Abstract
This study aims to uncover the heterogeneity of endothelial cells (ECs) in colorectal cancer (CRC) and their crucial role in angiogenesis, with a special focus on tip cells. Using single-cell RNA sequencing to profile ECs, our data suggests that CRC ECs predominantly exhibit enhanced angiogenesis and decreased antigen presentation, a shift in phenotype largely steered by tip cells. We also observed that an increase in the density and proportion of tip cells correlates with CRC occurrence, progression, and poorer patient prognosis. Furthermore, we identified endothelial cell-specific molecule 1 (ESM1), specifically expressed in tip cells, sustains a VEGFA-KDR-ESM1 positive feedback loop, promoting angiogenesis and CRC proliferation and migration. We also found the enrichment of KDR in tip cells and spotlight a unique long-tail effect in VEGFA expression: while VEGFA is primarily expressed by epithelial cells, the highest level of VEGFA expression is found in individual myeloid cells. Moreover, we observed that effective PD-1 blockade immunotherapy significantly reduced tip cells, disrupting the VEGFA-KDR-ESM1 positive feedback loop in the process. Our investigation into the heterogeneity of ECs in CRC at a single-cell level offers important insights that may contribute to the development of more effective immunotherapies targeting tip cells in CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is a prevalent and lethal type of cancer, ranking as the third most common malignancy and the second leading cause of cancer-related death worldwide [1]. Despite the recent advancements in immunotherapy, its efficacy in treating CRC remains limited, with only a small subset of patients with mismatch repair deficiency or high levels of microsatellite instability experiencing positive outcomes from immune checkpoint inhibitor (ICI) [2,3,4,5]. To address this challenge, it is crucial to gain a deeper understanding of the complex interactions between different cells and molecules in the CRC tumor microenvironment (TME) and identify new targets for immunotherapy.
Endothelial cells (ECs), known for their remarkable plasticity [6,7,8], constitute a single layer - the endothelium - lining the inner surfaces of blood vascular and lymphatic vessels. The anatomical position within the vasculature classifies ECs in resting tissues into arterial ECs, venous ECs, capillary ECs, and lymphatic ECs [7]. A specialized subcategory of ECs, tip cells, are integral to vessel sprouting, playing a leading role in initiating and guiding the sprout formation [7, 9, 10]. Existing single-cell RNA sequencing (scRNA-seq) studies substantiate the pivotal function of these tip cells across a wide range of human and mouse tumors, underscoring the critical role of angiogenesis in facilitating tumor growth and metastasis [11,12,13,14,15]. Tip cells are identified by a unique gene expression profile, such as APLN, a known marker, and other genes such as ESM1, PGF, NID2, COL4A1, COL4A2, KDR, PLVAP, HSPG2, and INSR, which have recently been found enriched in tip cells through single-cell studies [9, 13,14,15]. However, despite many CRC single-cell studies involving ECs, there is still a lack of systematic investigation into the heterogeneity of ECs in CRC, particularly with respect to the tip cells enriched in tumor ECs.
Currently, anti-angiogenic therapy (AAT) primarily targets the vascular endothelial growth factor (VEGF) pathway [12, 16]. These therapies are indiscriminately employed to prune the vasculature in a variety of cancer types, including CRC, but they often encounter insufficient efficacy and resistance [12, 14]. The distinct transcriptomic characteristics and phenotypes of tumor ECs across various types of cancer remain unclear, which could potentially result in varied responses to AAT. Additionally, the specific processes that tumor ECs employ to facilitate angiogenesis are yet to be fully comprehended. As such, it has become increasingly vital to delve into the heterogeneity of tumor ECs and further examine the mechanisms through which they contribute to angiogenesis in cancer research.
In this study, we utilized single-cell transcriptomic data to better understand how ECs influence CRC. Single-cell studies in lung and gastric cancers have revealed that, compared to normal ECs, tumor ECs display an enhanced angiogenesis yet a decreased antigen presentation [13, 15]. Through the analysis of transcriptomic features of different tumor EC subsets, we found that it is the tip cell that primarily drives the phenotype changes in CRC ECs, enhancing angiogenesis and reducing antigen presentation. Consequently, we narrowed our investigative focus to this EC subset. Intriguingly, our findings suggest the existence of a VEGFA-KDR-ESM1 positive feedback loop where VEGFA positively modulates ESM1 expression in tip cells, which in turn enhances VEGFA-mediated signaling. We also observed that effective anti-PD-1 immunotherapy significantly reduces the population of tip cells within the CRC TME, effectively disrupting this feedback loop. Delving into the heterogeneity of ECs and elucidating the angiogenesis-promoting mechanisms employed by tip cells could potentially pave the way for novel anti-angiogenic therapeutic targets and immunotherapy strategies.
Methods
Material
This study harnessed a comprehensive array of public datasets, including one spatial transcriptomics (ST) dataset, three scRNA-seq datasets, and 10 bulk RNA-sequencing (bulk RNA-seq) datasets (Table 1). The bulk RNA-seq datasets incorporated high-throughput sequencing data derived from the TCGA-CRC as well as nine additional GEO microarray datasets (GSE20842, GSE20916, GSE39582, GSE41258, GSE44076, GSE68468, GSE83889, GSE87211, and GSE106582). Transcriptome data and relevant clinical information from TCGA-CRC were sourced from UCSC Xena (http://xena.ucsc.edu/). For the TCGA pan-cancer cohort, data were collated from the National Cancer Institute Cancer Research Data Commons (https://gdc.cancer.gov/about-data/publications/pancanatlas) in conjunction with UCSC Xena (with cancer abbreviations represented in Table S1). The transcriptome data and associated clinical details from the GEO datasets were directly accessed from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The 5-cohorts used in this study consist of data from five CRC single-cell cohorts (CRC-SG1, CRC-SG2, KUL3, KUL5, and SMC) integrated in the study by Joanito et al. [17], with the data source detailed in Table 1, line 2. Human tissue specimens used in this study were obtained from Xijing Hospital with the approval of the Institutional Review Board and informed consent from the patients (Approval number: KY20212211-C-1).
Single-cell sequencing data processing
Data from three single-cell datasets, which were initially quality-controlled by the original data authors, were independently analyzed for this study. We normalized the expression of all cells using the “LogNormalize” function with a scale factor of 10,000. From this, we selected the top 2,000 highly variable genes based on their mean and dispersion, and regressed percent mitochondrial content during the scaling of these genes using the “var.to.regress” option. We zero-centered and scaled each gene to unit variance before principal component analysis (PCA) to minimize potential batch effects. This was followed by a dimensionality reduction process using PCA to generate results. Preliminary clustering and annotation were accomplished using the “FindClusters” function with 50 principal components and a resolution of 0.8. Further, we visualized cell clustering using the UMAP method for nonlinear dimensionality reduction. We then conducted a second round of clustering to delve deeper into the subpopulations of ECs. After this, certain EC subsets were filtered out. These included the proliferative EC subset (MKI67), doublet subsets such as Doublets-B (CD79A), Doublets-Myeloid (LYZ), and Doublets-T (CD3D), as well as other low-quality clusters (the details of which can be found in the accompanying code). It’s important to note that in the 5-cohorts, ribosome-related genes were previously filtered out by the original authors during preprocessing. As a result, we didn’t identify any immature EC subsets characterized by the enrichment of ribosome-related genes post-clustering in the 5-cohorts.
