Abstract
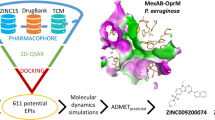
Pseudomonas aeruginosa is resistant to a wide range of antibiotics, thus making troublesome the infection treatment. Efflux systems are the main mechanisms involved; among these, MexAB-OprM is a tripartite efflux pump responsible for resistance to ciprofloxacin, aztreonam, gentamicin, tetracycline and tobramycin. In an attempt to contrast antibiotic efflux, databases of natural compounds were tested for their ability to bind MexB, the inner membrane channel, using a high-throughput virtual screening approach. The comparison of their common pharmacophoric features was the basis for inhibitor identification and selection process. In silico screening against the MexB protein was performed by Autodock/Vina and further refined using a minimization/focused docking protocol on the obtained complexes. The compounds showing the best docking and resulting potentially active at nanomolar concentration have been selected and used in combination with antibiotics usually exported by MexAB-OprM in antimicrobial in vitro synergy tests (checkerboard and time kill assays) against multidrug-resistant P. aeruginosa clinical isolates. The combinations morelloflavone and pregnan-20-one-derivative/ciprofloxacin showed a four-fold MIC decrease and 100-fold increase of the bacterial killing compared to the antibiotic alone. The two chosen hits were validated by ethidium bromide accumulation assay for their efflux inhibition potency. These compounds showed the ability to increase the accumulation of ethidium bromide inside the bacterial cells as evidenced by the increase of its fluorescence in the presence of the each of them. Finally, their toxicity has been preliminary tested through hemolysis assay. The observed good correlation between in silico docking and in vitro synergy tests, indicates these two compounds as promising drugs to be used in combination therapy against MDR and MexAB-OprM overexpressing P. aeruginosa.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antibiotic-resistant bacteria have increased in recent years and such resistance can compromise the efficacy of antimicrobial therapy (Taubes 2008). Multidrug-resistant (MDR) bacteria have been associated with infections characterized by a higher mortality than those caused by antibiotic-susceptible strains and nowadays MDR strains are cause of great concern. Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen that is frequently associated with life-threatening nosocomial infections, particularly in immune-compromised and cystic fibrosis (CF) patients. Intrinsically resistant to many drugs and readily developing high-level resistance during antimicrobial therapy, P. aeruginosa infections are often difficult to counteract. Efflux systems are among the major contributors to the emergence of MDR strains (Breidenstein et al. 2011; Ball et al. 2006; Ho-Fung Lau et al. 2014).
The physiological function of multidrug efflux pumps is to push solutes out of a cell, allowing the microorganisms to remove toxic substances, including antimicrobial agents, metabolites and quorum sensing signal molecules (Pearsons et al. 1999). Due to their ability to transport a wide range of antibiotics and to their wide diffusion, MDR pumps play a crucial role in the increasing prevalence of MDR bacterial strains (Lee et al. 2000). They occur as both single and multicomponent systems. The former extrude their substrates into the periplasmic space, whereas the latter crosses both the inner and the outer membrane and exports their substrates directly from the cytoplasm to the surrounding medium. The multicomponent systems include the multiple drug resistance MexAB-OprM pump of P. aeruginosa, which is closely related to AcrAB-TolC of E. coli, both belonging to the RND (resistance-nodulation-division) superfamily (Poole 2005).
They form tripartite complexes including outer and inner membrane channel and a wide periplasmic protein adaptor. Antibiotics are entrapped by MexB in the inner membrane, transferred to MexA (periplasmatic) and finally extruded by OprM, located in the outer membrane. Together these three proteins form a channel that allows the antibiotic to pass from the cytoplasm to the extracellular environment (Murakami et al. 2006; De Kievit et al. 2001).
MexAB-OprM is constitutionally expressed in Pseudomonas aeruginosa; however, its overexpression has been found in MDR clinical isolates, resulting from mutations in the regulator gene mex R (Nakashima et al. 2013; De Kievit et al. 2001).
Therefore, the MexAB-OprM pump can be considered an important target for the development of combination strategies using efflux-pump inhibitors (EPIs) in the treatment of high risk infections (Ohene-Agyei et al. 2014; Thai et al. 2015). The Rational drug design of targeting EPIs can develop successful multidrug resistance reversal agents (or chemosensitizers) that can be used, in clinical practice in combination with antibiotics, to restore antibiotic susceptibility.
A few EPIs have been already developed. Phenylalanine arginyl β-naphthylamide (PAβN; MC-207,110), a peptidomimetic compound exerting a competitive mechanism of inhibition, was the first described as effective for Pseudomonas strains overexpressing MexAB-OprM (Askoura et al. 2011; Lomovskaya et al. 2001), followed by Carbonyl cyanide m-chlorophenylhydrazone (CCCP), an energy-dependent EPI that de-energizes membranes so being less specific than PAbN (Nelson 2002). It can’t be considered a true EPI, due to its mechanism of action that reduce the proton motive force, thereby indirectly inhibiting the efflux mechanism of the RND pumps. However, both these compounds, as well as other derivatives, are not currently used in clinical practice due to their toxicity (Watkins et al. 2003; Askoura et al. 2011).
