Abstract
Trans-differentiation, or activation, of hepatic stellate cells (HSCs) is a hallmark event in liver fibrosis although the underlying mechanism is not fully appreciated. Serum response factor (SRF) is a pleiotropic sequence-specific transcription factor with a ubiquitous expression pattern. In the present study, we investigated the effect of HSC-specific ablation of SRF on liver fibrosis in vivo and the underlying mechanism. We report that SRF bound to the promoter regions of pro-fibrogenic genes, including collagen type I (Col1a1/Col1a2) and alpha smooth muscle actin (Acta2), with greater affinity in activated HSCs compared to quiescent HSCs. Ablation of SRF in HSCs in vitro downregulated the expression of fibrogenic genes by dampening the accumulation of active histone marks. SRF also interacted with MRTF-A, a well-documented co-factor involved in liver fibrosis, on the pro-fibrogenic gene promoters during HSC activation. In addition, SRF directly regulated MRTF-A transcription in activated HSCs. More importantly, HSC conditional SRF knockout (CKO) mice developed a less robust pro-fibrogenic response in the liver in response to CCl4 injection and BDL compared to wild-type littermates. In conclusion, our data demonstrate that SRF may play an essential role in HSC activation and liver fibrosis.
Key messages
• SRF deficiency decelerates activation of hepatic stellate cells (HSCs) in vitro.
• SRF epigenetically activates pro-fibrogenic transcription to promote HSC maturation.
• SRF interacts with MRTF-A and contributes to MRTF-A transcription.
• Conditional SRF deletion in HSCs attenuates BDL-induced liver fibrosis in mice.
• Conditional SRF ablation in HSCs attenuates CCl4-induced liver fibrosis in mice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fibrogenesis is an integral part of the wound healing process and plays key roles in host defense [1]. A myriad of cells and humoral factors participate in this process, which may result in either restoration of organ function or irreversible disruption of organ structure and organ failure. During fibrogenesis, the extracellular matrix (ECM) undergoes dynamic remodeling. Regardless of the etiologies, myofibroblasts are the primary source of ECM production and secretion in fibrotic organs [2]. The origins of myofibroblast have stirred much debate and remain controversial despite the development and application of sophisticated lineage-tracing techniques [3].
Liver fibrosis is a common pathophysiological event following exposure of the liver to a range of stimuli, including pathogens, toxins, nutrients, and medications [4]. The severity of liver fibrosis correlates with the prognosis of end-stage liver diseases, for which effective therapeutic solutions are still lacking. In the liver, activated hepatic stellate cells (HSCs) are considered the major reservoir of myofibroblasts [5]. Quiescent HSCs function as a storage site for vitamins and lipids; once stimulated by a pro-fibrogenic cue, HSCs trans-differentiate into myofibroblasts acquiring dramatically enhanced ability to synthesize ECM proteins, to proliferate and migrate, and to contract [6]. HSC activation is paralleled by a shift in the transcriptome, which is programmed by an array of transcriptional factors.
Serum response factor (SRF) is a pleiotropic transcription factor involved in the regulation of a wide range of pathophysiological processes [7]. SRF regulates transcription by recognizing and binding to the conserved CArG box located on its target promoters [8]. The ability of SRF to regulate transcription relies on its interaction with a group of co-factors that include myocardin, myocardin-related transcription factor A (MRTF-A), and MRTF-B [9]. Mounting evidence points to a role for SRF in tissue fibrogenesis [10]. Formulations of SRF inhibitors or SRF siRNAs have been shown to prevent conjunctival fibrosis [11,12,13] and lung fibrosis [14]. SRF protein levels correlate with HSC activation in vitro [15]. Of interest, Zheng et al. have recently shown that the long non-coding RNA HOTTIP can act as a competing endogenous RNA (ceRNA) to sequester miR-150, thereby enhancing the expression of SRF, which in turn promotes HSC activation [16]. You et al. have shown that SRF knockdown significantly antagonizes the miR-125b-induced α-SMA expression [17]. In addition, several independent investigations have implicated SRF in liver fibrosis in vivo [17, 18]. However, there is no direct evidence that links HSC-specific SRF deficiency to (attenuated) liver fibrosis. Here we report that SRF programs HSC activation by recruiting MRTF-A and by directly activating MRTF-A transcription. Importantly, HSC-specific SRF ablation attenuates liver fibrosis in vivo. Therefore, our data suggest that SRF may play an essential role in HSC activation and liver fibrosis.
Materials and methods
Animal studies
All the animal protocols were reviewed and approved by the intramural Ethics Committee on Humane Treatment of Experimental Animals. The Srfflox/flox strain [19] was crossbred with the GFAP-Cre strain [20, 21] to generate HSC-specific SRF knockout (CKO) mice. To induce liver fibrosis, 6–8-week-old male mice were subjected to bile duct ligation (BDL) or the sham procedure and sacrificed 2 weeks after surgery as previously described [22]. Alternatively, the mice were injected peritoneally with CCl4 (1.0 mL/kg body weight as 50%, vol/vol) or corn oil weekly for 4 weeks.