Differential gene expression analysis
To identify marker genes for each cluster or subset, we made use of the “FindAllMarkers” or “FindMarkers” functions provided by Seurat. Genes were considered as differentially expressed genes (DEGs) if they met the following criteria: log2 (fold-change of average expression) > 0.25, pct.1 (percentage of expressed cells) > 0.25, and P-value < 0.01. We subsequently used these identified DEGs for enrichment analysis.
Enrichment analysis
For the enrichment analysis of DEGs, we utilized Metascape (http://metascape.org/), a user-friendly portal designed for comprehensive gene annotation and analysis [18]. Additionally, we used LinkedOmics (http://www.linkedomics.org/) [19], a publicly accessible portal for multiomics data analysis, for performing Gene Set Enrichment Analysis (GSEA) on the ESM1 expression profiles in TCGA-CRC data (GO BP).
Analysis of cell to cell communication
To investigate the interactions between EC subsets and other major cell types, we utilized the Python-based software CellphoneDB [20]. Putative ligands and receptors were determined based on their expression on each cell. To accurately determine the extent of cell interactions, we performed a random sampling of 500 cells per population from the EC subsets and major cell types in GSE178341 dataset.
Defining phenotype scores
To characterize the differences between various EC subsets, we obtained phenotype scores using the “AddModuleScore” function in the “Seurat” package. These scores were calculated based on the average expression of genes related to a particular phenotype.
Angiogenesis score [21] were utilized to assess the angiogenesis characteristics of different major cell types. Additionally, MHC-I and MHC-II scores were used to characterize the antigen presentation capabilities of different EC subsets (Table S2).
Spatial transcriptomic analysis
The spatial transcriptomic data derived from the CRC sample was standardized by employing the “SCTransform” function within the “Seurat” package. We subsequently implemented dimensionality reduction and clustering strategies using the “RunPCA” and “RunUMAP” functions respectively, where we applied 15 principal components with a resolution of 0.8. After consolidating similar clusters, we identified normal, stromal, and tumor regions in the CRC sample.
CIBERSORTx
Cell composition deconvolution was conducted utilizing CIBERSORTx [22]. Our initial step was to generate a signature gene expression matrix using the CRC scRNA-seq dataset (GSE178341). We extracted raw count matrix data and cell type classifications from a subset of the Seurat object, which incorporated 500 samples each from various blood vascular EC subsets, including artery ECs, capillary ECs, immature ECs, tip cells, and vein ECs. This raw count matrix was introduced into CIBERSORTx and subsequently normalized. The signature matrix, was established with CIBERSORTx, utilizing all genes to create the signature gene expression matrix. In the final step, we evaluated the proportions of different blood vascular EC cells in each CRC sample using CIBERSORTx, based on the bulk RNA-seq data (GSE39582). To correct for cross-platform variation in the deconvolution of the GSE39582 RNA-seq data, we performed batch correction using S-mode.
Immunofluorescence staining
Two paired CRC specimens were secured within 30 min post-tumor resection and preserved in paraformaldehyde for 48 h. Standard procedures were employed for dehydration and paraffin embedding. The specimens were treated with 3% H2O2 for 25 min to quench endogenous peroxidase activity. To block nonspecific binding, the tissue sections were pre-incubated with 10% normal goat serum for 30 min. Subsequently, they were incubated overnight at 4 °C with primary antibodies in a humidified chamber. The primary antibodies included rabbit anti-human PECAM1 (CD31) (AbCam, Cat#ab76533, 1:200), rabbit anti-human ESM1 (AbCam, Cat#ab103590, 1:2000), rabbit anti-human KDR (VEGFR2) (Thermo Scientific, Cat#MA5-15157, 1:1000), rabbit anti-human VEGFA (AbCam, Cat#ab52917, 1:200), mouse anti-human pan-CK (Novus, Cat#NBP2-33200AF647, 1:200), and mouse anti-human CD68 (Proteintech, Cat#66231-2-Ig, 1:5000). Following thorough washing, the sections were mounted with an anti-fade reagent and covered with coverslips. Fluorescence images were captured using a NIKON ECLIPSE C1 microscope and further analysis was performed using CaseViewer software.
Cell culture
Human CRC cell lines HCT-8 and HCT-116 were obtained from Procell (Guangzhou, China) and are maintain in our lab. CRC cells were cultured in DMEM medium (Gibco, Thermo Fisher Scientific, Cambridge, MA, USA) supplemented with 10% fetal bovine serum (Oricell; Guangzhou, China), 100 µg/ml of streptomycin, and 100 U/ml of penicillin in the medium (HyClone; Logan, Utah, USA). The cells were incubated in a humidified incubator at 37℃ with 5% CO2 to provide optimal growth conditions. The cells were treated with different concentrations of recombinant ESM1 (Sinobiological, Beijing, China, 0-1000ng/ml) and subsequent in vitro experiments were conducted.
Total RNA extraction and qRT-PCR
Trizol reagent (Invitrogen, Waltham, MA, United States) was utilized for the isolation and extraction of total RNA from tissue samples and cell lines. The obtained RNA was subjected to reverse transcription using the PrimeScript RT Reagent Kit (TaKaRa, Tokyo, Japan) to generate cDNA. Subsequently, qRT-PCR was performed using the SYBR Premix Ex Taq II Kit (TaKaRa, Tokyo, Japan) to assess the expression levels of VEGFA and KDR. GAPDH was employed as an internal standard. The relative mRNA expression was calculated using the 2−△△Ct method. The primer sequences are provided below. VEGFA forward: 5’- ATCAGTTCGAGGAAAGGGAAAGG-3’; reverse: 5’-AAACAAATGCTTTCTCCGCTCTG − 3’; KDR forward: 5′-GGCCCAATAATCAGAGTGGCA-3′; reverse: 5′- CCAGTGTCATTTCCGATCACTTT-3′; GAPDH forward: 5′-GACAGTCAGCCGCATCTTCT-3′; reverse: 5′-GCGCCCAATACGACCAAATC-3′.
Western blot (WB) assay
The cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (Beyotime Biotechnology, Shanghai, China) supplemented with protease and phosphatase inhibitors from Millipore (Temecula, CA, USA) on ice. After centrifugation at 12,000 g for 15 min, the concentration of total proteins in the lysates was determined using a BCA Protein Assay Kit (Pierce, Rockford, IL, USA). Equal amounts of proteins were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (Millipore, Temecula, CA, USA). The membranes were blocked with 5% non-fat milk at 37℃ for one hour and then incubated with primary antibodies overnight at 4 °C. Subsequently, the membranes were incubated with HRP-labeled secondary antibodies against mouse IgG or rabbit IgG (Abcam, Cambridge, MA, USA, 1:5000) for one hour at 37℃. The following primary antibodies were applied to exam the proteins expression: anti-VEGFA (#ab46154, abcam, Cambridge, MA, USA, 1: 1000) and anti-β-tublin (#2148S, Cell Signaling Technology, Beverly, MA, USA, 1:1000). The protein bands were visualized using an enhanced chemiluminescence detection system (Bio-Rad, CA, USA) and quantified using ImageJ software.