Many experimental approaches in academic and industrial research, such as high-throughput screening ventures and bioassay-guided determination, have yielded to a number of promising EPIs in different pathogenic systems. This experimental strategy is however very expensive and can have a high impact in drug development costs. To reach the goal efficiently and at minor costs, our objective was to identify potential lead compounds to be used as EPI in synergistic combinations with antibiotics by an in silico strategy. On the bases of the recently published promising results obtained using bioinformatics and modeling protocols (Thai et al. 2015), we developed a combined Virtual Screening/microbiological testing protocol. It included an in silico screening of data bases of natural compounds based on their ability to interact with MexB, followed by a focused molecular docking/molecular dynamics protocol aimed to elucidate their binding interaction and evaluate the corresponding binding energy. The compounds showing the best potential activity were then tested by in vitro microbiological assays for their synergistic activity when used in combination with the antibiotics usually extruded from the bacterial cell by MexAB-OprM. In addition, their interaction with bacterial efflux pumps has been validated by the ethidium bromide accumulation assay and their toxicity evaluated by in vitro hemolysis assays.
Materials and methods
Chemicals, bacterial strain and growth media
The natural compounds, pregnan-20-one derivative (ZINC08382438), morelloflavone (ZINC26187321) and N-{2-(1,3-benzodioxol-5yl)-1-((6-oxo-7,11-diazatricyclo(7.3.1.0~2,7~)trideca-2,4-dien-11-yl)carbonyl)vinyl}benzamide (ZINC08382391) were purchased from SPECs (www.specs.net). All antibiotics (ciprofloxacin, ceftazidime, meropenem, piperacillin and tobramycin) were from Sigma (Sigma Aldrich SRL, Milano, IT).
Twenty-five P.aeruginosa strains isolated from cystic fibrosis sputum samples were obtained from the Microbiology Lab, AO “Ospedali Riuniti”, Ancona Italy. All of them resulted MDR by routine antimicrobial-susceptibility assay. Bacterial strains were routinely grown on Luria-Bertani agar plates for 24 h at 37 °C and maintained at room temperature up to 1 week or stored at −80 °C as stock cultures, supplemented with 20% glycerol.
In silico virtual screening
Protein preparation
The MexB (PDB: 2V50) protein structure was retrieved from Protein Data Bank. Its crystal structure for in silico studies was prepared using CHIMERA (Pettersen et al. 2004).
Protein ionization was settled out considering a pH of 7.4, which corresponds to that of the experimental conditions, and the following physical parameters: salinity 0.15, internal dielectric 6, external dielectric 80 (Bashford and Karplus 1990; Gordon et al. 2005, Myers et al. 2006).
Before proceeding with further studies, the macromolecule was minimized following a tested protocol (Gabbianelli et al. 2015; Galeazzi et al. 2014; Scirè et al. 2013) (Suite 2011 Macromodel V9.1, Mohamadi et al. 1990) using AMBER force field (Cornell et al. 1995; Duan et al. 2003) and conjugate gradient (PRCG) algorithm (Gilbert and Nocedal 1992) up to a heavy-atom RMSD of 0.30 Å.
A Site map prediction for MexB protein was then accomplished using AcrB binding cavity as starting point and the subsequent virtual screening was carried out extending the grid calculations to the surrounding regions, in order to better evaluate other hidden putative bonding regions.
Receptor grid generation
Receptor Grid Generation (MGLTools Autodock/Vina AMBER force field) (Sanner 1999; Morris et al. 2009) was used to specify the grid. With the absence of an inhibitor-MexB co-crystal PDB structure of P.aeruginosa, AcrB/MC-207110 complex structure (PDB ID: 1T9Y) from E. coli was used to identify the binding site information for P.aeruginosa MexB. Since a structural correlation (RMSD of 1.4 Å) exists between MexB of P. aeruginosa and AcrB of E. coli integral membrane proteins, the MC-207110 binding regions of AcrB were mapped on MexB (Aparna et al. 2014). Two MC-207110 molecules were bound to AcrB structure at two different sites. Pair-wise sequence alignment was performed using CHIMERA (Pettersen et al. 2004) for AcrB and MexB protein sequences. The comparison of the binding sites of MC-207110 of AcrB with MexB structure was done and the common amino acid residues at the binding site of MC-207110 were selected and used for Grid generation. More in details:
Site 1 (MC-207110 molecule number 7001) corresponds to Ala385, Phe386, Gly387, Phe388, and Arg468 having hydrophobic contacts with MC-207110.
Site 2 (MC-207110 molecule number 7002) corresponds to the binding of MC-207110 with Phe664, Val716, Arg717, Pro718, Leu828, and Gly829 of AcrB protein.
Natural compound database
Information on plant natural compounds with known antimicrobial and anticancer bioactivity was obtained from ZINC and associated Database (www.zinc.org); our plan was to start from pure natural origin compounds to better find out hits with different structure and to this purpose, we firstly screened the SPECnet ZINC database (about 1500 compounds). Their structures were retrieved from Pubchem database and used as starting point for ligand preparation. Compounds were minimized by AMBER force field to reach the convergence of 0.05 Å (Cornell et al. 1995). Charges were previously obtained using AM1-BCC Hamiltonian (Pettersen et al. 2004).
HTVS of phytochemicals against MexB integral membrane protein (molecular docking)
All molecular docking experiments were performed by AutoDock Vina version 1.1.2 (Trott and Olson 2010). The virtual screening software VINA was used to screen the collections of natural compounds against the two MC-207110 binding sites of MexB. The two active sites of MC-207110 from the crystal structure 1T9Y was defined as a box of size 40 × 40 × 40 Å with the pose of MC-207110 in its center. All the PDB structures were converted to PDBQT and then all the 1498 structures of the SPECnet database were docked in the identified binding sites of the two MexB receptors using the standard parameters of AutoDock Vina. The virtual screening workflow offers selective filtration of ligands with increased strictness on the bases of their efficiency to interact with the binding cavity residues (docking score function and binding energy). Before performing the docking, computational protocol was validated by evaluating the reproducibility of re-docking the co-crystallized ligand-receptor (RMSD 0.910 Å) complex (1T9Y).
