Abstract
Primary cilia are conserved organelles that mediate cellular communication crucial for organogenesis and homeostasis in numerous tissues. The retinal pigment epithelium (RPE) is a ciliated monolayer in the eye that borders the retina and is vital for visual function. Maturation of the RPE is absolutely critical for visual function and the role of the primary cilium in this process has been largely ignored to date. We show that primary cilia are transiently present during RPE development and that as the RPE matures, primary cilia retract, and gene expression of ciliary disassembly components decline. We observe that ciliary-associated BBS proteins protect against HDAC6-mediated ciliary disassembly via their recruitment of Inversin to the base of the primary cilium. Inhibition of ciliary disassembly components was able to rescue ciliary length defects in BBS deficient cells. This consequently affects ciliary regulation of Wnt signaling. Our results shed light onto the mechanisms by which cilia-mediated signaling facilitates tissue maturation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary cilia are microtubule-based organelles that protrude from the cellular membrane and are anchored by modified centrioles referred to as basal bodies. Cells use primary cilia as specific sensory organelles to detect external cues and mediate intracellular signaling during cell differentiation, organogenesis and tissue homeostasis. Cilia are involved in several signaling pathways including Wingless-related integration site (Wnt), Hedgehog (Hh) and platelet-derived growth factor receptor α (PDGFRα) [1,2,3,4], are evolutionally conserved and vital for diverse organisms ranging from metazoans to mammals. Although they are present on almost every cell type [5], not all cells retain their cilium throughout development and adulthood. In many tissues, for example the Organ of Corti in the mammalian cochlea [6], the luminal epithelial cells within mammary gland [7] and corneal endothelial cells in the eye [8], the cilium is vital during development but disassembles upon maturation. However, the physiological consequences of ciliary disassembly are largely unexplored. Cilia can disassemble in response to environmental stress [9], during differentiation or cell cycle progression [10, 11]. This shortening or absorption of the cilium will inevitably influence its signaling and functional role [12,13,14,15,16].
The importance of cilia during organogenesis and function of the vertebrate eye is well documented, particularly in photoreceptor cells [17, 18]. However, we only recently showed that the retinal pigment epithelium (RPE), a pigmented monolayer epithelium essential for photoreceptor development and visual function, also relies on the primary cilium for maturation through the regulation of canonical Wnt signaling [19]. Numerous reports have shown that RPE-derived cell lines are ciliated in vitro [20, 21] and although ciliary assembly and disassembly pathways have been studied in these models, the precise mechanisms are still being determined. The effect of these processes on downstream signaling pathways has not been established [22, 23]. Moreover, RPE ciliogenesis and ciliary disassembly have not been investigated in vivo. Although reports have suggested that cilia in the RPE are retained throughout adulthood [24], a report from Nishiyama suggested that they disappear in the adult rat RPE [25]. Therefore, we set out to investigate the extent of ciliation in developing vertebrate RPE and to understand the molecular mechanisms underlying ciliary disassembly during development.
Various ciliary proteins including Bardet–Biedl Syndrome (BBS) and Nephronophthisis (NPHP) proteins show ciliary localization [26] and regulate cilia length [27, 28]. BBS proteins have been shown to direct ciliary trafficking, whereas many of the NPHP proteins function as ciliary gatekeepers [26, 28,29,30,31]. Certain mutations in the genes encoding these proteins can cause severe ciliopathy phenotypes including retinal degeneration, cystic kidneys, central obesity and situs inversus [32, 33].
Ciliary disassembly is known to be regulated by various cell cycle regulators. This process is mediated by human enhancer of filamentation 1 (HEF1/NEDD9) [34], which translocates from focal adhesions to the basal body, a structure derived from the mother centriole at the base of the cilium. Translocation of the scaffold protein HEF1 activates Aurora A kinase (AurA), which in turn activates histone deacetylase 6 (HDAC6), destabilizing the microtubule axoneme and thereby causing ciliary disassembly [22]. Inversin (NPHP2) influences ciliary disassembly by regulating AurA [35] and Wnt signaling [36]. BBSome-interacting protein 1 (BBIP10/BBS18), an accessory BBS protein, has been shown to directly interact with HDAC6, thereby modulating acetylation and stabilization of cytoplasmic microtubules [37]. BBS proteins have also been shown to modulate Wnt signaling via degradation of β-catenin [19, 38, 39], which is controlled by precise coordination of phosphorylation and dephosphorylation events [40, 41].