CCK-8 assay
In each well of a 96-well plate, 1 × 103 cells in 100 µl of medium were seeded. At 0, 24, 48, 72, and 96 h, the original medium was replaced with a mixture of CCK-8 solution (TransDetect Cell Counting Kit, Transgene, Beijing, China) and fresh medium (without FBS) in a 1:9 ratio. The cells were then incubated at 37 °C for 3 hours. The absorbance of each well was measured at 450 nm using a microplate reader (Bio-Rad, CA, USA) to determine cell viability or proliferation.
In vitro migration and invasion assays
The in vitro migration and invasion abilities of cells were assessed using 24-well Transwells (8 μm pore size, Corning, Inc., NY, USA). A total of 4 × 104 cells in 200 µl fresh medium (without FBS) were seeded in the top chamber. For the invasion assay, each top chamber was coated with 200 mg/ml Matrigel (Corning, Inc., NY, USA), followed by plating 8 × 104 cells in the chamber. The lower chamber contained a 20% concentration of FBS. After 36 h, the invasive cells on the lower layer of the chamber were fixed with 4% paraformaldehyde for 30 min, stained by crystal violet (Beyotime Biotechnology, Shanghai, China) for another 30 min, and then counted. The migration experimental assay was conducted without Matrigel, following the same steps as the invasion assay.
Prognosis analysis
We performed an overall survival analysis in the GSE39582 CRC cohorts using the “survival” package, based on the capillary ECs and tip cells proportions computed by CIBERSORTx. To assess the prognostic role of CA4 and ESM1 expression across various cancer types, we utilized univariate Cox regression and the Kaplan-Meier model. We analyzed four types of prognosis data, including overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI). In the univariate Cox regression analysis, we used continuous expression data of CA4 and ESM1. Furthermore, we performed Kaplan-Meier curve analysis using bivariate expression levels, with the cutoff determined by the “surv-cutpoint” function of the R package “survminer”. We presented the results as a heatmap, including log-rank p value, hazard ratio (HR) with 95% confidence interval (95%CI).
Statistical analysis
R (version 4.2.2), GraphPad Prism (version 9), and Python (version 3.7) were used for statistical analysis. We employed the paired t-test for comparisons involving paired samples, while the Mann-Whitney U test was utilized for assessing differences between two groups in all other instances. The Spearman and Pearson methods were used for correlation tests. For survival analysis, we employed univariate Cox and Log-Rank methods, p value < 0.05 was considered statistically significant.
Results
Enhanced angiogenesis and reduced antigen presentation in CRC ECs
To characterize the transcriptomic heterogeneity of the vascular system and other cell types in human CRC tissues, we examined two large CRC single-cell datasets (GSE178341 and 5-cohorts). These datasets include 289 samples and a total of 743,173 cells, with 19,874 identified as ECs (Fig. 1a). We annotated major clusters based on defining marker genes and identified various cell types, including T cells (CD3D, CD7), B cells (CD79A, MS4A1), plasma cells (MZB1, JCHAIN), mast cells (TPSB2, TPSAB1, CPA3), myeloid cells (LYZ, CD68), ECs (PECAM1, VWF), epithelial cells (KRT8, EPCAM), and fibroblasts (COL1A1, DCN) (Fig. 1a and b, Table S3).
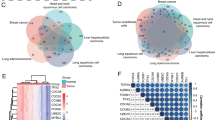
Decreased antigen presentation and enhanced angiogenesis in CRC ECs. (a) UMAP plots of major cell types in GSE178341 (n=100) and 5-cohorts (n=189). (b). Dot plots of marker genes for each major cell type in GSE178341. (c). Differential expressed genes (DEGs) between normal ECs and tumor ECs in GSE178341 and 5-cohorts. Top 10 significant genes are displayed. The criteria for screening DEGs are depicted within the dotted lines. (d). Venn plots of downregulated (left) and upregulated (right) DEGs in CRC ECs in GSE178341 and 5-cohorts. (e). Enrichment analysis of 37 common downregulated DEGs in CRC ECs by metascape. Top 10 terms are displayed. (f). Enrichment analysis of 1,488 common upregulated DEGs in CRC by metascape. Top 10 terms are displayed
In order to investigate the alterations in gene expression and functionality of ECs during CRC tumorigenesis, we subsequently compared the DEGs between tumor ECs and adjacent normal ECs in both GSE178341 and 5-cohorts. Notably, the top 10 DEGs from each cohort showed significant overlap, highlighting the potential key markers in CRC EC transformation. For instance, genes associated with extracellular matrix (ECM) remodeling, such as COL4A1, COL4A2, COL15A1, MMP2, and SPARC, were markedly upregulated in tumor ECs (Fig. 1c, Table S4). Conversely, genes such as FABP5, CD36, and LIFR were more prevalent in adjacent normal ECs.
To understand the functional implications of the observed gene expression changes, we carried out an enrichment analysis of the overlapping DEGs from both cohorts using Metascape (Fig. 1d). This analysis disclosed that the most significant functional change corresponding to the downregulated DEGs (n = 37) was linked to “antigen processing and presentation of exogenous peptide antigen via MHC class II” (Fig. 1e), indicating a decrease in antigen presentation in CRC ECs. On the other hand, for the upregulated DEGs (n = 1488), the term “VEGFA-VEGFR2 signaling pathway” emerged as the most significant (Fig. 1f), suggesting an increase in angiogenic activity within the CRC ECs. Collectively, these findings indicate that during the course of CRC tumorigenesis, ECs exhibit a dual adaptation characterized by enhanced angiogenesis and reduced antigen presentation.
Heterogeneity of CRC ECs and diminished antigen presentation in tip cells
Single-cell analysis has offered unprecedented insights into the phenotypic transformations of ECs in CRC, especially when viewed through the lens of EC heterogeneity. In our study, we classified the 13 EC clusters in GSE178341 into 7 EC subsets, based on characteristic genes (Fig. 2a). These subsets encompass five blood vascular EC subsets: artery ECs (GJA5, FBLN5), capillary ECs (CA4, CD36, and BTNL9), immature ECs (enriched with ribosome-related genes), tip cells (ESM1, PGF, and NID2), and vein ECs (ACKR1, SELP). Additionally, we classified lymphatic ECs (LYVE1, CCL21, and PROX1) and mural cells, which display features of both pericytes (RGS5) and vascular smooth muscle cells (ACTA2) (Fig. 2a-c). Mural cells were broadly defined as ECs in this study. To aid fellow researchers in identifying these EC subsets within CRC, we compiled a summary table of representative genes primarily referencing the top 10 DEGs in GSE178341 and 5-cohorts (Fig. 2c and Table S5).