After the identification of the best scored poses in both sites, Autodock 4.2.1 (AD4) was used to refine the binding pose and relative energy (Morris et al. 1998; Chang et al. 2010). The number of AD4 GA runs was increased from 10 to 100 and the grid spacing kept at 0.375; the size of the docking grid was firstly increased to 80 × 80 × 80 Å that encompassed the entire periplasmatic receptor structure. Then the best scored cluster re-docked using a smaller grid size (40 × 40 × 40 Å), focusing inside the identified binding site. All the 100 independent GA runs from AD4 were processed using the built-in clustering analysis with a 2.0 Å cutoff.
Molecular dynamics of the MexB docked complexes in membrane
The MexB-inhibitor models have been oriented in membrane through OPM server (http://opm.phar.umich.edu/server.php), which generates the coordinates along the Z axis, and we used CHARMM GUI (www.charmm-gui.org) to build a membrane composed by 800 palmitoyloleyl phosphaditylcholine (POPC) molecules. Using these coordinates, we got a MexB-trimer (with docked ligands) system properly surrounded by the lipid matrix, that has been appropriately solvated with water (about 10,000) and ions (to reach up 0.15M NaCl, adding 397 Na ions and 361 Cl ions also to balance the trimer charge). We used AMBER99SB-ILDN force field parameters (Guy et al. 2012) for the protein and lipids, the TIP3P (Jorgensen 1998) model for solvent as implemented in GROMACS 5 (Hess et al. 2008). The models were minimized, and after then, six equilibration phases and molecular dynamics simulations were carried out. The Force field used for MD simulation was CHARMM 36 as implemented in GROMACS 5, with the overall time of simulation of 15 ns. The time-step used was 0.002 ps, and coordinates were written out every 10 ps, while energy data were collected every 2 ps. Periodic boundary conditions were applied in all directions using a neighbor searching grid type, and also setting at 1.4 nm the cut-off distance for the short-range neighbor list. Electrostatic interactions were taken into account implementing a fast smooth particle-mesh Ewald algorithm, with a 1.4 nm distance for the Coulomb cut-off (Essmann et al. 1995).
Microbiological assays
Bacterial strains
Twenty-five clinical isolates of P. aeruginosa resulted resistant by standard clinical assays (Sensititre AIM™ Automated Inoculation Delivery System, Thermo Scientific) to two or more antibiotics expelled by MexAB-OprM (i.e., ciprofloxacin, ceftazidime, piperacillin, meropenem and/or tobramycin) were collected (see Table 3).
Minimal inhibitory concentration (MIC) and efflux pump genes detection
Antibiotic MIC was determined by agar dilution following the CLSI guidelines (CLSI-M07-A10 2015), using P. aeruginosa ATCC 27853 as reference strain.
The mexB gene (coding MexB, the inner membrane protein channel of MexAB-OprM) was sought by PCR, using the primer pair mexB-F (5′-CAAGGGCGTCGGTGACTTCCAG-3′) and mexB-R (5′-ACCTGGGAACCGTCGGGATTGA-3′) (Oh et al. 2003) and 5 µl of genomic bacterial DNA extracted as previously described (Hynes et al. 1992); P. aeruginosa PAO1 was used as positive control (Huseyin et al. 2014).
Synergy tests
Three different laboratory methods, i.e., disk diffusion, checkerboard arrays and time killing curve, to assess the activity of antimicrobial combinations, were used.
Disk agar diffusion
It was performed as recommended by CLSI (CLSI-M02-A12 2015). Briefly, a Mueller-Hinton (MH) agar plate was uniformly inoculated with the bacterial culture containing 1 × 108 CFU/m. Disks containing standard amount of antibiotic was then applied on the plate (CLSI-M02-A12 2015), and beside this, an additional disk containing different concentrations (from 0,06 to 160 µg/ml) of the selected natural compounds (2,5,6), at a distance corresponding to half of the radium of the antibiotic inhibition zone, as determined in preliminary assays. Plates were incubated at 37 °C for 24 h and examined for variations in the inhibition halo.
Checkerboard assays
The checkerboard assay was performed as previously described (Isenberg 1992a), using 2-fold increasing concentrations of the antibiotic (from 1/64 to 16X MIC) and natural compounds (from 2 to 320 µg/ml).
Since the selected natural compound did not show any antibacterial activity (neither bactericidal nor bacteriostatic) the result of the association couldn’t be tested by FIC index determination. Synergy was then evaluated on the bases of the ability of natural compounds to decrease the MIC of the antibiotic. A 3–4 fold MIC decrease was considered a synergistic effect.
Killing curves
Time-kill curve analysis was performed as described by Isenberg (Isenberg 1992b). Antibiotic concentrations ranging from ¼x to 2x MIC were used alone and in combination with 40 µg/ml of pregnan-20-one derivative or morelloflavone. The bacterial inoculum was standardized at 105 CFU/ml. The dynamic of the bactericidal effect of the antibiotic-natural compound combination was evaluated by CFU counts at 2, 4, 6, 8, and 24 h.
As for the checkerboard assay, the results couldn’t be evaluated according to CLSI standards for drugs combinations, and any increase of the bactericidal power of the association compared to that of the antibiotic alone, was considered as synergy.
In vitro validation assays
Ethidium bromide accumulation assay
The ability to accumulate ethidium bromide by P. aeruginosa in the presence and in the absence of pregna-20-one derivative and morelloflavone was tested as described by Aparna et al. (2014) using the compound concentration resulted the most effective in checkerboard assays.