Cell isolation, viral infection, and transient transfection
Primary hepatic stellate cells were isolated and maintained as previously described [23]. The cells were infected with adenovirus carrying GFP or Cre (Biowit, China) and harvested 2 days after infection. Small interfering RNA targeting SRF (GAUGGAGUUCAUCGACAACAA) was transfected with Lipofectamine RNAiMax (Thermo) per vendor’s recommendation. Immortalized rat hepatic stellate cells (HSC-T6) were maintained in DMEM supplemented with 10% FBS. SRF expression constructs [24] and human MRTF-A promoter-luciferase construct [25] have been previously described. Transient transfection was performed with Lipofectamine 2000. Cells were harvested 48 h after transfection and reporter activity was measured using a luciferase reporter assay system (Promega) as previously described [26]. Briefly, cells were plated in 12-well culture dishes (~ 60,000 cells/well). The next day, 0.1 μg of reporter construct and 0.1–0.3 μg of effector construct (SRF WT or SRF DN) were transfected into each well. DNA content was normalized by the addition of an empty vector (pcDNA3). For monitoring transfection efficiency and for normalizing luciferase activity, 0.02 μg of GFP construct was transfected into each well.
Protein extraction, immunoprecipitation, and Western blot
Whole cell lysates were obtained by re-suspending cell pellets in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100) with freshly added EDTA-free protease inhibitor tablet (Roche) as previously described [27]. Western blot analyses were performed with anti-SRF (sc-13029, Santa Cruz), anti-collagen type I (600-403-103, Rockland), anti-β-actin (A1978, Sigma), anti-α-SMA (ab5694, Abcam), and anti-MRTF-A (sc-32,909, Santa Cruz) antibodies. All experiments were repeated at least three times.
RNA isolation and real-time PCR
RNA was extracted with the RNeasy RNA isolation kit (Qiagen). Reverse transcriptase reactions were performed as previously described using a SuperScript First-strand Synthesis System (Invitrogen) [28]. Data were normalized with 18S rRNA as an internal control according to manufacturer’s protocol and expressed as fold change over the control group. All experiments were repeated at least three times.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described before [29,30,31]. Briefly, chromatin in control and treated cells were cross-linked with 1% formaldehyde. Cells were incubated in lysis buffer (150 mM NaCl, 25 mM Tris pH 7.5, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholate) supplemented with protease inhibitor tablet and PMSF. DNA was fragmented into ∼ 500 bp pieces using a Branson 250 sonicator. Aliquots of lysates containing 200 μg of protein were used for each immunoprecipitation reaction with anti-acetyl histone H3 (06-599, Millipore), anti-acetyl histone H4 (06-866, Millipore), anti-trimethyl H3K4 (07-473, Millipore), anti-p300 (sc-585, Santa Cruz), anti-ASH2 (A300-489A, Bethyl Laboratories), anti-WDR5 (A302-429A, Bethyl Laboratories), anti-SRF (sc-13,029, Santa Cruz), and anti-MRTF-A (sc-32,909, Santa Cruz) antibodies. Precipitated genomic DNA was amplified by real-time PCR with the following primers: for Col1a1, 5′-ATTTGAAGTCCCAGAAAG-3′ and 5′-AGAAACTCCCGTCTGCTC-3′; for Col1a2, 5′-CTTCGTGCATGACTTCAGCTTT-3′ and 5′-CGTCCTTTAGCATGGCAAGAC-3′; for Acta2, 5′-CCTGTTTCGGGAGCAGAA-3′ and 5′-GGTTATATAGCCCCCTGG-3′; for Col3a1, 5′-GACTCTGGCAAAACTCAAAGTATCA-3′ and 5′-TAGGAATGTGCTTTGTGATAGCCT-3′; for Lox, 5′-ACGTTTCCAATCACATTACG-3′ and 5′-ACGGTCCTCCTCTCCCCTTT-3′; for Gapdh, 5′-ATCACTGCCACCCAGAAGACTGTGGA-3′ and 5′-CTCATACCAGGAAATGAGCTTGACAAA-3′. All experiments were repeated at least three times.
Histology
Histological analyses were performed essentially as described before [32,33,34]. Briefly, paraffin sections were stained with picrosirius red (Sigma) or Masson’s trichrome (Sigma) according to standard procedures. Pictures were taken using an Olympus IX-70 microscope.
Statistical analysis
One-way ANOVA with post hoc Scheffe analyses were performed using an SPSS package. Unless otherwise specified, p values smaller than .05 were considered statistically significant (*).
Results
SRF regulates pro-fibrogenic transcription in HSCs
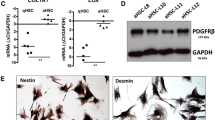
We first examined whether SRF depletion might dampen HSC activation in vitro. We isolated primary HSCs from Srff/f mice and deleted SRF by infecting these cells with adenovirus carrying Cre enzyme. As shown in Fig. S1, when the cells were harvested at day 8, they typically had a myofibroblast-like morphology and expressed large amount of α-SMA indicative of successful trans-differentiation. Compared to HSCs infected with GFP adenovirus, Cre adenovirus significantly downregulated SRF levels. Consequently, expression of pro-fibrogenic genes, including collagen type I (Col1a1/Col1a2), collagen type III (Col3a1), and α-SMA (Acta2), was decreased at both mRNA (Fig. 1a) and protein (Fig. 1b) levels. We also tried an alternative strategy by depleting SRF with siRNA. Two separate pairs of siRNAs comparably decreased SRF expression. Concomitantly, SRF siRNAs repressed the expression of pro-fibrogenic genes (Fig. 1c, d).