Heterogeneity of CRC ECs and diminished antigen presentation in tip cells. (a) Log ratio of average fraction per EC clusters in tumor to normal tissue (top). Mann-Whitney U test, **: p < 0.01, ***: p < 0.001. Dot plot showing the scaled average expression and the percentage of expression of marker genes per EC cluster (bottom). (b) UMAP plots of 6,785 ECs colored by EC subset (left) and tissue type (center) in GSE178341 dataset. Bar charts show the proportion of EC subsets in different tissues (right). (c) The table to exhibit the representative genes of EC subsets. (Representative genes are mainly referred from the intersection of the top 10 genes of each subgroup, see Table S2 for details). (d) UMAP plot to show the distribution of RGCC expression (a microvascular EC marker) across ECs. The inset violin plot in the top right corner presents the expression level of RGCC across different blood vascular ECs. (e) UMAP plot to show the distribution of FBLN2 expression (a large vessel EC marker) across ECs. The inset violin plot in the top right corner presents the expression level of FBLN2 across different blood vascular ECs. (f) Dot plot to show the expression of MHC-I and MHC-II genes across different blood vascular ECs. (g-h). The violin plot to show the MHC-I score (g) and MHC-II score (h) across different blood vascular EC subsets
We differentiated ECs into microvascular (composed of capillary ECs, immature ECs, and tip cells) and large vessel ECs (comprising artery and vein ECs) using RGCC [23], a microvascular marker, and FBLN2 [24], a large vessel EC marker, respectively (Fig. 2d-e). Interestingly, we observed a pronounced increase in the proportion of tip cells, accompanied by a significant decline in capillary ECs in tumor ECs when compared to normal ECs (Fig. 2b). However, the proportions of artery and vein ECs remained comparatively stable.
Some EC subtypes are considered semi-professional antigen presenting cells (APCs) as they express genes involved in antigen capture, processing and presentation [25, 26]. To probe the cause behind the weakened antigen presentation in tumor ECs, we further compared the expression of MHC-I (HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, and HLA-G) and MHC-II (HLA-D) genes across different EC subsets. The expression of MHC-I genes was found to be highest in capillary ECs, while MHC-II genes were notably expressed in both capillary and vein ECs (Fig. 2f). Notably, the expression of both MHC-I and MHC-II genes was significantly reduced in tip cells (Fig. 2f). Further, the MHC-I and MHC-II scoring results showed the reduced antigen presentation feature of tip cells in ECs (Fig. 2g-h), indicating that tip cells may be the crucial EC subset contributing to the attenuated antigen presentation in tumor ECs. These findings were affirmed in the 5-cohorts (Figure S1a-g).
In summary, tip cells, which are enriched in tumor ECs, appear to be the key cell type contributing to the impaired antigen presentation in tumor ECs.
Tip cells are associated with CRC tumorigenesis, progression, and poor prognosis
Recognizing the potential value of tip cells in the clinical diagnosis and treatment of CRC, our subsequent research aimed to identify the most specific biomarkers of tip cells and explore their correlation with CRC tumorigenesis, progression, and prognosis. By comparing the differential genes between capillary ECs and tip cells, we confirmed that tip cell markers, such as ESM1 and PGF, along with capillary EC markers like CD4 and CD36, exhibit a high degree of specificity (Fig. 3a, Fig. S2a, and Table S6). Notably, ESM1 expression is almost exclusive to tip cells (Fig. 3b-c, Fig. S2b-c), leading us to consider ESM1 as the most representative marker of tip cells.
Tip cells are associated with CRC tumorigenesis, progression, and poor prognosis. (a). Volcano plot to illustrate differential genes between capillary ECs and tip cells in GSE178341 dataset. (b). Dot plot to display the expression of ESM1 across different major cell types and EC subsets. (c). UMAP visualization shows the expression of ESM1 in all cells and ECs. (d). Heatmap to show the expression difference of tip cell markers (ESM1 and PGF) and capillary markers (CA4 and CD36) between NC and CRC in nine bulk RNA-seq cohorts. The color gradient of each tile signifies changes in the marker expression level, where red represents an increase in CRC, blue represents a decrease, and the depth of the color reflects the statistical significance. NC, normal colorectum or adjacent colorectum. (e). Line graph to depict the proportion change of tip cells and capillary ECs across different stages in the GSE39582 dataset, as evaluated by the CIBERSORTx algorithm. (f). Kaplan-Meier analysis of the overall survival curve in GSE39582 CRC patients with a high or low capillary ECs proportion (based on the 50% cut-off). Log-rank test. (g). Kaplan-Meier analysis of the overall survival curve in GSE39582 CRC patients with a high or low Tip cells proportion (based on the 50% cut-off). Log-rank test. (h). Summary of the prognostic role of the expression of CA4 and ESM1 in TCGA pan-cancer. Numbers represent the count of cancers associated with CA4 or ESM1 expression regarding prognosis (see Figure S3 for details). Two-sample proportion test for ESM1
Compared to normal colorectum (NC), ESM1 and PGF expression levels are significantly higher in CRC across nine bulk RNA-seq datasets (Fig. 3d), while the expression of CD4 and CD36 notably decreased, indicating an increased density of tip cells and decreased density of capillaries during tumorigenesis. In a pan-cancer analysis from TCGA, tumor tissues also showed elevated expression of tip cell markers (Fig. S2d) and decreased capillary markers (Fig. S2e) compared to normal tissues. Additionally, using the CIBERSORTx method based on single-cell data (GSE178341), we calculated the proportion of different blood vascular EC subsets in each CRC sample from a bulk RNA-seq dataset (GSE39582). The results showed an increase in the proportion of tip cells and a decrease in capillary ECs as the cancer stage progressed (Fig. 3e). This finding was supported by analyses of both markers and CIBERSORTx for TNM staging (Fig. S3a-b), suggesting an increase in tip cell density and a decline in capillary density as the tumor progresses.
Next, we explored the relationship between tip cells and CRC prognosis. The results based on the CIBERSORTx approach showed that a high proportion of capillary ECs was associated with a better prognosis (log-rank, p = 0.027) (Fig. 3f), while a high proportion of tip cells correlated with a worse prognosis (log-rank, p = 7.1e-04) (Fig. 3g). This observation is also applicable in a pan-cancer context, where we analyzed the relationship between CA4 and ESM1 expression and the prognosis of 32 types of cancer within TCGA. CA4 demonstrated a protective effect in 8 cancer types and a risk effect in 1 type, whereas ESM1 displayed a protective effect in 3 cancer types and a risk effect in 11 types (p = 1.4e-04, two-sample proportion test) (Fig. 3h, and Fig. S3c-d). These results suggest that, in comparison to capillary ECs, which exhibit protective characteristics in CRC patients, tip cells display significantly malignant traits.
In conclusion, tip cells are associated with the initiation, progression, and poor prognosis of CRC.
The relationship between tip cells and angiogenesis
Tip cells are integral to angiogenesis, as they spearhead new vessel growth by sensing gradients of angiogenic signals such as VEGFA and guiding the direction of the new sprout [12, 16]. To understand the relationship between tip cells and angiogenesis in CRC more deeply, we examined the expression of growth factors (GFs) such as VEGFA, VEGFB, VEGFC, PGF, PDGFA, PDGFB, ANGPT2, and APLN [16], and growth factor receptors (GFRs) including FLT1, KDR, FLT4, and APLNR across various major cell types and EC subsets. We found that most of these GFs and their receptors are enriched in ECs (Fig. 4a and Fig. S4a). Notably, compared with other blood vascular ECs, the majority of these GFs (e.g., PGF, PDGFA, PDGFB, ANGPT2, and APLN) and GFRs such as KDR, FLT1 (VEGFR1), and APLNR are most highly expressed in tip cells, indicating a pro-angiogenic phenotype of tip cells in CRC. In addition, the VEGFC-VEGFR3 pair, closely related to lymphangiogenesis [12], demonstrated distinct distribution: VEGFC was primarily observed in artery ECs, while FLT4 (VEGFR3) was prevalent in lymphatic ECs (Fig. 4a).