Briefly, an overnight culture of P. aeruginosa C24 was diluted 1:50 in MH broth and incubated at 37 °C for 4 h with shaking. After centrifugation at 10000 x g for 10 min, the pellet was washed in phosphate buffered saline (PBS) and resuspended until to OD of 0.1 at 600 nm. 170 μl of this standardized culture, 20 μl of ethidium bromide (10 mg/ml), final concentration 2.5 μM, and 10 μl of P1 or P2, final concentration 40 μg/ml, were then added to each well of a flat bottomed, black 96-well microtiter plate. The plate was incubated at 37 °C and the kinetic of the intracellular accumulation of ethidium bromide was evaluated immediately and after further 24 h incubation by reading at an excitation of 530 nm and at an emission of 590 nm wavelengths for 30 min at 5 min intervals, using the Synergy HT MicroPlate Reader Spectrophotometer (BioTek, Winooski, VT, USA). Each condition was tested in triplicate and PBS was used as blank.
The percentage increase in fluorescence, indicating the ability of the compounds to accumulate ethidium bromide by the inhibition of the efflux activity, was calculated as below (Wang and Joseph 1999):
where Fta is the fluorescence emitted at times 5, 10, 15, 20, 25 and 30 min and Ft0 the fluorescence at time 0 min.
Hemolysis assays
Hemolysis assays were performed as described by Chongsiriwatana et al. (2008). Briefly, four ml of freshly drawn, heparanized human blood were diluted with 25 ml of phosphate buffered saline (PBS), pH 7.4. After 3x washing in 25 ml PBS by spinning at 1000 g for 10 min the red blood cells, the pellet was resuspended in PBS to ~20 vol%. One-hundred µl of erythrocyte suspension was added to 100 µl of different concentrations of the compound to be tested (1:2 serial dilutions in PBS from 80 to 20 µg/ml). The negative and the positive control were 100 µl of PBS and 100 µl of 0.2 vol% Triton X-100, respectively. Each sample was tested in triplicate. After 1 h incubation at 37 °C each well was supplemented with 150 µl of PBS and the plate centrifuged at 1.200 x g for 15 min. The supernatant (150 µl) was transferred in a new plate and its OD350 measured using the Synergy HT MicroPlate Reader Spectrophotometer (BioTek, Winooski, VT, USA).
The hemolysis (%) was determined as follows
where A is the absorbance of the test well, A0 the absorbance of the negative control, and Atotal the absorbance of the positive control; the mean value of the three replicates of each sample was recorded.
Results and discussion
Considering previous evidence that natural products may act as EPIs in both Gram-positive and Gram-negative bacteria (Aparna et al. 2014; Medeiros Barreto et al. 2014), in this work we have screened in silico a library of natural compounds to evaluate their ability to inhibit MexAB-OprM efflux system of P. aeruginosa by binding specific efflux proteins. The ability of the in silico selected compounds to decrease the MIC of the antibiotics extruded from the bacterial cell by MexAB-OprM was then validated by in vitro microbiological assay.
The computational investigation started from retrieving the crystal structures of MexB (PDB code 2V50) and AcrB (PDB code 1T9Y). In the last, MC–207,110 that acts as a competitive inhibitor for both MexB and AcrB efflux systems, is co-crystallized with AcrB (Yu et al. 2005).
The MC-207,110 molecule binds at two regions in the AcrB structure: the binding site 1, involving residues Phe664, Arg717, Pro718, Leu828, Ser715 and the binding site 2 (central cavity), the antibiotic binding site, by interacting with residues Phe386, Phe388, and Phe459. Since sequence alignment and 3D model analysis assays (Pettersen et al. 2004) have previously showed that these binding sites were conserved in the MexB protein sequence, we easily identified the binding sites 1 and 2 also in MexB.
Then, we employed HTVS docking using Autodock/Vina protocol, to identify novel hits from ZINC natural compounds database subset (SPECs net). A total of 1489 natural compounds were shortlisted against AcrB and MexB considering both sites of interaction. Suitable efflux pumps inhibitory compounds were selected on the bases of the predicted binding energy and cluster population.
Before performing the docking, the reproducibility of the modeling protocol was validated by re-docking the co-crystallized ligand in 1T9Y (RMSD 0.910 Å).
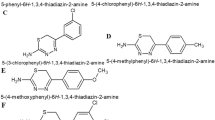
As regards MexB, we found out 368 compounds (223 for the site 1 and 145 for the site 2) showing a binding energy lower than −9.0 kcal/mol (which corresponds to a micromolar activity). The top ten for each site were selected according to their predicted binding energy. The structure of the first six showing high affinity for both sites are reported in Fig. 1. In Tables 1 and 2, the list of the selected compounds is reported together with their binding interactions and energies.