SRF regulates pro-fibrogenic transcription in HSCs. a, b Primary hepatic stellate cells were isolated from Srff/f mice and allowed to undergo spontaneous activation. At day 5, the cells were infected with adenovirus carrying GFP or Cre. Expression of pro-fibrogenic gene expression was measured by qPCR (a) and Western (b). c, d Primary hepatic stellate cells were isolated from C57/BL6 mice and allowed to undergo spontaneous activation. At day 5, the cells were transfected with siRNA targeting SRF or scrambled siRNA (SCR). Expression of pro-fibrogenic gene expression was measured by qPCR (c) and Western (d). e, f Primary hepatic stellate cells were isolated from WT and SRF CKO mice and allowed to undergo spontaneous activation for 7 days. Expression of pro-fibrogenic gene expression was measured by qPCR (e) and Western (f). All experiments were repeated three times
We also crossed the SRF-Flox mouse strain [19] with a GFAP-Cre strain [20, 21] to specifically delete SRF in HSCs. Primary HSCs were isolated from SRF conditional knockout (CKO) mice or wild-type (WT) mice and allowed to undergo spontaneous activation. As shown in Fig. 1e, f, expression of pro-fibrogenic genes were significantly downregulated in HSCs isolated from CKO mice compared to those from WT mice.
SRF recruits MRTF-A to activate pro-fibrogenic transcription during HSC activation
We then assessed the activity of SRF during HSC activation by examining its recruitment to pro-fibrogenic gene promoters. ChIP assays showed that SRF did not bind to the promoter regions of Col1a1, Col1a2, Col3a1, and Acta2, all of which contain at least one verified CArG box [23, 35,36,37], with significant affinity relative to the IgG control during the initial phase of HSC activation (Fig. 2a). At day 4 after HSC activation, SRF started to occupy the pro-fibrogenic gene promoters, and by day 8, SRF bound to the promoters with even higher affinity. By comparison, SRF did not bind to the Lox promoter either before or after HSC activation (Fig. 2a).
SRF recruits MRTF-A to activate pro-fibrogenic transcription during HSC activation. a Primary hepatic stellate cells were isolated from C57/BL6 mice and allowed to undergo spontaneous activation. The cells were harvested at indicated time points and ChIP assays were performed with anti-SRF or IgG. b Primary hepatic stellate cells were isolated from C57/BL6 mice and allowed to undergo spontaneous activation. The cells were harvested at indicated time points and Re-ChIP assays were performed with indicated antibodies. c Primary hepatic stellate cells were isolated from C57/BL6 mice and allowed to undergo spontaneous activation. At day 5, the cells were transfected with siRNA targeting SRF or scrambled siRNA (SCR). ChIP assays were performed with anti-MRTF-A. All experiments were repeated three times
Myocardin-related transcription factor A (MRTF-A) is a co-factor for SRF and a well-documented pro-fibrogenic protein [23, 35, 38,39,40]. We asked whether SRF might recruit MRTF-A to activate pro-fibrogenic transcription during HSC activation. Indeed, Re-ChIP assay showed that an SRF-MRTF-A complex could be detected on the pro-fibrogenic promoters in activated HSCs as opposed to the quiescent HSCs (Fig. 2b). In addition, SRF knockdown by siRNA significantly weakened the binding of MRTF-A to target promoters (Fig. 2c). Combined, these data suggest that SRF may play a role in pro-fibrogenic transcription by recruiting MRTF-A during HSC activation.
SRF regulates pro-fibrogenic transcription by recruiting epigenetic co-factors
We have previously shown that MRTF-A recruits histone-modifying enzymes to activate the transcription of pro-fibrogenic genes, thereby promoting HSC maturation [23]. We asked whether SRF deficiency would cripple the interaction between the histone-modifying enzymes with pro-fibrogenic gene promoters. Indeed, SRF depletion via Cre adenovirus infection significantly downregulated the enrichment of acetylated H3 (Fig. 3a) and H4 (Fig. 3b) surrounding the pro-fibrogenic gene promoters. In accordance, occupancies of p300, a key acetyltransferase responsible for H3 and H4 acetylation, were reduced by SRF deletion (Fig. 3c). In addition, we also found that SRF deletion attenuated the accumulation of trimethylated H3K4 surrounding the pro-fibrogenic gene promoters (Fig. 3d). H3K4 trimethylation is catalyzed by the COMPASS complex in mammals [41]. Congruent with decreased trimethyl H3K4 levels, SRF deletion also weakened the binding of ASH2 (Fig. 3e) and WDR5 (Fig. 3f), two core components of COMPASS, to the pro-fibrogenic gene promoters. Of note, SRF deletion did not alter the expression levels of these histone-modifying proteins (Fig. 3g).