The relationship between tip cells and angiogenesis. (a). Dot plots showing the expression of growth factors and growth factor receptors in different major cell types (left) and EC subsets (right) in GSE178341. (b). UMAP plot displaying the expression of VEGFA and KDR in GSE178341. (c). Violin plot displaying the expression of angiogenesis signature of different major cells in GSE178341. (d). Bar graphs comparing the expression difference of VEGFA between tumor and adjacent normal tissue in myeloid cells (left) and epithelial cells (right). Each point represents the mean expression of VEGFA in myeloid or epithelial cells within the respective sample. (e). Bar graph comparing the expression of VEGFA between tumor ECs and normal tissue ECs. (f). Spearman correlation analysis of VEGFA expression in myeloid cells and KDR expression in ECs. Blue dots represent adjacent normal tissue samples, while red dots represent CRC samples. (g). Spearman correlation analysis between VEGFA expression in myeloid cells and KDR expression in ECs. (h). Spatial plots of H&E staining (column 1), tissue regions (column 2), ECs (PECAM1) (column 3), tip cells (ESM1) (column 4), and KDR expression (column 5) in a CRC sample. (i). Comparison of the proportion of KDR positive expression (exp>0) between PECAM1+ESM1+ and PECAM1+ESM1- spatial plots. (j). Dot plots display the expression of regional marker genes, cell type marker genes, and the expression of VEGFR and VEGF-related genes in different tissue regions. (k). Diagram of the long-tail effect of VEGFA expression in CRC. (l). Immunofluorescence staining of human CRC tissue. VEGFA (red), macrophages (CD68, green), cytokeratin-positive tumor cells (pan-CK, pink), DAPI (blue), Bar, 100 μm. (m). Immunofluorescence staining of human CRC tissue. VEGFA (red), ECs (CD31, green), DAPI (blue), Bar, 100 μm
VEGF-A, ubiquitously expressed in virtually all malignant tumors, is widely recognized as the principal factor propelling tumor angiogenesis [27]. The in vivo angiogenic response to VEGFA is predominantly orchestrated through the activation of VEGFR2 [28]. Notably, our single-cell results indicated that VEGFA expression was the highest in myeloid cells, followed by epithelial cells (Fig. 4a-b, Fig. S4a-b, and Table S3). Moreover, the highest angiogenesis score was identified in myeloid cells (Fig. 4c and Fig. S4c), suggesting their pivotal role in fostering angiogenesis. We found that VEGFA expression is significantly higher in tumor myeloid cells compared to normal myeloid cells, and epithelial cells express more VEGFA than normal epithelial cells (Fig. 4d and Fig. S4d). Similarly, tumor ECs express significantly more KDR expression than normal ECs (Fig. 4e), indicating elevated VEGFA and KDR expression levels in the TME. Although KDR expression in ECs is strongly positively correlated with VEGFA from both myeloid cells (r = 0.45, p < 0.001) (Fig. 4f) and epithelial cells (r = 0.54, p < 0.001) (Fig. 4g), the correlation is higher with VEGFA from epithelial cells. This suggests that, given their greater abundance, epithelial cells are the primary cell type expressing VEGFA.
To further investigate the relationship between the spatial distribution of tip cells and angiogenesis, we used spatial transcriptomics data to analyze the distribution of ECs in CRC tissues. We classified 3,138 spots into three regions - normal, stromal, and tumor regions, based on their transcriptomic expression and Hematoxylin and Eosin (HE) tissue data (Fig. 4h). Our findings revealed that the proportion of spots showing KDR expression (expression > 0) was significantly higher in PECAM + ESM1 + spots (tip cells) compared to PECAM + ESM1- spots (other ECs) (40/93 vs. 102/756, p = 4.60e-15) (Fig. 4i). A dot plot illustrated the concentration of myeloid/macrophage cells, endothelial/tip cells, and most VEGF/VEGFRs in the stromal region (Fig. 4j). Interestingly, VEGFA expression was significantly higher in areas rich in epithelial cells, possibly due to dilution of VEGFA derived from myeloid cells in stromal spots by other cell types. In conclusion, the spatial transcriptomics data highlighted the enrichment of KDR in tip cells and emphasized the long-tail effect of VEGFA: While VEGFA expression was highest in individual myeloid cells, epithelial cells constituted the majority of VEGFA-expressing cells (Fig. 4k).
Immunofluorescence results demonstrated a significant accumulation of VEGFA protein in regions positive for the EC marker (CD31), as compared to regions positive for epithelial cell marker (pan-CK) and macrophage marker (CD68) (Fig. 4l and m). This suggests that VEGFA protein may be efficiently recruited to its target cells, namely ECs, thereby promoting angiogenesis.
ESM1 maintains VEGFA-KDR-ESM1 positive feedback loop and promotes CRC proliferation and migration
Next, we sought to confirm ESM1’s role in perpetuating the VEGFA-KDR-ESM1 positive feedback loop in CRC. GSEA of ESM1 in TCGA-CRC, conducted using LinkedOmics, affirmed ESM1’s involvement in the angiogenesis pathway (Fig. 5a). By analyzing 60 pairs of CRC tissues and corresponding para-cancer tissues, we noted higher expression levels of ESM1, KDR, and VEGFA in CRC tissues (Fig. 5b). ESM1 correlated significantly with both VEGFA (r = 0.6585, p < 0.001) and KDR (r = 0.8649, p < 0.001) in these patients (Fig. 5c). A parallel strong correlation between ESM1 with KDR (r = 0.51, p < 0.001) and VEGFA (r = 0.4, p < 0.001) was observed in TCGA-CRC (Fig. 5d). Immunofluorescence results further revealed heightened expression and spatial colocalization of ESM1 and KDR in CRC (Fig. 5e).
ESM1 promotes VEGFA-KDR-ESM1 positive feedback loop and enhances CRC proliferation and migration. (a). Significantly enriched Gene Ontology Biological Process (GO BP) terms in genes coexpressed with ESM1 in the TCGA-CRC cohort. FDR denotes the false discovery rate. (b). qRT-PCR analysis to show ESM1, VEGFA, and KDR expression levels in 60 pairs of human CRC tissues and paired NC tissue (Paired t test). (c). Correlation analysis between the expression of ESM1 and VEGFA, as well as between the expression of ESM1 and KDR in 59 CRC patients (Pearson test). An outlier was identified and excluded from the initial set of 60 samples. (d). Correlation analysis between the expression of ESM1 and VEGFA, as well as between the expression of ESM1 and KDR in the TCGA-CRC cohort (Pearson test, n=367). (e). Immunofluorescence staining of human CRC tissue and paired adjacent normal tissue. ESM1 (green), KDR (pink), CD31 (red), DAPI (blue). (f). qRT-PCR analysis to show VEGFA mRNA expression level in HCT-116 after manipulating different concentrations of ESM1 protein. (g). WB analysis to show VEGFA protein expression level in HCT-116 after manipulating different concentrations of ESM1 protein. (h). CCK-8 assay to illustrate the alteration of CRC cells’ proliferative capacity after manipulating different concentrations of ESM1 protein. The optical density (OD) was measured daily for 5 days. The bar graph in the lower part displays the statistical results for day five shown in the upper part. (i). Transwell analysis to illustrates the migrative and invasive capability of CRC cells after manipulating different concentrations of ESM1 protein. The bar graph in the lower part displays the statistical differences depicted in the upper part. All data are shown as the mean ± SD. *: p < 0.05, **: p < 0.01, ***: p < 0.001
Subsequently, we conducted in vitro experiments to evaluate the impact of ESM1 on VEGFA expression in CRC cells. Recombinant ESM1 protein was added to the culture medium at varying concentrations for this purpose. As the concentration of ESM1 protein increased, both qRT-PCR and WB analysis showed a corresponding increase in VEGFA expression within CRC cells (Fig. 5f and g). The above evidence collectively supports that ESM1 maintains the VEGFA-KDR-ESM1 positive feedback loop in CRC.
