0086990
Li J, Lei H, Xu Y, Tao ZZ (2015) miR-512-5p suppresses tumor growth by targeting hTERT in telomerase positive head and neck squamous cell carcinoma in vitro and in vivo. PLoS ONE 10(8):e0135265. https://doi.org/10.1371/journal.pone.0135265
Xia W, Ni J, Zhuang J, Qian L, Wang P, Wang J (2016) MiR-103 regulates hepatocellular carcinoma growth by targeting AKAP12. Int J Biochem Cell Biol 71:1–11. https://doi.org/10.1016/j.biocel.2015.11.017
Chen L, Lü MH, Zhang D, Hao NB, Fan YH, Wu YY, Wang SM, Xie R, Fang DC, Zhang H et al (2014) miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis 5(1):e1034. https://doi.org/10.1038/cddis.2013.553
Zhang X, Bai J, Yin H, Long L, Zheng Z, Wang Q, Chen F, Yu X, Zhou Y (2020) Exosomal miR-1255b-5p targets human telomerase reverse transcriptase in colorectal cancer cells to suppress epithelial-to-mesenchymal transition. Mol Oncol 14(10):2589–2608. https://doi.org/10.1002/1878-0261.12765. (RetractionpublishedMolOncol.202317(7):1454)
Guzman H, Sanders K, Idica A, Bochnakian A, Jury D, Daugaard I, Zisoulis DG, Pedersen IM (2018) miR-128 inhibits telomerase activity by targeting TERT mRNA. Oncotarget 9(17):13244–13253. https://doi.org/10.18632/oncotarget.24284
Choi JE, Jeon HS, Wee HJ, Lee JY, Lee WK, Lee SY, Yoo SS, Choi SH, Kim DS, Park JY (2023) Epigenetic and genetic inactivation of tumor suppressor miR-135a in non-small-cell lung cancer. Thorac Cancer 14(11):1012–1020. https://doi.org/10.1111/1759-7714.14838
Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T et al (2011) The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia 25(8):1324–1334. https://doi.org/10.1038/leu.2011.81
Dinami R, Ercolani C, Petti E, Piazza S, Ciani Y, Sestito R, Sacconi A, Biagioni F, le Sage C, Agami R et al (2014) miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res 74(15):4145–4156. https://doi.org/10.1158/0008-5472.CAN-13-2038
Dinami R, Pompili L, Petti E, Porru M, D’Angelo C, Di Vito S, Rizzo A, Campani V, De Rosa G, Bruna A et al (2023) MiR-182–3p targets TRF2 and impairs tumor growth of triple-negative breast cancer. EMBO Mol Med 15(1):e16033. https://doi.org/10.15252/emmm.202216033
Li T, Luo Z, Lin S, Li C, Dai S, Wang H, Huang J, Ma W, Songyang Z, Huang Y (2020) miR-185 targets POT1 to induce telomere dysfunction and cellular senescence. Aging 12(14):14791–14807. https://doi.org/10.18632/aging.103541
Dinami R, Petti E, Porru M, Rizzo A, Ganci F, Sacconi A, Ostano P, Chiorino G, Trusolino L, Blandino G et al (2022) TRF2 cooperates with CTCF for controlling the oncomiR-193b-3p in colorectal cancer. Cancer Lett 533:215607. https://doi.org/10.1016/j.canlet.2022.215607
Ohira T, Naohiro S, Nakayama Y, Osaki M, Okada F, Oshimura M, Kugoh H (2015) miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells. Sci Rep 5:8201. https://doi.org/10.1038/srep08201
Farhana L, Dawson MI, Fontana JA (2015) Down regulation of miR-202 modulates Mxd1 and Sin3A repressor complexes to induce apoptosis of pancreatic cancer cells. Cancer Biol Ther 16(1):115–124. https://doi.org/10.4161/15384047.2014.987070
Nakamura M, Hayashi M, Konishi H, Nunode M, Ashihara K, Sasaki H, Terai Y, Ohmichi M (2020) MicroRNA-22 enhances radiosensitivity in cervical cancer cell lines via direct inhibition of c-Myc binding protein, and the subsequent reduction in hTERT expression. Oncol Lett 19(3):2213–2222. https://doi.org/10.3892/ol.2020.11344