SRF regulates pro-fibrogenic transcription by recruiting epigenetic co-factors. a–g Primary hepatic stellate cells were isolated from Srff/f mice and allowed to undergo spontaneous activation. At day 5, the cells were infected with adenovirus carrying GFP or Cre. ChIP assays were performed with anti-acetylated histone H3 (a), anti-acetyl histone H4 (b), anti-p300 (c), anti-trimethylated H3K4 (d), anti-ASH2 (e), and anti-WDR5 (f). g Protein expression was examined by Western. All experiments were repeated three times
Similarly, SRF silencing by siRNA dampened the deposition of acetyl H3 (Fig.S2A), acetyl H4 (Fig.S2B), and trimethyl H3K4 (Fig.S2C) surrounding the pro-fibrogenic gene promoters. The decrease in histone modifications was probably due to the weakened recruitment of histone-modifying enzymes such as p300 (Fig.S2D), ASH2 (Fig.S2E), and WDR5 (Fig.S2F). Taken together, these data suggest that SRF might contribute to pro-fibrogenic transcription by recruiting epigenetic co-factors to influence locus-specific histone modifications.
SRF directly regulates MRTF-A transcription during HSC activation
Of interest, we found that SRF depletion by siRNA in HSCs led to a decrease in MRTF-A expression at both mRNA (Fig. 4a) and protein (Fig. 4b) levels. Similarly, Cre-mediated SRF ablation in HSCs also reduced MRTF-A expression (Fig. 4c, d), indicating that SRF might directly regulate MRTF-A transcription to promote HSC activation.
SRF directly regulates MRTF-A transcription during HSC activation. a, b Primary hepatic stellate cells were isolated from C57/BL6 mice and allowed to undergo spontaneous activation. At day 5, the cells were transfected with siRNA targeting SRF or scrambled siRNA (SCR). MRTF-A expression was measured by qPCR and Western. c, d Primary hepatic stellate cells were isolated from Srff/f mice and allowed to undergo spontaneous activation. At day 5, the cells were infected with adenovirus carrying GFP or Cre. MRTF-A expression was measured by qPCR and Western. e An MRTF-A promoter luciferase construct was transfected into HSC-T6 cells with or without increasing doses of SRF expression construct. Luciferase activities were normalized by both GFP fluorescence and protein concentration. f An MRTF-A promoter luciferase construct was transfected into HSC-T6 cells with or without increasing doses of dominant negative SRF expression construct. Luciferase activities were normalized by both GFP fluorescence and protein concentration. All experiments were repeated three times
We next transfected into immortalized rat HSC cells (HSC-T6) an MRTF-A promoter-luciferase construct with or without an SRF expression construct (Fig. 4e). SRF over-expression dose-dependently activated the MRTF-A promoter activity. On the contrary, over-expression of a dominant negative (DN) SRF construct repressed the MRTF-A promoter activity in a dose-dependent manner (Fig. 4f). These data collectively suggest that SRF may regulate HSC phenotype by directly activating MRTF-A transcription.
SRF ablation attenuates liver fibrosis in mice
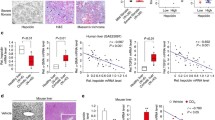
In order to probe the role of SRF in HSC activation and liver fibrosis in vivo, we exploited two classic animal models of liver fibrosis. Western blotting showed that SRF expression was decreased in primary HSCs, but not hepatocytes, isolated from CKO (Srff/f; GFAP-Cre) mice compared to those isolated from WT (Srff/f) mice (Fig. 5a). Both the CKO and the WT mice were subjected to weekly CCl4 injection for 4 weeks. CCl4 injection in WT and CKO mice inflicted comparable liver damages, as evidenced by plasma ALT (Fig. 5b) and AST (Fig. 5c) levels and by H&E staining of liver sections (Fig. 5d). Several lines of evidence indicate that liver fibrosis was attenuated in CKO mice compared to WT mice. First, qPCR measurements showed that expression levels of pro-fibrogenic genes were collectively downregulated in the CKO livers (Fig. 5e). Second, both picrosirius red staining and Masson’s trichrome staining showed that liver fibrosis was less severe in CKO mice than in WT mice (Fig. 5f). Finally, hepatic hydroxyproline quantification confirmed that fibrillar collagen levels were decreased in CKO livers (Fig. 5g).
SRF ablation attenuates CCl4-induced liver fibrosis in mice. a Primary hepatocytes and HSCs were isolated from WT and SRF CKO mice. SRF expression was examined by Western blotting. b–g WT and SRF CKO mice were injected with CCl4 to induce liver fibrosis as described in “Materials and methods.” b Plasma ALT levels. c Plasma AST levels. d H&E staining. e Hepatic expression of pro-fibrogenic genes was examined by qPCR. f Picrosirius red and Masson’s trichrome stainings. g Hepatic hydroxyproline levels. N = 8~9 mice for each group. Error bars represent SD (*p < 0.05, two-tailed t test)
Next, we exploited an alternative mouse model of liver fibrosis in which the mice were subjected to bile duct ligation (BDL) and sacrificed 2 weeks after the surgery. Both the WT mice and the CKO mice displayed similar levels of liver injury induced by the BDL procedure as gauged by plasma ALT (Fig. 6a) and AST (Fig. 6b) as well as hepatic necrotic areas identified by H&E staining (Fig. 6c). Quantitative PCR showed that SRF deficiency in HSCs reduced the expression of pro-fibrogenic genes (Fig. 6d). In accordance, picrosirius red and Masson’s trichrome stainings indicate that liver fibrosis was less widespread in CKO livers than in WT livers (Fig. 6e). Consistently, fewer fibrillar collagens were detected by hepatic hydroxyproline quantification (Fig. 6f). Together, these data support an essential for HSC-specific SRF in liver fibrosis.