In this study, we investigated the influence of ciliary disassembly components in the developing RPE in vivo and in vitro. Furthermore, we sought to elucidate how BBS proteins mediate ciliary disassembly and how this influences downstream signaling cascades involved in tissue morphogenesis.
Results
Primary cilia are transiently expressed in the developing RPE
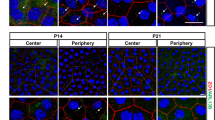
We characterized primary cilia expression in mouse flatmount RPE at different stages of development using immunohistochemistry, RNA expression and electron microscopy. An antibody against ADP Ribosylation Factor-Like GTPase 13B (Arl13b) was used to identify primary cilia. Co-staining with other cilia (acetylated α-tubulin) and basal body markers (Pericentrin2, GT335) confirmed Arl13b as a reliable cilia marker in the RPE (Fig. 1a, b; Supp. Figure 1a–d). Tight junction marker Zonula occludens-1 (ZO-1) (Fig. 1a, b; Supp. Figure 1b, c) and β-catenin were used as membrane markers to visualize cell borders (Supp. Figure 1d). We performed stainings at various time points from E14.5 onward and observed that the expression of primary cilia in the developing RPE diminishes as the RPE matures (Fig. 1c–f). E14.5 was the earliest time point at which cilia were reliably observed and E18.5 was chosen instead of post-natal stages in order to circumvent embryonic lethality. The highest percentage of ciliated cells was observed at E14.5 and E16.5 (69.8 and 64%, respectively). By E18.5, only 26% of cells in the RPE were ciliated, suggesting that the primary cilium had retracted in the majority of the cells (Fig. 1f). Changes in cilia length accompanied cilia retraction, decreasing from 2.63 µm at E16.5 to 0.95 µm at E18.5 (Fig. 1g–j). In support of developmentally dependent transient expression of the primary cilium, we also observed differences in ciliation between the center and periphery of the RPE (compare Fig. 1c–e and Supp. Figure 1e–h). Ciliated cells were rarely identified after E18.5. In P1 and adult RPE, only a few cells could be identified with short stumpy Arl13b and acetylated α-tubulin positive cilia (Fig. 2a; Supp. Figure 1i–k). As previously documented [42], we also observed a developmental increase in cell size (Fig. 1c–e, Supp. Figure 1e–g). Because many cells were observed with cilia at embryonic timepoints, these data suggest that the primary cilium retracts as the RPE develops.
The primary cilium is transiently expressed during RPE development. Representative high-resolution immunofluorescence images of E16.5 mouse RPE flatmounts labeled with antibodies against ciliary structures show co-localization of Arl13b and acetylated α-tubulin extending from the basal body (a, b). Arl13b (axoneme marker, red); Pericentrin 2 (PCNT2, basal body marker, cyan); acetylated α-tubulin (Ac. tubulin, axoneme marker, cyan); Zona Occludens (ZO-1, cell junctions, green). Low magnification immunofluorescence images show ciliation (number of ciliated cells) at three embryonic timepoints (c–e). Boxplots show a significant decrease in the number of ciliated cells from E14.5 to E18.5 (f). E14.5 n = 1700 cells, E16.5 n = 750 cells, E18.5 n = 650 cells. Boxplots of cilia length demonstrate that mouse RPE cilia are longest at E16.5 (g). n = 25 for each age group. High-resolution immunofluorescence images of cilia (Arl13b, red) highlight differences in ciliary length between E14.5 and E18.5 (h–j) as quantified in g. Three or more animals were used per data set. Statistics were done using the Dunnett’s multiple comparison test ***p ≤ 0.001; ns not significant. Scale bars: a, b, h, i, j 2 µm; c–e 10 µm
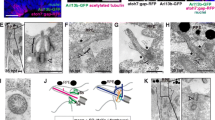
Primary cilia disassemble during mouse RPE development. Quantification of ciliation in mouse RPE at E16.5, post-natal day 1 (P1) and adult show that the number of ciliated cells drastically decreased after birth (a). Transmission electron micrographs of basal bodies or ciliary axoneme profiles (marked by red asterisks) of adult mouse RPE (b). Schematics show classification of ciliary structures into different classes (I, II and III), depending on the absence (class I) or presence of a membranous attachment (the ciliary vesicle) (class II) or ciliary axoneme (class III). Quantification of class I-III profiles shows that the number of class III profiles decrease as the RPE matures (c). Gene expression as measured by quantitative real-time PCR from isolated primary mouse RPE cells show altered expression of ciliary (Arl13b and Ift88) and ciliary disassembly markers (Hef1, AurA and Hdac6) (d–h). Fold changes and significance were calculated relative to E16.5. Values represent data from three or more independent animals (biological repeats) each with three technical repeats per data set. Statistics were done using the Dunnett’s multiple comparison test *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns not significant. Scale bars: 2 µm