Li M, Chen SM, Chen C, Zhang ZX, Dai MY, Zhang LB, Wang SB, Dai Q, Tao ZZ (2015) microRNA-299-3p inhibits laryngeal cancer cell growth by targeting human telomerase reverse transcriptase mRNA. Mol Med Rep 11(6):4645–4649. https://doi.org/10.3892/mmr.2015.3287
Bai L, Wang H, Wang AH, Zhang LY, Bai J (2017) MicroRNA-532 and microRNA-3064 inhibit cell proliferation and invasion by acting as direct regulators of human telomerase reverse transcriptase in ovarian cancer. PLoS ONE 12(3):e0173912. https://doi.org/10.1371/journal.pone.0173912
Jung HM, Phillips BL, Chan EK (2014) miR-375 activates p21 and suppresses telomerase activity by coordinately regulating HPV E6/E7, E6AP, CIP2A, and 14–3-3ζ. Mol Cancer 13:80. https://doi.org/10.1186/1476-4598-13-80
Cimino-Reale G, Gandellini P, Santambrogio F, Recagni M, Zaffaroni N, Folini M (2017) miR-380-5p-mediated repression of TEP1 and TSPYL5 interferes with telomerase activity and favours the emergence of an “ALT-like” phenotype in diffuse malignant peritoneal mesothelioma cells. J Hematol Oncol 10(1):140. https://doi.org/10.1186/s13045-017-0510-3
Vinchure OS, Whittemore K, Kushwah D, Blasco MA, Kulshreshtha R (2021) miR-490 suppresses telomere maintenance program and associated hallmarks in glioblastoma. Cell Mol Life Sci 78(5):2299–2314. https://doi.org/10.1007/s00018-020-03644-2
Yan T, Ooi WF, Qamra A, Cheung A, Ma D, Sundaram GM, Xu C, Xing M, Poon L, Wang J et al (2018) HoxC5 and miR-615-3p target newly evolved genomic regions to repress hTERT and inhibit tumorigenesis. Nat Commun 9(1):100. https://doi.org/10.1038/s41467-017-02601-1
Kaul Z, Cheung CTY, Bhargava P, Sari AN, Yu Y, Huifu H, Bid H, Henson JD, Groden J, Reddel RR (2021) Functional characterization of miR-708 microRNA in telomerase positive and negative human cancer cells. Sci Rep 11(1):17052. https://doi.org/10.1038/s41598-021-96096-y
Wen N, Wang Y, Wen L, Zhao SH, Ai ZH, Wang Y, Wu B, Lu HX, Yang H, Liu WC et al (2014) Overexpression of FOXM1 predicts poor prognosis and promotes cancer cell proliferation, migration and invasion in epithelial ovarian cancer. J Transl Med 12:134. https://doi.org/10.1186/1479-5876-12-134
Pan H, Zhu Y, Wei W, Shao S, Rui X (2018) Transcription factor FoxM1 is the downstream target of c-Myc and contributes to the development of prostate cancer. World J Surg Oncol 16(1):59. https://doi.org/10.1186/s12957-018-1352-3
Katzenellenbogen B, Guillen VS, Katzenellenbogen JA (2023) Targeting the oncogenic transcription factor FOXM1 to improve outcomes in all subtypes of breast cancer. Breast Cancer Res 25(1):76. https://doi.org/10.1186/s13058-023-01675-8
Zhao J, Xu Y (2023) PITX1 plays essential functions in cancer. Front Oncol 13:1253238. https://doi.org/10.3389/fonc.2023.1253238
Gehring S, Rottman S, Menkel AR, Mertsching J, Krippner-Heidenreich A, Lücher B (2000) Inhibition of proliferation and apoptosis by the transcriptional repressor Mad1. Repression of Fas-induced caspase-8 activation. J Biol Chem 275(14):10413–10420. https://doi.org/10.1074/jbc.275.14.10413
Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, Eng H, Nair MG, Makvandi P, Geoerger B et al (2023) PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer 22(1):138. https://doi.org/10.1186/s12943-023-01827-6
Hemmings BA, Restuccia DF (2012) PI3K-PKB/Akt Pathway. Cold Spring Harb Perspect Biol 4(9):a011189. https://doi.org/10.1101/cshperspect.a011189