Structure of the selected six compounds to be used in association with antibiotics in in vitro synergy assays against P.aeruginosa. 1, Alpha-viniferin; 2, 2-(3,4-dihydroxyphenyl)-2′-(4-hydroxyphenyl)-3′-methyl-2′,3′-dihydro-3′,8-bis(5,7-dihydroxy-4H-chromen-4-one) (morelloflavone); 3, 3-acetyl-3,5,10,12-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydro-1-naphthacenyl 3-((anilinocarbothioyl)amino)-2,3,6-trideoxyhexopyranoside; 4 Glycyrrhizic acid derivative; 5, [(5S,10S,13S,14S,17S)-17-acetyl-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta-[α]phenanthren-3-yl] 2-(4-benzoylphenyl)acetate (pregna-20-one derivative); 6, N-{2-(1,3-benzodioxol-5-yl)-1-((6-oxo-7,11 diazatricyclo(7.3.1.0~2,7~)trideca-2,4-dien-11-yl)carbonyl)vinyl}benzamide
Focused docking and binding energy evaluation
In order to evaluate the binding energy and the intermolecular interactions with a higher level of accuracy, we considered the top ten hits compounds both for MexB site 1 and site 2 showing a predicted nanomolar activity. These products were further re-docked using a focused docking protocol (Autodock 4.0) extending the binding cavity surface (Gabbianelli et al. 2015). Among them, three compounds targeting MexB were chosen for further testing their ability to counteract efflux pumps by in vitro assays (Fig. 2 ) since the focused docking poses corresponds to a bonding energy even more negative than the starting ones. Two of them (2 and 5) showed a high score for both sites whilst the third (6) gains a high score only for site 2. The refined binding interactions and energies are reported in Table 2.
As a result, we can point out common features of the top 10 hits for MexB: all of them show the presence of clusters of poses located in both sites of MexB, even if with a different extent of affinity (Table 2). More in details, morelloflavone (2) shows two main highly populated clusters in close proximity to the binding site 1 and site 2 respectively (Figs. 3a and b), whereas the pregna-20-one (5) has a clear preference for MexB site 1, showing for this site very high populated clusters (Fig. 3c). On the contrary, there is only one cluster referring to site 2, and it has very low populated even if it has a good binding affinity.
Finally, the third tested compound (6) has a different preferred site of interaction that doesn’t resemble neither site 1 nor the antibiotic site 2.
However, high energy clusters which can be referred to these sites were identified but they gain a too high energy and very low population to be considered (Fig. 4). This different behavior of 6 with respect 2 and 5 suggests a different interaction of this compound with MexB, that can justify its observed less efficient inhibitory activity (Table 2).
Microbiological assays
Screening of MDR P. aeruginosa clinical isolates for the presence of MexAB-OprM and antibiotic susceptibility tests
The twenty-five collected P. aeruginosa strains were analysed for the presence of the MexAB-OprM cluster by PCR assays targeting mexB. All gave positive results and were then analyzed by agar dilution for their susceptibility to the antibiotics expelled by MexAB-OprM i.e. ciprofloxacin, ceftazidime, piperacillin, meropenem, and tobramycin. All but one resulted resistant to ciprofloxacin and four to all tested antibiotics (Table 3). Despite the uniform presence of mexB, most isolates exhibited a different pattern of resistance, in agreement with Webber and Piddock (Webber and Piddock 2003), who reported the ancestral physiological role of this efflux pump, constitutively present in P. aeruginosa, in expelling toxic compounds, and the possibility that the selective pressure exerted by specific antibiotics, such as ciprofloxacin, shaped it to extrude additional drugs. Although the presence of efflux pumps typically confers low-level resistance, mutants exhibiting their overexpression (in particular in the first steps of biofilm development), are involved in multi-drug, high-level resistant infections (Soto 2013; Visaggio et al. 2015).
Agar disk diffusion assays
Antibiotic double disk diffusion assays on agar plates did not produce significative results, except for a bare enlargement of the inhibitory zone halo against 4/25 P. aeruginosa strains around ciprofloxacin disks in the presence of 10 µg/ml of two (2,5) of the three selected natural compounds. However, the halo enlargement (2–3 mm) was too limited to indicate synergy. This behavior might be explained by a limited ability of the tested compounds, characterized by hydrophobic nature and low solubility, to spread in the agar medium. Additional experiments are in progress to overcome this drawback.
Checkerboard assays
The synergistic activity of the selected natural compounds with antibiotics extruded by MexAB-OprM was further analysed by cherckerboard assays. Two of them (2,5), indicated by in silico assays as possible antibiotic competitors for MexB, at a concentration of 40 µg/ml showed a synergistic effect when used in combination with ciprofloxacin, resulting in a four-eight fold MIC decrease against most P. aeruginosa (Table 4). They did not improve the activity of ceftazidime, piperacillin, meropenem or tobramycin. No synergy was observed for the third chosen compound (6). Interestingly, the P. aeruginosa strains against which we did not detect any improvement of ciprofloxacin activity when used in combination with compounds 2 and 5 were all pyoverdine producers. It has been reported that pyoverdine acts as signal molecule for the expression of metabolic pathways (Beare et al. 2003; Mueller et al. 2007), virulence factors and extracellular polysaccharides. We might thus hypothesize that pyoverdine can also influence the expression of factors binding and sequestering (or destroying) the tested compounds, thus preventing synergy. Further studies are in progress to verify this hypothesis and to better understand the discrepancies observed with different P. aeruginosa strains and different antibiotics extruded by MexAB-OprM.
Both compounds seem to represent valid candidates for a combined anti-Pseudomonas therapy in association with ciprofloxacin. However, their activity as EPIs needs further investigations, to explain their inefficacy when combined with ceftazidime, piperacillin, meropenem and tobramycin, excreted by the same efflux system. The presence of additional resistance mechanisms (making the inhibition of the efflux pump activity insufficient to increase antibiotic susceptibility) is under investigation as well as the enhancement of MexAB-OprM expression in a reference strains, susceptible to the five selected antibiotics, by either genetic modification or induction through the addiction of toxic compounds, such as Pentachlorophenol (Mueller et al. 2007), to the growing medium. A further decrease of ciprofloxacin MIC when combined with morelloflavone (2) or pregnan-20-one derivative (5) will support their involvement in the MexAB-OprM inhibition. On the other hand our data are in agreement with other studies involving different EPIs, that report synergistic combinations mainly with fluoroquinolones (Lomovskaya et al. 2001; Goli et al. 2016), and synergy results consistent with ours.