SRF ablation attenuates BDL-induced liver fibrosis in mice. a–f WT and SRF CKO mice were subjected to BDL to induce liver fibrosis as described in “Materials and methods.” a Plasma ALT levels. b Plasma AST levels. c H&E staining. d Hepatic expression of pro-fibrogenic genes was examined by qPCR. eN = 6 mice for each group. Error bars represent SD (*p < 0.05, two-tailed t test). g A schematic model. In wild-type HSCs, SRF recruits MRTF-A and possibly other co-factors (CoF), which in turn may help bring various histone-modifying enzymes to remodel the chromatin surrounding and activate the transcription of the pro-fibrogenic gene promoters. SRF may also directly activate MRTF-A transcription and maintain MRTF-A levels in HSCs. In SRF null HSCs, transcription of both the pro-fibrogenic genes and MRTF-A is disrupted leading to impaired HSC activation and attenuation of liver fibrosis
Discussion
Recent investigations strongly argue for a role for SRF in liver fibrosis. Here we provide direct evidence that links SRF deficiency in hepatic stellate cells to attenuated liver fibrosis in mice. Herrmann et al. previously have shown that SRF expression is upregulated by TGF-β in activated HSCs [15]. Moreover, SRF levels are found to be higher in the livers of Long-Evans Cinnamon rats that develop hepatitis and hepatocellular carcinoma, both of which are preceded or followed by liver fibrosis [42]. Of note, ablation of SRF in HSCs did not appear to affect either hepatotoxic substance (CCl4)-induced liver injury or cholestatic liver injury (BDL). These observations contrast a previous report wherein hepatocyte-specific deletion of SRF leads to partial lethality and postnatal growth retardation owing to increased apoptosis of hepatocyte [43], indicating that SRF may play cell type-dependent, distinctive roles in liver injury and liver fibrosis. It alludes to one of main weaknesses of the present study. We focused on the role of HSC-specific SRF in liver fibrosis because HSCs are thought to be the predominant source of myofibroblasts in the liver. Other cell types, including endothelial cells [44] and portal fibroblast cells [45], also contribute to this process. In addition, the specificity of the Cre driver (GFAP) used in this study to delete SRF in HSCs has been called into question recently [5]. The potential effect of SRF deletion on HSC development cannot be ignored since SRF is absent the entire time even when HSCs are quiescent. Clearly, further studies are warranted to determine the role of SRF in liver fibrosis.
We and others have previously shown that MRTF-A is a key regulator of liver fibrosis [23, 38, 46]. Here we show that SRF recruited MRTF-A to the pro-fibrogenic gene promoters. Moreover, SRF deficiencies are synonymous with a repressed chromatin structure surrounding the promoter regions of the genes involved in fibrogenesis. The ability to engage the epigenetic machinery is considered a paradigm in SRF-dependent regulation of smooth muscle phenotypic modulation [47]. More recently, Rosen and colleagues have performed extensive ChIP-seq analysis to correlate specific chromatin structure with activation of genes key to adipogenesis, which consequently leads to the identification of SRF as a novel transcription factor that bridge epigenetic factors to locus-specific gene transcription [48]. It is possible that SRF may regulate HSC trans-differentiation by the same virtue. Alternatively, SRF may rely on MRTF-A to recruit various histone-modifying enzymes to activate transcription because MRTF-A has been found to make extensive dialogs with the epigenetic machinery [25, 49,50,51,52,53]. We note that there is a caveat regarding this model because SRF can directly regulate MRTF-A transcription (Fig. 3). The observation that MRTF-A recruitment was impaired in the absence of SRF might be due to decreased MRTF-A expression (Fig. 2). In the same vein, attenuation of histone-modifying enzymes on the SRF target promoters in SRF-deficient cells may also be attributed to lower MRTF-A levels (Fig. 4). We propose that SRF regulates pro-fibrogenic transcription via several inter-dependent mechanisms, by recruiting co-factors (e.g., MRTF-A), by directly controlling the availability (expression) of co-factors (e.g., MRTF-A), and by engaging histone-modifying enzymes. In addition to MRTF-A, other SRF co-factors have been found to bridge SRF to the epigenetic machinery. BRG1, for instance, has been shown to act as a co-factor for SRF-dependent transcription of smooth muscle-specific genes by forming a complex with both SRF and MRTF-A [51]. We have reported previously that BRG1 can interact with and recruit several different histone-modifying enzymes to regulate transcription [54,55,56,57,58]. SRF is absolutely required for the integrity of this complex; without SRF, the stability of these binding factors are affected so that pro-fibrogenic transcription is essentially shut down (Fig. 6g). The lingering issues regarding this model as highlighted above must be resolved to clarify the epigenetic mechanism whereby SRF regulates liver fibrosis.
An interesting finding in the present study is that SRF may directly regulate MRTF-A transcription in activated HSCs. Although the activation of MRTF-A is thought to be determined predominantly by its sub-cellular localization, MRTF-A expression levels are sensitive to various cues attributable to both transcriptional and post-transcriptional regulation. We have previously shown that during HSC activation, MRTF-A proteins are upregulated via a post-transcriptional mechanism whereby the histone deacetylase HDAC4 represses miR-206 to stabilize MRTF-A messages [46]. Our data suggest that multiple mechanisms contribute to the maintenance of MRTF-A levels in HSCs to sustain fibrogenesis. It remains to be determined whether SRF may directly bind to the MRTF-A promoter and activate its transcription.