To confirm retraction of the primary cilium in mature RPE tissues, we prepared transmission electron micrographs (TEM) of mouse RPE at multiple time points and imaged all identifiable basal body or ciliary axoneme profiles. All profiles were classified into one of the three categories that we defined as follows: basal bodies or centrioles with no attached membrane vesicles were categorized as Class I, whereas Class II profiles were identified as basal bodies containing a membranous attachment (the ciliary vesicle). Class III profiles were those with an extended ciliary axoneme into the extracellular space (Fig. 2b). Class I profiles are likely over-represented as an artifact of sectioning through a basal body of a Class II and III structure that had an attached membrane or axoneme in a different cutting plane. Consistent with an increase in ciliary disassembly, the number of Class III profiles decreased with RPE maturation (Fig. 2c). Combined, these data are consistent with our observations on immunohistochemistry and imply that primary cilia disassemble upon maturation of the RPE.
To further support retraction of the primary cilium, we measured changes in gene expression of two ciliary genes, Arl13b and Ift88 in isolated mouse RPE cells using quantitative real-time PCR (qRT-PCR). We developed a method to isolate pure RPE cells from embryonic mouse eyes and confirmed cell purity by determining lack of choroidal or retinal-specific gene expression (Supp. Figure 2a–c). Gapdh was taken as the housekeeping control as its expression remained stable from E14.5 through to P7 in control and knockout mouse RPE (Supp. Figure 2d, e). Expression of both cilia genes peaked at E16.5 and decreased as the tissue matured (Fig. 2d, e). Because ciliary retraction in the RPE may be actively controlled via ciliary disassembly, we also analyzed the gene expression of Hef1, AurA and Hdac6 [22, 43, 44], which are known regulators of ciliary disassembly. Expression of ciliary disassembly components changed dynamically as the cilium retracted (Fig. 2f–h). Hef1 expression increased gradually from E14.5 to E18.5. Expression of AurA and Hdac6 was decreased by ~ 50% between E16.5 and E18.5. This suggests that AurA and Hdac6 expressions are highest just before ciliary disassembly is observed and decline by E18.5, at which point the majority of cilia have disassembled.
Taken together, these data demonstrate that the primary cilium is disassembled upon maturation of the RPE. Temporal and spatial patterning of ciliation across the RPE matches the known mechanism of maturation for this tissue. The present data and our previous work [19] support the hypothesis that the transient expression of the primary cilium is required for RPE maturation, which occurs in an organized pattern. To further understand the mechanism of ciliary disassembly, we next sought to investigate how ciliary proteins contribute to the process of ciliary disassembly.
Loss of BBS proteins affects ciliated RPE
We focused on Bardet–Biedl syndrome (BBS) proteins in view of our prior observation that BBS proteins regulate primary cilia length and ciliary trafficking [27]. BBS6 (MKKS) is a component of the BBS chaperonin complex thought to be required for BBSome assembly. BBS8 (TTC8) is a component of the BBSome complex required for ciliary trafficking. We observed that RPE tissue from Bbs6 knockout (Bbs6−/−) mouse embryos displayed fewer ciliated cells at E16.5. The knockout cilia were shorter (0.87 ± 0.67 µm) compared to WT (2.25 ± 0.94 µm) littermates (Fig. 3a–d). In addition to changes in cilia length, we also observed changes in cell size in Bbs6 knockout mouse embryos. In WT, we saw little variation in cell size between neighboring cells. However, in Bbs6−/− we observed an increase in average cell size and broader distribution of cell size (Fig. 3a; Supp. Figure 3a, b). This abnormal patterning reflects disrupted RPE maturation as we had seen for Bbs8−/− in our previous paper [19]. Abnormal patterning is possibly a consequence of dysregulation of cell junction components such as Occludin and Epithelial Cadherin (Cadherin 1). Occludin is an integral membrane protein found at tight junctions [45], and Epithelial Cadherin [46] is a major component of adherens junctions, both of which are essential for the regulation of RPE intercellular junction integrity and function, and apical basal polarity. Ras-related protein Rab-27A (Rab27) and Myosin VIIA (Myo7a) are both required for melanosome transport in the RPE, dysfunction of which leads to defective visual cycle [47]. Expression of these genes is seen as a marker and benchmark for RPE maturation and their disruption can contribute to aberrant epithelial morphology. We observed a downregulation of Cdh1, Ocln, Rab27a and Myo7a as quantified by qRT-PCR (Supp. Figure 3c). Bbs8 knockout (Bbs8−/−) embryos also showed reduced number of ciliated RPE cells at E16.5 compared to that of WT (Fig. 3e, f, Supp. Figure 3c) although less significant changes in gene expression were observed.