Further in vitro experiments were conducted to explore the effects of ESM1 on CRC cell proliferation and migration. Specifically, as the concentration of ESM1 increased, there was a corresponding rise in the proliferative and migratory capabilities of CRC cells (Fig. 5h and i). These results suggest that ESM1 can stimulate the expression of VEGFA in CRC cells, possibly thereby promoting the proliferative and migratory properties of CRC cells.
Intercellular communication unveils NRP1/2 assisting VEGFA in activating KDR in tip cells
In this section, our aim was to uncover potential mechanisms for intercellular communication involving VEGFA and angiogenesis promotion within the CRC TME. To do this, we utilized GSE178341 dataset and conducted a CellphoneDB analysis. We discovered that tip cells had a higher number of interactions with other cell types when compared to other blood vascular ECs (Fig. 6a). Looking specifically at VEGFA-related cellular communication, we observed strong interactions between VEGFA in myeloid and epithelial cells and NRP1/2 in tip cells (Fig. 6b). NRP1/2 act as co-receptors for VEGFA and have been shown to promote angiogenesis by strengthening the binding of VEGFA and VEGFR [29,30,31]. Additionally, NRP2 has been reported to be critical for VEGFC-VEGFR3-induced lymphatic sprouting [32]. Our single-cell analysis further highlighted that NRP1/2 expression was enriched in tip cells, and that NRP2 expression was highest in lymphatic ECs (Fig. 6c). Results from TCGA-CRC also confirmed a strong positive correlation between NRP1 (Spearman r = 0.83, p < 0.001) and NRP2 (Spearman r = 0.67, p < 0.001) with KDR (Fig. 6d). Taken together, these findings suggest that the NRP1/2 enriched in tip cells assists in activating the VEGFA-KDR signaling pathway (Fig. 6e).
Intercellular communication unveils NRP1/2 Assisting VEGFA in activating KDR in tip cells. (a). Heatmap generated by CellphoneDB showing the potential ligand-receptor interactions between blood vascular EC subsets and other major cell types in CRC (GSE178341). Numbers indicate the number of potential ligand-receptor pairs. (b). Dot plots of VEGFA-related cellular interactions between blood vascular EC subsets and myeloid cells and epithelial cells. (c). Dot plots to show average expression of NRP1 and NRP2 across different major cell types and EC subsets. (d). Correlation analysis of KDR with NRP1 and NRP2 expression in TCGA-CRC (Spearman test, n=367). (e). Schematic diagram of NRP1/2 assisting VEGFA in activating KDR in tip cells
Effective PD-1 blockade treatment significantly reduces tip cells in CRC
In this study, we leveraged the GSE205506 dataset [33], which includes information on the response to PD-1 blockade, enabling us to investigate the impact of ICI therapy on ECs within the CRC TME (Fig. 7a). From a total of 324,030 cells, we identified 18,151 ECs (Fig. 7b, Fig. S5a and Table S3).
Effective PD-1 blockade treatment significantly reduces tip cells in CRC. (a). Schematic diagram of tissue samples before and after immunotherapy. (b). UMAP plot of 18,151 ECs colored by sample types in GSE205506. (c). UMAP plot of ESM1 expression in ECs. (d). Average ESM1 expression in ECs among patient groups. Each point represents the mean ESM1 expression in the ECs of that sample. Mean ± SD, Mann-Whitney U test. (e). Volcano plot of DEGs between ECs in the -ICI group and +ICI/pCR group. (f). Enrichment analysis of the 269 upregulated DEGs in the -ICI group by Metascape. Top 5 terms are displayed. (g). Enrichment analysis of the 938 upregulated DEGs in the +ICI/pCR group by Metascape. Top 5 terms are displayed. (h). Dot plot comparing the expression of MHC-I and MHC-II genes among patient groups. (i). Dot plot comparing the expression of tip marker, growth factors, and growth factor receptor-related genes among patient groups. (j). Average KDR expression in ECs among patient groups. Each point represents the mean KDR expression in the ECs of that sample. Mean ± SD, Mann-Whitney U test
We found that, in comparison to the treatment naïve (-ICI) group (n = 10) and the ICI treated non-pathological complete response (+ ICI/non-pCR) group (n = 4), the ECs in the ICI treated pathological complete response (+ ICI/pCR) group (n = 13) showed a significant decrease in ESM1 expression (Fig. 7c-d), as well as other tip cell markers (Fig. 7i). This suggests that an effective PD-1 blockade considerably reduces the number of tip cells within the TME.
When analyzing the DEGs between the -ICI and + ICI/pCR groups, we found that the + ICI/pCR group displayed an increase in “MHC class II protein complex” within the ECs, and a decrease in “VEGFA-VEGFR2 signaling” (Fig. 7e-g and Table S7). This implies that effective immune therapy enhances antigen presentation in ECs, while reducing the traits of angiogenesis. Supporting this observation, the + ICI/pCR group exhibited increased expression of MHC-I and MHC-II genes (Fig. 7h) and decreased expression of GFs and GFRs (Fig. 7i and Fig. S5b), compared to the -ICI group and the + ICI/non-pCR group.
In addition, we observed a notable decrease in the expression of KDR in ECs (Fig. 7j) and VEGFA in epithelial cells (Fig. S5c) in the + ICI/pCR group, implying that effective immune therapy disrupts the positive feedback loop of VEGFA-KDR-ESM1 within the CRC TME.
Discussion
The purpose of this research is to provide an in-depth characterization of the heterogeneity of ECs in CRC and to elucidate the mechanisms by which ECs promote angiogenesis. The insights gained from this study are as follows:
Our study demonstrates that the phenotypic shift in tumor ECs is primarily governed by tip cells. Capillaries, acting as microvessels and displaying enhanced transcriptional plasticity [23], are known to express genes associated with antigen processing and presentation, thereby bolstering immune responses [9]. Recent advancements in single-cell research have unveiled a trend within tumor ECs where antigen presentation diminishes while angiogenesis intensifies, compared to their normal counterparts [9, 23, 26]. In our study, we find this trait to be chiefly driven by tip cells, which display a significant reduction in antigen presentation and enhanced angiogenesis. Moreover, tip cells substantially increase in the CRC TME, replacing the ecological niche originally occupied by capillary ECs in normal tissue (Fig. 8a).