In summary, we provide evidence to directly link HSC-specific SRF to liver fibrosis both in vitro and in vivo. Future studies exploiting additional mouse models and transcriptomic/epigenomic techniques will hopefully solidify the role for SRF as a key regulator of fibrogenesis and pave the way for targeting SRF in the intervention of liver fibrosis.
Abbreviations
- HSC:
-
Hepatic stellate cell
- SRF:
-
Serum response factor
- BDL:
-
Bile duct ligation
- MRTF-A:
-
Myocardin-related transcription factor A
- CKO:
-
Conditional knockout
- ChIP:
-
Chromatin immunoprecipitation
- α-SMA:
-
Alpha smooth muscle actin
References
Zhang X, Hu M, Lyu X, Li C, Thannickal VJ, Sanders YY (2017) DNA methylation regulated gene expression in organ fibrosis. Biochim Biophys Acta 1863:2389–2397
Bochaton-Piallat ML, Gabbiani G, Hinz B (2016) The myofibroblast in wound healing and fibrosis: answered and unanswered questions. F1000Research 5. https://doi.org/10.12688/f1000research.8190.1
Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, Lin JSC, Aronow BJ, Tallquist MD et al (2016) Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun 7:12260
Seki E, Schwabe RF (2015) Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology 61:1066–1079
Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF (2013) Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun 4:2823
Friedman SL (2010) Evolving challenges in hepatic fibrosis. Nat Rev Gastroenterol Hepatol 7:425–436
Miano JM (2010) Role of serum response factor in the pathogenesis of disease. Lab Investig 90:1274–1284
Miano JM (2003) Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol 35:577–593
Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN (2002) Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci U S A 99:14855–14860
Small EM (2012) The actin-MRTF-SRF gene regulatory axis and myofibroblast differentiation. J Cardiovasc Transl Res 5:794–804
Tagalakis AD, Madaan S, Larsen SD, Neubig RR, Khaw PT, Rodrigues I, Goyal S, Lim KS, Yu-Wai-Man C (2018) In vitro and in vivo delivery of a sustained release nanocarrier-based formulation of an MRTF/SRF inhibitor in conjunctival fibrosis. J Nanobiotechnol 16:97
Fernando O, Tagalakis AD, Awwad S, Brocchini S, Khaw PT, Hart SL, Yu-Wai-Man C (2018) Development of targeted siRNA nanocomplexes to prevent fibrosis in experimental glaucoma filtration surgery. Mol Ther 26:2812–2822
Yu-Wai-Man C, Tagalakis AD, Manunta MD, Hart SL, Khaw PT (2016) Receptor-targeted liposome-peptide-siRNA nanoparticles represent an efficient delivery system for MRTF silencing in conjunctival fibrosis. Sci Rep 6:21881
Sisson TH, Ajayi IO, Subbotina N, Dodi AE, Rodansky ES, Chibucos LN, Kim KK, Keshamouni VG, White ES, Zhou Y, Higgins PDR, Larsen SD, Neubig RR, Horowitz JC (2015) Inhibition of myocardin-related transcription factor/serum response factor signaling decreases lung fibrosis and promotes mesenchymal cell apoptosis. Am J Pathol 185:969–986
Jiang M, Ku WY, Fu J, Offermanns S, Hsu W, Que J (2013) Gpr177 regulates pulmonary vasculature development. Development 140:3589–3594
Zheng J, Mao Y, Dong P, Huang Z, Yu F (2019) Long noncoding RNA HOTTIP mediates SRF expression through sponging miR-150 in hepatic stellate cells. J Cell Mol Med 23:1572–1580
You K, Li SY, Gong J, Fang JH, Zhang C, Zhang M, Yuan Y, Yang J, Zhuang SM (2018) MicroRNA-125b promotes hepatic stellate cell activation and liver fibrosis by activating RhoA signaling. Mol Ther Nucleic Acids 12:57–66
Takata A, Otsuka M, Kishikawa T, Yamagami M, Ishibashi R, Sekiba K, Suzuki T, Ohno M, Yamashita Y, Abe T et al (2017) RASAL1 is a potent regulator of hepatic stellate cell activity and liver fibrosis. Oncotarget 8:64840–64852
Guo B, Lyu Q, Slivano OJ, Dirkx R, Christie CK, Czyzyk J, Hezel AF, Gharavi AG, Small EM, Miano JM (2018) Serum response factor is essential for maintenance of podocyte structure and function. J Am Soc Nephrol 29:416–422
Li M, Hong W, Hao C, Li L, Wu D, Shen A, Lu J, Zheng Y, Li P, Xu Y (2018) SIRT1 antagonizes liver fibrosis by blocking hepatic stellate cell activation in mice. FASEB J 32:500–511
Li M, Hong W, Hao C, Li L, Xu H, Li P, Xu Y (2017) Hepatic stellate cell-specific deletion of SIRT1 exacerbates liver fibrosis in mice. Biochim Biophys Acta 1863:3202–3211