Absence of BBS proteins decreases RPE ciliation in vivo and in vitro. Representative immunofluorescence images of E16.5 mouse RPE flatmounts from wildtype and Bbs6−/− littermates, labeled with antibodies against the primary cilium (Arl13b; red, acetylated α-tubulin (Ac. tubulin); cyan) and cell junctions (Zona Occludens (ZO-1); green) (a, c). Boxplots of ciliation (b) and cilia length (d) show a significant decrease in cilia number and cilia length in Bbs6 knockout animals. Representative immunofluorescence images of E16.5 mouse RPE flatmounts from wildtype and Bbs8−/− littermates, labeled with antibodies against Arl13b; red and ZO-1; green show a reduced number of ciliated cells in Bbs8 knockout mice (e). Boxplots confirmed the significant reduction of ciliated cells in Bbs8 knockout RPE (f). Representative immunofluorescence images of primary cilia labeled with antibodies against Arl13b; green and polyglutamylated tubulin (GT335); red in BBS8 and BBS6 KD hTERT-RPE1 compared to non-targeting control (NTC) (g). Graphical representation of percentage of ciliated cells (NTC n = 250, BBS8 KD n = 150 and BBS6 KD n = 200) (h) and cilia length (i) of control in comparison to KD cells. White asterisks (*) label cells lacking cilia. Three or more individual animals were used per sample set. Statistical analyses in b, d and f were performed using two-tailed Mann–Whitney U test, where ***p < 0.001. For dn = 40 cells per genotype. Statistical analyses in h and i were done using the Dunnett’s multiple comparison test ***p ≤ 0.001. Scale bars: a, e 10 µm; b 2 µm; g 10 µm, magnified images 5 µm. KD Knockdown
To further elucidate the mechanisms by which BBS molecules regulate ciliary disassembly, we utilized an in vitro culture system by knocking down (KD) BBS8 or BBS6 using short interfering RNA (siRNA) in hTERT-RPE1 cells. This cell line has previously been used to characterize ciliary disassembly and is derived from human RPE tissue. Knockdown was validated by RT-qPCR and Western blotting (Supp. Figure 4a–d). Cells were serum-starved to induce ciliogenesis. Similar to the in vivo mouse RPE, we observed fewer ciliated cells upon BBS8 and BBS6 KD (24.9 ± 11.6 and 24.1 ± 9.62%) compared to the non-targeting siRNA control (NTC, 74.7 ± 9.77%) (Fig. 3g, h). In the remaining cells that retained cilia, cilia length was also significantly reduced compared to NTC (NTC: 3.58 μm, BBS8 KD: 2.26 μm, BBS6 KD: 2.62 μm) (Fig. 3i).
Interaction of BBS proteins with mediators of ciliary disassembly
Next, we sought to identify whether additional BBS proteins interact with Inversin, a protein known to be involved in AurA mediated ciliary disassembly [35]. Since we had previously shown that BBS8 directly interacts with Inversin during regulation of Wnt signaling in development of the RPE [19], we wanted to see if this interaction extended to other BBS proteins. Using GFP-traps, we were able to pull down overexpressed BBS6-myc and BBS2-myc with Inversin-eGFP in HEK293T cells (Fig. 4a, b). To confirm the physical interaction between Inversin and BBS6 in situ, a proximity ligation assay (PLA) was performed using antibodies against endogenous Inversin and overexpressed myc-BBS6. Positive PLA events shown as distinct fluorescent foci confirmed the interaction (Fig. 4c).