Killing curves
To further support the effectiveness of the combination of morelloflavone 2 or pregnan-20-one derivative 5 with ciprofloxacin against P. aeruginosa, we analyzed the bactericidal activity of the two associations by killing curve analysis. The antibiotic alone, at a concentration corresponding to the MIC, resulted bactericidal after 6 h, while when used at a concentration corresponding to ½ MIC exhibited an overall bacteriostatic effect. These effects were enhanced by the presence of either or morelloflavone or pregnan-20-one derivative. In particular, it is to note that the combination pregnan-20-one derivative (40 µg/ml )–ciprofloxacin (2 µg/ml, i.e., 1/2 MIC) showed 1 log decrease of CFU at 6 h and about 2 log at 24 h. The increased killing of both combinations compared to ciprofloxacin alone becomes more evident with combinations containing the same concentration as above (i.e., 40 µg/ml) of the two compounds and 4 µg/ml (the MIC) of ciprofloxacin. The use of 8 µg/ml (2xMIC) of ciprofloxacin did not show any remarkable difference between the bactericidal activity of the antibiotic alone and combined with either pregnan-20-one derivative or morelloflavone (Fig. 5).
The time curve analysis showed a different killing dynamic of the tested combinations: when using compound 5, the synergy was detectable earlier (Figs. 5 a and b) than when using compound 2. However, after 24 h, both combinations showed comparable activities. This behavior is in agreement with data obtained by molecular dynamics (see more below) showing a faster stabilization for compound 5 in the MexB binding site rather than compound 2.
Overall, our data confirm the possible role of morelloflavone or pregnan-20-one derivative as inhibitors of P. aeruginosa efflux pump MexAB-OprM; further studies are in progress in order to better understand and improve their competitive interaction with MexAB-OprM and to assess their effectiveness combined with the other antibiotics usually expelled by this efflux mechanism.
Intracellular ethidium bromide accumulation
The synergy of the combination ciprofloxacin and morelloflavone (2) or pregnan-20-one derivative was further confirmed by ethidium bromide accumulation assays, performed in absence/presence of the two selected compounds. Since both showed the best synergistic activity at 40 μg/ml, their effect on ethidium bromide accumulation was evaluated at the same concentration. Both compounds showed the ability to inhibit the MexAB-OprM efflux activity, as indicated by the fluorescence percentage increase during the 30 min kinetics (Fig. 6): In particular, the pregnan-20-one derivative exhibited an earlier inhibition activity compared to morelloflavone, (22 and 16% fluorescence increase respectively after 15 min and 36, and 22.4% after 30 min). However, after further 24 h exposure to the EPIs, the morelloflavone showed a greater inhibition of the efflux pump than pregnan-20-one (fluorescence increase of 24.2 and 13%, respectively). These data suggest that while Pregnan-20-one interacts with the MexB protein more directly and in a shorter time, after 30 min morelloflavone causes a more efficient inhibition of the efflux pump, probably as a consequence of its efficient interaction with the target protein. This is in agreement with both data obtained from time-killing assays and molecular dynamics previsions.
Hemolytic activity
Preliminary toxicity information about the two chosen hits was retrieved from previous studies [Nongporn et al. 2007; Xiufeng et al. 2009; www.specs.net ZINC08382438]. Nevertheless, in order to better evaluate their lack of toxicity, Morelloflavone and Pregnan-20-one derivative were further tested for their citotoxicity by hemolysis assays, performed using 20, 40 and 80 μg/ml of each compound. No hemolytic activity was visibly observed with all tested concentrations of both compounds; however Pregna-20-one derivative showed an OD350 increase of 0,8% (±0.005 and ±0.004, respectively) at 20 and 40 μg/ml and of 1.6 % (±0.005) at 80 μg/ml; Morelloflavone showed an 2.4 % (± 0.002) OD350 increase only when used at a concentration of 80 μg/ml; 20 and 40 μg/ml of the same compound didn’t show any detectable hemolytic activity.
Molecular dynamics of MexB inhibitors complexes in membrane
On the basis of the encouraging microbiological results, we assessed the binding stability of the two natural compound (2, 5) against the efflux pump MexAB-OprM. To do this, the ligand-bound monomer receptor MexB is rebuilt in its trimeric structural form in analogy with the trimeric structure of AcrB (1T9Y) and inserted into a lipid bilayers matrix, mimicking the periplasmic membrane.
Four molecular systems were settled out: (1) trimeric MexB complexed with morelloflavone in site 2 positioned as in the highest affinity cluster from the previous molecular docking simulations (Model1); (2) trimeric MexB complexed with two molecules of morelloflavone (one in the first and one in the second site of the MexB monomer) (Model2); (3) trimeric MexB complexed with pregnan-20-one in the only binding site highlighted during the previous docking (Model3); (4) MexB alone to be used as a control (Model4). The four models underwent a complete minimization protocol prior to proceed with MD simulations in membrane using AMBER99SB force field as implemented in GROMACS software package (Hess et al. 2008). Then, a molecular dynamic simulation up to 15 ns has been carried out and the overall RMSD evaluated. After stabilization of the ligands inside their binding site, the interactions with the surrounding protein residues have been analyzed.