Zhou B, Zeng S, Li L, Fan Z, Tian W, Li M, Xu H, Wu X, Fang M, Xu Y (2016) Angiogenic factor with G patch and FHA domains 1 (Aggf1) regulates liver fibrosis by modulating TGF-beta signaling. Biochim Biophys Acta 1862:1203–1213
Tian W, Hao C, Fan Z, Weng X, Qin H, Wu X, Fang M, Chen Q, Shen A, Xu Y (2015) Myocardin related transcription factor A programs epigenetic activation of hepatic stellate cells. J Hepatol 62:165–174
Yang Y, Chen D, Yuan Z, Fang F, Cheng X, Xia J, Fang M, Xu Y, Gao Y (2013) Megakaryocytic leukemia 1 (MKL1) ties the epigenetic machinery to hypoxia-induced transactivation of endothelin-1. Nucleic Acids Res 41:6005–6017
Fang F, Yang Y, Yuan Z, Gao Y, Zhou J, Chen Q, Xu Y (2011) Myocardin-related transcription factor A mediates OxLDL-induced endothelial injury. Circ Res 108:797–807
Li Z, Chen B, Dong W, Xu W, Song M, Fang M, Guo J, Xu Y (2018) Epigenetic activation of PERP transcription by MKL1 contributes to ROS-induced apoptosis in skeletal muscle cells. Biochim Biophys Acta Gene Reg Mech 1861:905–915
Li Z, Chen B, Weng X, Yu L, Song M, Fang M, Guo J, Xu Y (2018) The histone methyltransferase SETD1A regulates thrombomodulin transcription in vascular endothelial cells. Biochim Biophys Acta Gene Reg Mech 1861:752–761
Zeng S, Wu X, Chen X, Xu H, Zhang T, Xu Y (2018) Hypermethylated in cancer 1 (HIC1) mediates high glucose induced ROS accumulation in renal tubular epithelial cells by epigenetically repressing SIRT1 transcription. Biochim Biophys Acta Gene Reg Mech 1861:917–927
Weng X, Zhang Y, Li Z, Yu L, Xu F, Fang M, Hou L, Ge J, Xu Y (2019) Class II transactivator (CIITA) mediates IFN-gamma induced eNOS repression by enlisting SUV39H1. Biochim Biophys Acta Gene Reg Mech 1862:163–172
Shao J, Weng X, Zhuo L, Yu L, Li Z, Shen K, Xu W, Fang M, Xu Y (2019) Angiotensin II induced CSF1 transcription is mediated by a crosstalk between different epigenetic factors in vascular endothelial cells. Biochim Biophys Acta Gene Reg Mech 1862:1–11
Li Z, Zhang X, Liu S, Zeng S, Yu L, Yang G, Guo J, Xu Y (2018) BRG1 regulates NOX gene transcription in endothelial cells and contributes to cardiac ischemia-reperfusion injury. Biochim Biophys Acta Mol basis Dis 1864:3477–3486
Li N, Kong M, Zeng S, Hao C, Li M, Li L, Xu Z, Zhu M, Xu Y (2019) Brahma related gene 1 (Brg1) contributes to liver regeneration by epigenetically activating the Wnt/beta-catenin pathway in mice. FASEB J 33:327–338
Li N, Li M, Hong W, Shao J, Xu H, Shimano H, Lu J, Xu Y (2018) Brg1 regulates pro-lipogenic transcription by modulating SREBP activity in hepatocytes. Biochim Biophys Acta Mol Basis Dis 1864:2881–2889
Li N, Kong M, Zeng S, Xu Z, Li M, Hong W, Chu X, Sun X, Zhu M, Xu Y (2018) The chromatin remodeling protein BRG1 regulates APAP-induced liver injury by modulating CYP3A11 transcription in hepatocyte. Biochim Biophys Acta Mol basis Dis 1864:3487–3495
Luchsinger LL, Patenaude CA, Smith BD, Layne MD (2011) Myocardin-related transcription factor-A complexes activate type I collagen expression in lung fibroblasts. J Biol Chem 286:44116–44125
Small EM, Thatcher JE, Sutherland LB, Kinoshita H, Gerard RD, Richardson JA, Dimaio JM, Sadek H, Kuwahara K, Olson EN (2010) Myocardin-related transcription factor-a controls myofibroblast activation and fibrosis in response to myocardial infarction. Circ Res 107:294–304
McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK (2006) Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest 116:36–48
Fan Z, Hao C, Li M, Dai X, Qin H, Li J, Xu H, Wu X, Zhang L, Fang M, Zhou B, Tian W, Xu Y (2015) MKL1 is an epigenetic modulator of TGF-beta induced fibrogenesis. Biochim Biophys Acta Gene Reg Mech 1849:1219–1228
Tian W, Fan Z, Li J, Hao C, Li M, Xu H, Wu X, Zhou B, Zhang L, Fang M, Xu Y (2016) Myocardin-related transcription factor A (MRTF-A) plays an essential role in hepatic stellate cell activation by epigenetically modulating TGF-beta signaling. Int J Biochem Cell Biol 71:35–43
Xu H, Wu X, Qin H, Tian W, Chen J, Sun L, Fang M, Xu Y (2015) Myocardin-related transcription factor a epigenetically regulates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol 26:1648–1660
Shilatifard A (2012) The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81:65–95
Noubade R, Wong K, Ota N, Rutz S, Eidenschenk C, Valdez PA, Ding J, Peng I, Sebrell A, Caplazi P, DeVoss J, Soriano RH, Sai T, Lu R, Modrusan Z, Hackney J, Ouyang W (2014) NRROS negatively regulates reactive oxygen species during host defence and autoimmunity. Nature 509:235–239