Mislocalization of Inversin upon loss of BBS proteins. HEK293T cells transiently co-transfected with myc-tagged BBS and Inversin-GFP plasmids. Cell lysates subjected to GFP-TRAP pulldown followed by Western blotting show that Inversin-GFP interacts with myc-tagged BBS6 (a) and myc-tagged BBS2 (b). Proximity ligation assays (PLA) of HEK cells overexpressing myc-BBS6 were performed using antibodies against myc and endogenous Inversin. Positive PLA foci (green) indicate interaction between Inversin and BBS6. Empty-myc transfected control cells did not display positive PLA foci. TRITC-Phalloidin (F-actin, red) was used to visualize cell outlines (c). Representative immunofluorescence images of kidney medullary (KM) cells labeled with antibodies against the primary cilium (acetylated α-tubulin (Ac. tubulin); red) and Inversin (green) show diminished localization of Inversin to the base of the cilium in Bbs6−/− cells as compared to the wildtype control (d). ROI linear profile represents fluorescence intensity of corresponding cyan line on the merged image. Peaks indicated by asterisk represent area of co-localization of Inversin and Ac. tubulin in wildtype KM cells (e). Bbs6+/+n = 215 cilia, Bbs6−/−n = 140 cilia. Boxplots show a reduction in percentage of Inversin positive Ac. tubulin in Bbs6−/− KM cells compared to control cells (f). Statistical analyses in e were performed from three independent experiments using two-tailed Mann–Whitney U test, where ***p < 0.001. Scale bars: c, d 10 µm
In an immortalized murine kidney medullary (KM) cell line, Inversin is localized to the base of the cilium as seen by Inversin co-localization with acetylated α-tubulin in the majority of ciliated cells (Fig. 4d, e). These cells were used as endogenous Inversin is better detectable in these cells upon immunocytochemistry. Furthermore, the Wnt signaling and ciliary phenotype are recapitulated in these cells [27]. In KM cells derived from Bbs6 knockout (Bbs6−/−) mice, significantly fewer cilia showed Inversin localization at the base, and Inversin expression was more dispersed throughout the cytoplasm (Fig. 4d, f). Combined these data suggest that BBS6 and BBBS2 proteins interact with Inversin and regulate its expression at the ciliary base.
BBS proteins regulate key mediators of primary ciliary disassembly
In mammalian cells, ciliary disassembly is mediated by recruitment of HEF1 at the basal body, leading to downstream phosphorylation and activation of AurA and HDAC6, and subsequent destabilization of ciliary tubulin [22, 48]. We wanted to determine whether BBS proteins are involved in cilia maintenance by protecting the cilium against AurA-HDAC6-mediated disassembly. Gene expression analysis showed an increase of ciliary disassembly genes HDAC6 and HEF1 upon knockdown (KD) of BBS8 or BBS6, consistent with reduced ciliary length (Figs. 3 g–i; 5a). Western blot analysis also showed elevated levels of HDAC6 protein expression in hTERT-RPE1 cells (Fig. 5b, c). Expression of total AurA mRNA and AurA protein was decreased in KD cells, as analyzed by qPCR, Western blot and flow cytometry (Fig. 5a, d, e, f, g). In contrast, the level of active phosphorylated AurA (pAurA) was unchanged as shown by Western blot with an antibody specific for pAurA (Thr288) (Fig. 5d, e). Therefore, the ratio of pAurA/AurA was elevated in the KD cells compared to control even without ciliary disassembly via serum activation. Previous reports showed that levels of pAurA peak when cilia disassemble [23]. When we do combined serum activation with BBS KD, we observed a similar trend in that active pAurA is retained upon BBS8 or BBS6 KD (Supp. Figure 5a). As quantified by qPCR, HEF1 transcript expression was elevated upon both BBS8 and BBS6 KD (Fig. 5a). In an attempt to verify these data in vivo, we analyzed mRNA expression of Hef1, AurA and Hdac6 in knockout mouse tissue using unpurified RPE (RPE and choroid). Expression of ciliary disassembly genes Hef1 and Hdac6 was increased in Bbs8−/− relative to wildtype control littermate tissue. No significant change in expression level could be seen for AurA. In Bbs6−/− mice, Hef1 was significantly increased, AurA significantly decreased and Hdac6 remained unchanged (Supp. Figure 3d). These gene expression patterns show a similar trend to the in vitro data. Although HEF1 mRNA expression levels were elevated in vitro and in vivo, Western blot analysis showed a reduction in protein upon loss of BBS8 (Supp. Figure 5b). We hypothesized that in the absence of BBS8, HEF1 is actively phosphorylated and targeted for proteasomal degradation. To test this, we treated serum-starved cells with the proteasome inhibitor MG132. This resulted in ~ 8-fold recovery of HEF1 expression in treated KD cells compared to untreated KD cells which only had a 3.7-fold recovery (Fig. 5h). Therefore, over 50% more HEF1 was recovered in KD cells compared to control (0.1–0.8 vs 1–3.7). Although we saw a mild recovery of total AurA upon inhibiting proteasomal activity in these cells (Fig. 5i), there was little difference between KD and control. Based on these observations, we propose that ciliary disassembly components are differentially regulated by BBS proteins. While loss of BBS8 results in increased levels of HDAC6, it also results in the proteasomal degradation of HEF1 and to some extent of AurA. We wanted to further elucidate whether the ciliary length defect observed in BBS deficient cells could be attributed to dysfunction of ciliary disassembly components.