Changes of microvascular ECs in CRC TME. (a). During the occurrence and progression of CRC, the proportion of tip cells in ECs increases, while the proportion of capillary ECs decreases. This suggests that in the tumor microenvironment, tip cells with high expression of ESM1, PGF, and NID2 occupy the microvascular niche that belonged to capillary ECs with high expression of CA4, CD36, and BTNL9 in normal tissue. (b). Tip cells, characterized by diminished antigen presentation and enhanced angiogenesis, shift the overall microvascular EC profile from the anti-tumor traits of capillary ECs to a pro-tumor phenotype. Furthermore, high ESM1 expression in tip cells potentially sustains the VEGFA-KDR-ESM1 feedback loop through interactions with VEGFA from myeloid and epithelial cells, promoting angiogenesis and CRC progression
Our research highlights the potential clinical translational value of tip cells as biomarkers. Through various methodologies and multiple datasets, we have confirmed that the number and proportion of tip cells increase with the onset and progression of CRC, correlating with poorer prognoses. This contrasts significantly with capillary ECs. These findings are also validated across pan-cancer studies. We speculate that this might be related to the diminished antigen presentation and increased angiogenic traits of tip cells within CRC. Tip cells, characterized by these traits, shift the overall microvascular EC profile from the anti-tumor properties of capillary ECs towards a pro-tumor phenotype. Furthermore, we observe that effective immunotherapy can significantly reduce tip cells, indicating the potential of tip cells as a marker for assessing the effectiveness of immunotherapy and anti-angiogenic therapy. In summary, these findings suggest that tip cells and their markers, such as ESM1, could serve as critical malignancy indicators in CRC, guiding the diagnosis, disease assessment, and prognostic treatment of CRC.
Our study also reveals the potential mechanisms by which tip cells promote angiogenesis. Although tip cells are recognized for their role in angiogenesis, the specific mechanisms driving this process remain unclear [6, 16]. We discovered that most GFs and their receptors (GFRs), including KDR, are enriched in tip cells. This suggests that tip cells represent a key cellular subset driving angiogenesis within CRC. Importantly, our research identifies ESM1 as not just a marker gene for tip cells, but as a crucial molecule that drives angiogenesis. Recent studies in cervical squamous cell carcinoma and ovarian cancer have demonstrated that ESM1 can enhance VEGFA expression [34, 35], and correspondingly, VEGFA has been reported to increase ESM1 expression via VEGFR2 activation [36, 37]. Our results echo these findings, suggesting a positive feedback loop where VEGFA upregulates ESM1 expression in tip cells, which in turn enhancing VEGFA-mediated signaling in CRC (Fig. 8b). Additionally, through intercellular communication analysis, we unveil NRP1/2 assisting VEGFA in activating KDR in tip cells. These findings support that the VEGFA-KDR-ESM1 feedback loop may be a key pathway by which tip cells stimulate angiogenesis. Interestingly, we also observe a long-tail effect in VEGFA expression; VEGFA is primarily expressed by epithelial cells, but the highest level of VEGFA expression is found in individual myeloid cells.
This study offers insights into potential new strategies for immunotherapy that are specifically oriented towards ECs. Currently, approved AATs primarily focus on blocking essential pro-angiogenic targets such as VEGF [12, 16, 38]. Bevacizumab, a monoclonal antibody that neutralizes VEGFA, has demonstrated effectiveness in the treatment of metastatic CRC, thereby improving patient outcomes [27, 28]. Despite these advances, the performance of VEGF blockade therapies can still be limited by factors such as resistance [11, 39]. Our research raises the prospect of fine-tuning these approaches. Through scRNA-seq analysis, we identified novel tumor EC targets that may potentially enhance the efficacy of immunotherapies. Specifically, our findings propose that strategies aimed at modulating ESM1 expression or selectively eliminating ESM1 + tip cells could be beneficial. Moreover, our observations indicate that effective anti-PD-1 therapy can decrease the number of tip cells within the CRC TME, disrupting the VEGFA-KDR-ESM1 feedback loop - an important mechanism through which immunotherapy could potentially exert its effects. Supporting this notion, recent preclinical studies have shown that anti-VEGF treatment can enhance the effectiveness of anti-PD-L1 therapy, promoting an increased infiltration of cytotoxic T lymphocytes and subsequent tumor cell destruction [40]. Other research has demonstrated that a dual anti-angiogenesis approach, employing dual-specificity anti-VEGF/Ang2 antibodies, can amplify the effect of anti-PD-L1 therapy in various models [41]. Therefore, future investigations could explore the potential of innovative combinations of AAT and immunotherapy to potentially further enhance remission rates in cancer patients.
In conclusion, our study highlights the pivotal role of tip cells in driving the angiogenesis enhancement and decreased antigen presentation observed in tumor ECs. We found ESM1, a molecule uniquely expressed in tip cells, to facilitate a self-enhancing loop with VEGFA, thereby stimulating angiogenesis and promoting CRC tumor growth. Moreover, we observed that successful immunotherapy significantly reduces the number of tip cells in CRC tissue, interrupting the VEGFA-KDR-ESM1 positive feedback loop. Our investigation into the heterogeneity of ECs in CRC at a single-cell level offers important insights that may contribute to the development of more effective immunotherapies targeting tip cells in CRC.
Data availability
The data used in this study are all from public datasets, which are detailed in the Methods section of the manuscript. The codes used in this study are available on Github (https://github.com/LPC19970117/Endo_tip).