Méndez-Pertuz M, Martínez P, Blanco-Aparicio C, Gómez-Casero E, Belen García A, Martínez-Torrecuadrada J, Palafox M, Cortés J, Serra V, Pastor J et al (2017) Modulation of telomere protection by the PI3K/AKT pathway. Nat Commun 8(1):1278. https://doi.org/10.1038/s41467-017-01329-2
Tolomeo M, Cascio A (2021) The multifaced role of STAT3 in cancer and its implication for anticancer therapy. Int J Mol Sci 22(2):603. https://doi.org/10.3390/ijms22020603
Sakamuro D, Prendergast GC (1999) New Myc-interacting proteins: a second Myc network emerges. Oncogene 18(19):2942–2954. https://doi.org/10.1038/sj.onc.1202725
Li H (2022) Physiologic and pathophysiologic roles of AKAP12. Sci Prog 05(3):368504221109212. https://doi.org/10.1177/00368504221109212
Garus A, Autexier C (2021) Dyskerin: an essential pseudouridine synthase with multifaceted roles in ribosome biogenesis, splicing, and telomere maintenance. RNA 27(12):1441–1458. https://doi.org/10.1261/rna.078953.121
Kim YM, Watanabe T, Allen PB, Kim YM, Lee SJ, Greengard P, Nairn AC, Kwon YG (2003) PNUTS, a protein phosphatase 1 (PP1) nuclear targeting subunit. Characterization of its PP1- and RNA-binding domains and regulation by phosphorylation. J Biol Chem 278(16):13819–13828. https://doi.org/10.1074/jbc.M209621200
Okamoto K, Iwano T, Tachibana M, Shinkai Y (2008) Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J Biol Chem 283(35):23981–23988. https://doi.org/10.1074/jbc.M802395200
van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92(3):401–413. https://doi.org/10.1016/S0092-8674(00)80932-0
Holwerda SJ, de Laat W (2013) CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos Trans R Soc Lond B Biol Sci 368(1620):20120369. https://doi.org/10.1098/rstb.2012.0369
Chen LY, Redon S, Lingner J (2012) The human CST complex is a terminator of telomerase activity. Nature 488(7412):540–544. https://doi.org/10.1038/nature11269
Lim CJ, Cech TR (2021) Publisher Correction: Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat Rev Mol Cell Biol 22(4):299. https://doi.org/10.1038/s41580-021-00353-x
Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8(1):37–45. https://doi.org/10.1038/ncb1337
Oh J, Symington LS (2018) Role of the Mre11 Complex in Preserving Genome Integrity. Genes 9(12):589. https://doi.org/10.3390/genes9120589
Rosen EM (2013) BRCA1 in the DNA damage response and at telomeres. Front Genet 4:85. https://doi.org/10.3389/fgene.2013.00085
Acknowledgements
The authors thank the Systematic Review group of the BIOTEC program, Faculdades Pequeno Príncipe, for the support of this study. L.R.C. is recipient of a productivity research grant by Conselho Nacional de Desenvolvimento Científico e Tecnológico-Brasil (CNPQ), (PQ: 309501/2021-0).
Funding
No specific funding was utilized for this study.
Author information
Authors and Affiliations
Contributions
Conceptualization, SMB, ENS, RM, and LRC.; Methodology, SMB, ENS, and RM.; Formal Analysis, SMB, ENS, RM, and MOR; Writing-Original Draft Preparation, SMB, ENS, and RM; Image Development, LMO, and MVT; Writing-Review & Editing, ASF, AC, DR, and LRC; Supervision and Project Administration, LRC. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
N/A.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bortoletto, S., Nunes-Souza, E., Marchi, R. et al. MicroRNAs role in telomere length maintenance and telomerase activity in tumor cells. J Mol Med 102, 1089–1100 (2024). https://doi.org/10.1007/s00109-024-02467-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-024-02467-z