The RMSD values obtained in Model1 showed that the flavonoid 2 competes with the antibiotic binding site (site 2); it reached stability in a short time (1 ns), and then keeps interactions with protein residues constant throughout the course of the dynamic (Fig. 7). For Model2 carrying the same molecule (morelloflavone) in both sites, we observed a different behavior: a major reorientation of the two ligands, especially inside the antibiotic cleft (see Fig. 8). More in details, the molecule in site 1 partially rotates and in addition morelloflavone inside site 2 undergoes a dynamic reorientation and oscillates between two poses that can be directly related with the docking predicted cluster 1 (lower in energy but less populated) and cluster 2 (higher in energy but more populated). Finally, also in the Model3, pregnan-20-one derivative 5 undergoes a major reorientation and reaches the RMSD stabilization only at the end of the simulation (15 ns) (Fig. 9). These different behaviors observed for the two active compounds can be put in relation with the killing curve analysis (Fig. 5). In fact, from the microbiological data the pregna-20-one derivative results active from the beginning of the experiments whilst morelloflavone show a major comparable activity only after a delay. Indeed, from the MD results, compound 5 shows a different site of action with respect compound 2; moreover the first is correctly positioned and does not vary its orientation. On the contrary, morelloflavone must reorient after docking inside the clefts. These observations support a different mode of action at molecular level that is strongly supported by the microbiological data. In Fig. 10, the relative binding poses of both compounds (2, 5) in the trimeric protein in membrane are shown.
Conclusions
Molecular docking calculations identified morelloflavone (2) and the pregna-20-one derivative (5) as good candidates to inhibit the MexAB-OprM efflux pump of P. aeruginosa. These findings were supported by in vitro microbiological assays that showed their synergism with ciprofloxacin increasing both the inhibitory (4 times, 2 fold dilutions) and the killing (1–2 log) activity of the antibiotic; these results are further validated by ethidium bromide accumulation assays. As the incidence of drug resistant P. aeruginosa is alarmingly increasing, a combination therapy based on ciprofloxacin and one of these phytochemicals might be a promising approach to counteract MDR P. aeruginosa infections.
References
Aparna V, Dineshkumar K, Mohanalakshmi N, Velmurugan D, Hopper W (2014) Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PLOs One 9(7):e101840
Askoura M, Mottawea W, Abujamel T, Taher I (2011) Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa. Libyan J Med 6 doi:10.3402/ljm.v6i0.5870
Ball AR, Casadei G, Samosorn S, Bremner JB, Ausubel FM, Moy TI, Lewis K (2006) Conjugating berberine to a multidrug efflux pump inhibitor creates an effective antimicrobial. ACS Chem Biol 1:594–600
Bashford D, Karplus M (1990) pKa of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry 29:10219–10225
Beare PA, For RJ, Martin LW, Lamont IL (2003) Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol Microbiol 47(1):195–207
Breidenstein EB, de la Fuente-Nunez C, Hancock RE (2011) Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426
Chang MW, Ayeni C, Breuer S, Torbett BE (2010) Virtual screening for HIV protease inhibitors: a comparison of AutoDock 4 and Vina. PLoS One 5(8):e11955
Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE (2008) Peptoids that mimic the structure, function and mechanism of helical antimicrobial peptides. Proc Natl Acad Sci U.S.A. 105(8):2794–2799
CLSI-M02-A12 (2015) Clinical and Laboratory Standard Institute: Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard—Twelfth Edition.
CLSI-M07-A10 (2015) Clinical and Laboratory Standard Institute: Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Tenth Edition. January 2015
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz Jr KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117:5179
De Kievit TR, Parkins MD, Gillis RJ, Srikumar R, Ceri H, Poole K, Iglewski BH, Storey DG (2001) Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 45:1761–1770
Duan WC, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chemistry 24(16):1999–2012
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Gabbianelli R, Carloni M, Marmocchi F, Nasuti C, Fedeli D, Laudadio E, Massaccesi L, Galeazzi R (2015) Permethrin and its metabolites affects Cu/Zn Superoxide conformation: fluorescence and in silico evidences. Mol Biosyst 11:208–217
Galeazzi R, Massaccesi L, Piva F, Principato G, Laudadio E (2014) Insights into the Influence of 5-HT2c aminoacidic variants with the inhibitory action of Serotonin inverse agonists and antagonists. J Mol Model 20(3):2120. doi:10.1007/s00894-014-2120-0
Gilbert JC, Nocedal J (1992) Global convergence properties of conjugate gradient methods for optimization. SIAM J Optim 2:21–42
Goli HR, Nahaei MR, Rezaee MA, Hasani A, Kafil HS, Aghazadeh M, Sheikhalizadeh V (2016) Contribution of mexAB-oprM and mexXY (-oprA) efflux operons in antibiotic resistance of clinical Pseudomonas aeruginosa isolates in Tabriz, Iran. Infect Genet Evol 45(2016):75–82
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A (2005) H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 33:W368–W371
Guy AT, Piggot TJ, Khalid S (2012) Single-stranded DNA within nanopores: conformational dynamics and implications for sequencing; a molecular dynamics simulation study. Biophys J 103:1028–1036
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, andscalable molecular simulation. J Chem Theory Comput 4:435–447
Ho-Fung Lau C, Hughes D, Poole K, Hirotani A, Kunishima H, Yamamoto N, Hatta M, Kitagawa M, Kohno S, Kaku M (2014) MexY-promoted aminoglycoside resistance in Pseudomonas aeruginosa: involvement of a putative proximal binding pocket in aminoglycoside recognition. mBio 5(2):e01068–14