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72(1):7–33
Fan A, Wang B, Wang X, Nie Y, Fan D, Zhao X, Lu Y (2021) Immunotherapy in colorectal cancer: current achievements and future perspective. Int J Biol Sci 17(14):3837–3849
Zaanan A, Shi Q, Taieb J, Alberts SR, Meyers JP, Smyrk TC, Julie C, Zawadi A, Tabernero J, Mini E et al (2018) Role of deficient DNA mismatch repair status in patients with stage III colon cancer treated with FOLFOX adjuvant chemotherapy: a pooled analysis from 2 randomized clinical trials. JAMA Oncol 4(3):379–383
Nordholm-Carstensen A, Krarup PM, Morton D, Harling H, Danish Colorectal Cancer Group (2015) Mismatch repair status and synchronous metastases in colorectal cancer: a nationwide cohort study. Int J Cancer 137(9):2139–2148
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D et al (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372(26):2509–2520
Pasut A, Becker LM, Cuypers A, Carmeliet P (2021) Endothelial cell plasticity at the single-cell level. Angiogenesis 24(2):311–326
Potente M, Mäkinen T (2017) Vascular heterogeneity and specialization in development and disease. Nat Rev Mol Cell Biol 18(8):477–494
Trimm E, Red-Horse K (2023) Vascular endothelial cell development and diversity. Nat Rev Cardiol 20(3):197–210
Zeng Q, Mousa M, Nadukkandy AS, Franssens L, Alnaqbi H, Alshamsi FY, Safar HA, Carmeliet P (2023) Understanding tumour endothelial cell heterogeneity and function from single-cell omics. Nat Rev Cancer
Naito H, Iba T, Takakura N (2020) Mechanisms of new blood-vessel formation and proliferative heterogeneity of endothelial cells. Int Immunol 32(5):295–305
Becker LM, Chen S-H, Rodor J, de Rooij LPMH, Baker AH, Carmeliet P (2023) Deciphering endothelial heterogeneity in health and disease at single-cell resolution: progress and perspectives. Cardiovasc Res 119(1)
Apte RS, Chen DS, Ferrara N (2019) VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 176(6):1248–1264
Goveia J, Rohlenova K, Taverna F, Treps L, Conradi L-C, Pircher A, Geldhof V, de Rooij LPMH, Kalucka J, Sokol L et al (2020) An Integrated Gene Expression Landscape Profiling Approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell 37(1)
Geldhof V, de Rooij LPMH, Sokol L, Amersfoort J, De Schepper M, Rohlenova K, Hoste G, Vanderstichele A, Delsupehe A-M, Isnaldi E et al (2022) Single cell atlas identifies lipid-processing and immunomodulatory endothelial cells in healthy and malignant breast. Nat Commun 13(1):5511
Kang B, Camps J, Fan B, Jiang H, Ibrahim MM, Hu X, Qin S, Kirchhoff D, Chiang DY, Wang S et al (2022) Parallel single-cell and bulk transcriptome analyses reveal key features of the gastric tumor microenvironment. Genome Biol 23(1):265
Lugano R, Ramachandran M, Dimberg A (2020) Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci 77(9):1745–1770
Joanito I, Wirapati P, Zhao N, Nawaz Z, Yeo G, Lee F, Eng CLP, Macalinao DC, Kahraman M, Srinivasan H et al (2022) Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat Genet 54(7):963–975
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10(1):1523
Vasaikar SV, Straub P, Wang J, Zhang B (2018) LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 46(D1):D956–963
Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R (2020) CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 15(4):1484–1506
Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O’Brien SA, He Y, Wang L, Zhang Q, Kim A et al (2020) Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181(2):442–459e429
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D et al (2019) Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37(7):773–782
Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen L-A, Veys K et al (2020) Single-cell transcriptome atlas of murine endothelial cells. Cell 180(4)
Li Q, Zhu Z, Wang L, Lin Y, Fang H, Lei J, Cao T, Wang G, Dang E (2021) Single-cell transcriptome profiling reveals vascular endothelial cell heterogeneity in human skin. Theranostics 11(13):6461–6476
Huinen ZR, Huijbers EJM, van Beijnum JR, Nowak-Sliwinska P, Griffioen AW (2021) Anti-angiogenic agents - overcoming tumour endothelial cell anergy and improving immunotherapy outcomes. Nat Rev Clin Oncol 18(8):527–540
Amersfoort J, Eelen G, Carmeliet P (2022) Immunomodulation by endothelial cells - partnering up with the immune system? Nat Rev Immunol 22(9):576–588
Sitohy B, Nagy JA, Dvorak HF (2012) Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Res 72(8):1909–1914
Claesson-Welsh L, Welsh M (2013) VEGFA and tumour angiogenesis. J Intern Med 273(2):114–127
Sarabipour S, Mac Gabhann F (2018) VEGF-A121a binding to Neuropilins - A concept revisited. Cell Adh Migr 12(3):204–214
Zhao L, Chen H, Lu L, Wang L, Zhang X, Guo X (2021) New insights into the role of co-receptor neuropilins in tumour angiogenesis and lymphangiogenesis and targeted therapy strategies. J Drug Target 29(2):155–167
Islam R, Mishra J, Bodas S, Bhattacharya S, Batra SK, Dutta S, Datta K (2022) Role of Neuropilin-2-mediated signaling axis in cancer progression and therapy resistance. Cancer Metastasis Rev 41(3):771–787
Xu Y, Yuan L, Mak J, Pardanaud L, Caunt M, Kasman I, Larrivée B, Del Toro R, Suchting S, Medvinsky A et al (2010) Neuropilin-2 mediates VEGF-C-induced lymphatic sprouting together with VEGFR3. J Cell Biol 188(1):115–130
Li J, Wu C, Hu H, Qin G, Wu X, Bai F, Zhang J, Cai Y, Huang Y, Wang C et al (2023) Remodeling of the immune and stromal cell compartment by PD-1 blockade in mismatch repair-deficient colorectal cancer. Cancer Cell
Li Y-K, Zeng T, Guan Y, Liu J, Liao N-C, Wang M-J, Chen K-X, Luo X-Y, Chen C-Y, Quan F-F et al (2023) Validation of ESM1 related to Ovarian Cancer and the Biological function and prognostic significance. Int J Biol Sci 19(1):258–280
Huang Y, Li D, Yao L, Yu D, Wang L, Tang X-b (2022) Su X-m: ESM1/VEGFα/ERK signaling axis augments cell proliferation and tumor angiogenesis in human cervical squamous cell carcinoma
Rocha SF, Schiller M, Jing D, Li H, Butz S, Vestweber D, Biljes D, Drexler HCA, Nieminen-Kelhä M, Vajkoczy P et al (2014) Esm1 modulates endothelial tip cell behavior and vascular permeability by enhancing VEGF bioavailability. Circ Res 115(6):581–590
Yazici SE, Gedik ME, Leblebici CB, Kosemehmetoglu K, Gunaydin G, Dogrul AB (2023) Can endocan serve as a molecular hepatostat in liver regeneration? Mol Med 29(1):29
Marziano C, Genet G, Hirschi KK (2021) Vascular endothelial cell specification in health and disease. Angiogenesis 24(2):213–236
De Bock K, Mazzone M, Carmeliet P (2011) Antiangiogenic therapy, hypoxia, and metastasis: risky liaisons, or not? Nat Rev Clin Oncol 8(7):393–404
Allen E, Jabouille A, Rivera LB, Lodewijckx I, Missiaen R, Steri V, Feyen K, Tawney J, Hanahan D, Michael IP et al (2017) Combined antiangiogenic and anti-PD-L1 therapy stimulates tumor immunity through HEV formation. Sci Transl Med 9(385)
Schmittnaegel M, Rigamonti N, Kadioglu E, Cassará A, Wyser Rmili C, Kiialainen A, Kienast Y, Mueller H-J, Ooi C-H, Laoui D et al (2017) Dual angiopoietin-2 and VEGFA inhibition elicits antitumor immunity that is enhanced by PD-1 checkpoint blockade. Sci Transl Med 9(385)
Zhenyu Xie LN, Kunli, Du et al (2023) Single-cell analysis reveals endothelial cell heterogeneity in colorectal cancer: tip cells drive enhanced angiogenesis and reduced antigen presentation. Res Square
Acknowledgements
We acknowledge the researchers who contributed to the public databases and those who provided data related to human medicine. This manuscript has been released as a pre-print at ResearchSquare [42].
Funding
This work was supported by the National Natural Science Foundation of China (82072655), Scientific and technological innovation team of Shaanxi Innovation Capability Support Plan (2023-CX-TD-67), and Key R&D Plan of Shaanxi Province (2022SF-603).
Author information
Authors and Affiliations
Contributions
ZYX and LRN designed the study. KLD and LC collected the tissue samples. ZYX and LRN collected the data. ZYX, LRN, FF and GZZ analyzed the data. ZYX, KLD, HJD, LLD, and XYD visualized the data. ZYX and LRN drafted the manuscript. SCD, JZ and JYZ revised the manuscript. All authors have read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Ethical approval
The research was approved by the medical ethics committee of the first affiliated hospital of the Fourth Military Medical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xie, Z., Niu, L., Du, K. et al. Endothelial cell heterogeneity in colorectal cancer: tip cells drive angiogenesis. Cell. Mol. Life Sci. 81, 365 (2024). https://doi.org/10.1007/s00018-024-05411-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-024-05411-z