Yang Y, Yu X (2003) Regulation of apoptosis: the ubiquitous way. FASEB J 17:790–799
Ribera J, Pauta M, Melgar-Lesmes P, Cordoba B, Bosch A, Calvo M, Rodrigo-Torres D, Sancho-Bru P, Mira A, Jimenez W et al (2017) A small population of liver endothelial cells undergoes endothelial-to-mesenchymal transition in response to chronic liver injury. Am J Physiol Gastrointest Liver Physiol 313:G492–G504
Iwaisako K, Jiang C, Zhang M, Cong M, Moore-Morris TJ, Park TJ, Liu X, Xu J, Wang P, Paik YH, Meng F, Asagiri M, Murray LA, Hofmann AF, Iida T, Glass CK, Brenner DA, Kisseleva T (2014) Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A 111:E3297–E3305
Han X, Hao C, Li L, Li J, Fang M, Zheng Y, Lu J, Li P, Xu Y (2017) HDAC4 stimulates MRTF-A expression and drives fibrogenesis in hepatic stellate cells by targeting miR-206. Oncotarget 8:47586–47594
McDonald OG, Owens GK (2007) Programming smooth muscle plasticity with chromatin dynamics. Circ Res 100:1428–1441
Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED (2010) Comparative epigenomic analysis of murine and human adipogenesis. Cell 143:156–169
Hanna M, Liu H, Amir J, Sun Y, Morris SW, Siddiqui MA, Lau LF, Chaqour B (2009) Mechanical regulation of the proangiogenic factor CCN1/CYR61 gene requires the combined activities of MRTF-A and CREB-binding protein histone acetyltransferase. J Biol Chem 284:23125–23136
Lockman K, Taylor JM, Mack CP (2007) The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res 101:e115–e123
Zhang M, Fang H, Zhou J, Herring BP (2007) A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem 282:25708–25716
Yu L, Fang F, Dai X, Xu H, Qi X, Fang M, Xu Y (2017) MKL1 defines the H3K4Me3 landscape for NF-kappaB dependent inflammatory response. Sci Rep 7:191
Liu L, Wu X, Xu H, Yu L, Zhang X, Li L, Jin J, Zhang T, Xu Y (2018) Myocardin-related transcription factor A (MRTF-A) contributes to acute kidney injury by regulating macrophage ROS production. Biochim Biophys Acta Mol Basis Dis 1864:3109–3121
Liu L, Mao L, Wu X, Wu T, Liu W, Yang Y, Zhang T, Xu Y (2019) BRG1 regulates endothelial-derived IL-33 to promote ischemia-reperfusion induced renal injury and fibrosis in mice. Biochim Biophys Acta Mol Basis Dis 1865:2551–2561
Liu L, Hong W, Li M, Ren H, Wang J, Xu H, Shi X, Xu Y (2019) A cross talk between BRG1 and males absent on the first contributes to reactive oxygen species production in a mouse model of nonalcoholic steatohepatitis. Antioxid Redox Signal 30:1539–1552
Weng X, Yu L, Liang P, Li L, Dai X, Zhou B, Wu X, Xu H, Fang M, Chen Q, Xu Y (2015) A crosstalk between chromatin remodeling and histone H3K4 methyltransferase complexes in endothelial cells regulates angiotensin II-induced cardiac hypertrophy. J Mol Cell Cardiol 82:48–58
Zhang X, Liu S, Weng X, Wu T, Yu L, Xu Y, Guo J (2018) Brg1 trans-activates endothelium-derived colony stimulating factor to promote calcium chloride induced abdominal aortic aneurysm in mice. J Mol Cell Cardiol 125:6–17
Wang Y, Zhang L, Li Y, Chen L, Wang X, Guo W, Zhang X, Qin G, He SH, Zimmerman A, Liu Y, Kim IM, Weintraub NL, Tang Y (2015) Exosomes/microvesicles from induced pluripotent stem cells deliver cardioprotective miRNAs and prevent cardiomyocyte apoptosis in the ischemic myocardium. Int J Cardiol 192:61–69
Funding
This work was supported, in part, by grants from the National Natural Science Foundation of China (81725001, 81700554, 81770487), from the Nanjing Municipal Administration of Health and Human Services (YKK17061), from the Fundamental Research Funds for Central Universities (021414380323), and from the Haihan Province R&D Fund Key Project (ZDYF2018102).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
All the animal protocols were reviewed and approved by the intramural Ethics Committee on Humane Treatment of Experimental Animals.
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 558 kb)
Rights and permissions
About this article
Cite this article
Kong, M., Hong, W., Shao, Y. et al. Ablation of serum response factor in hepatic stellate cells attenuates liver fibrosis. J Mol Med 97, 1521–1533 (2019). https://doi.org/10.1007/s00109-019-01831-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-019-01831-8