Huseyin AT, Kulah C, Ciftci IH (2014) The effects of active efflux pumps on antibiotic resistance in Pseudomonas aeruginosa. World J Microbiol Biotechnol 30(10). doi:10.1007/s11274-014-1692-2
Hynes WL, Ferretti JJ, Gilmore MS, Segarra RA (1992) PCR amplification of streptococcal DNA using crude cell lysate. FEMS Microbiol Lett 73(1–2):139–142
Isenberg HD (1992a) Clinical Microbiology Procedures Handbook. American Society for Microbiology Press, Washington, DC, vol. 1, 5 (18): 1–23 (A)
Isenberg HD (1992b) Clinical Microbiology Procedures Handbook. American Society for Microbiology Press, Washington, DC, vol. 1, 5 (16): 14–33 (B)
Jorgensen WL (1998) Temperature dependence of TIP3P, SPC, and TIP4P water from NPT Monte Carlo simulations: seeking temperatures of maximum density. J Comput Chemistry 19:1179–1186
Lee A, Mao W, Warren MS, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O (2000) Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol 182:3142–3150
Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Lee VJ (2001) Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother 45(1):105–116
Medeiros Barreto H, Cerqueira Fontinele F, Pereira de Oliveira A, Arcanjo DD, Cavalcanti Dos Santos BH, de Abreu AP,Douglas Melo Coutinho H, Alves Carvalho da Silva R, Oliveira de Sousa T, Freire de Medeiros MD, Lopes Citó AM, Dantas Lopes JA (2014) Phytochemical prospection and modulation of antibiotic activity in vitro by Lippia origanoides H.B.K. in methicillin resistant Staphylococcus aureus. Biomed Res Int 2014:305610
Mohamadi F, Richard NG, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC (1990) MacroModel - an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem 11(4):440–467
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Morris GM, Huey Rì, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comput Chem 16:2785–2789
Mueller JF, Stevens AM, Craig J, Love NG (2007) Transcriptome analysis reveals that Multidrug Efflux Genes are upregulated to protect Pseudomonas aeruginosa from Pentachlorophenol Stress. Appl Environ Microbiol 73(14):4550–4558
Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A (2006) Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443(7108):173–179
Myers J, Grothaus G, Narayanan S, Onufriev A (2006) A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins 63:928–938
Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, Onodera Y, Nishino K, Yamaguchi A (2013) Structural basis for the inhibition of bacterialmultidrug exporters. Nature 500:102–106
Nelson ML (2002) Modulation of antibiotic efflux in bacteria. Curr Med Chem—Anti-Infect Agents 1:35–54
Nongporn HT, Suyanee K, Wilawan M (2007) Inhibition of human lipoprotein oxidation by morelloflavone and camboginol from Garcinia dulcis. Nat Prod Res 21(7):655–662
Oh H, Stenhoff J, Jalal S, Wretlind B (2003) Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone- resistant Pseudomonas aeruginosa strains. Microb Drug Resist 9:323–328
Ohene-Agyei T, Mowla R, Rahman T, Venter H (2014) Phytochemicals increase the antibacterial activity of antibiotics by acting on a drug efflux pump. Microbiologyopen 3(6):885–896
Pearson JP, Van Delden C, Iglewski BH (1999) Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol 181:1203–1210
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera- a visualization system for exploratory research and analysis. J Comput Chem 13:1605–1612
Poole K (2005) Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56(1):20–51
Sanner MF (1999) Python: a programming language for software integration and development. J Mol Graph Mod 17:57–61
Scirè A, Baldassarre M, Galeazzi R, Tanfani F (2013) Fibrillation properties of human α(1)-acid glycoprotein. Biochimie 95:158–166
Soto SM (2013) Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4(3):223–229
Suite (2011) MacroModel, Version 9.9. Schrödinger, LLC, New York, NY
Taubes G (2008) The bacteria fight back. Science 321:356–361
Thai KM, Ngo TD, Phan TV, Tran TD, Nguyebm NV, Nguyen TH, Le MT (2015) Med Chem 11:135–155
Thai KM, Ngo TD, Phan TV, Tran TD, Nguyen NV, Nguyen TH, Le MT (2015) Virtual screening for novel Staphylococcus ureus NorA efflux pump inhibitors from natural products. Med Chem 11(2):135–155
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Visaggio D, Pasqua M, Bonchi C, Kaever V, Visca P, Imperi F (2015) Cell aggregation promotes pyoverdine-dependent iron uptake and virulence in Pseudomonas aeruginosa. Front Microbiol 6:902
Wang H, Joseph JA (1999) Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic Biol Med 27:612–616
Watkins WJ, Landaverry Y, Léger R, Litman R, Renau TE, Williams N, Yen R, Zhang JZ, Chamberland S, Madsen D, Griffith D, Tembe V, Huie K, Dudley MN (2003) The Relationship between Physicochemical Properties, In Vitro Activity and Pharmacokinetic Profiles of Analogues of Diamine-Containing Efflux Pump Inhibitors. Bioorg Med Chem Lett 13(2003):4241–4244
Webber MA, Piddock LJV (2003) The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother 51:9–11
Xiufeng P, Tingfang Y, Zhengfang Y, Sung Gook C, Weijing Q, Decha P, Ken F, Mingyao L (2009) Morelloflavone, a Biflavonoid, Inhibits Tumor Angiogenesis by Targeting Rho GTPases and Extracellular Signal-Regulated Kinase Signaling Pathways. Cancer Res 69(2):518–525
Yu EW, Aires JR, McDermott G, Nikaido H (2005) A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J Bacteriol 187(19):6804–6815
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Mangiaterra, G., Laudadio, E., Cometti, M. et al. Inhibitors of multidrug efflux pumps of Pseudomonas aeruginosa from natural sources: An in silico high-throughput virtual screening and in vitro validation. Med Chem Res 26, 414–430 (2017). https://doi.org/10.1007/s00044-016-1761-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-016-1761-1