BBS proteins regulate key mediators of cilia disassembly. Quantitative real-time PCR shows increased gene expression of cilia disassembly components (HEF1 and HDAC6) and decreased expression of AurA relative to non-targeting control (NTC, red line) in serum-starved BBS8 or BBS6 knockdown (KD) hTERT-RPE1 cells. GAPDH was used as housekeeping control (a). Western blots show a significant increase in protein levels of HDAC6 upon KD of BBS8 and BBS6 in serum-starved hTERT-RPE1 cells (b, c). Conversely, AurA protein levels were decreased upon KD of BBS8 and BBS6 in serum-starved hTERT-RPE1 cells, although pAurA levels were retained (d, e) suggesting an increased ratio of active over total AurA. Flow cytometry analysis was used to further quantify AurA expression in serum-starved BBS8 and BBS6 KD hTERT-RPE1 cells (f, g). Representative flow cytometry dot plots show the AurA-positive cell population (P3, blue) and AurA-negative cell population (P2, red) (f). Quantification of the AurA-positive cell population confirmed a significant decrease in the BBS8 and BBS6 KD cells compared to NTC (g). Western blots show that KD of BBS8 in hTERT-RPE1 cells leads to decreased level of HEF1, which was partially restored by treatment with proteasome inhibitor MG132 (h). An 8-fold increase in HEF1 protein expression was observed upon BBS8 KD compared to 3.7-fold in NTC. Decreased protein levels of AurA in BBS8 KD hTERT-RPE1 cells were also partially restored by treatment with proteasome inhibitor MG132 (i). Increased levels of HDAC6 in BBS8 KD hTERT-RPE1 cells were concomitant with an increase in acetylated α-tubulin and were reduced upon treatment with HDAC6 inhibitor tubacin, as quantified by Western blot (j). Quantification of Western blot data was normalized to GAPDH levels. Bar charts show relative protein expression in arbitrary units (AU). Data are expressed as mean ± SD, n = 3 separate experiments for (a-g), while n = 2 for h-j. Statistical analyses in c, g were done using the Dunnett’s multiple comparison test and e using Shidak’s multiple comparison test. In c, e and g *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns not significant. KD Knockdown, NTC non-targeting control, SS serum-starved, FC fold change, FSC-A forward scatter area
Ciliary disassembly component inhibition rescues the ciliary length defect caused by BBS knockdown
To confirm that ciliary defects upon BBS KD can be attributed in part to an increase in HDAC6 expression, we treated serum-starved NTC and BBS KD cells with tubacin, a specific HDAC6 inhibitor [22, 49]. Tubacin treatment caused a decrease in HDAC6 and a concomitant increase in the protein level of acetylated tubulin in both NTC and KD cells compared to the vehicle (DMSO) alone, as seen by Western blot (Fig. 5j). This corresponded to a rescue in cilia length (Fig. 6a, b). A significant change in the number of ciliated cells was not observed after treatment with tubacin (Fig. 6c). Next, we used a potent and selective inhibitor of AurA, AurA Inhibitor I [35, 50]. Similarly, this also ameliorated the cilia length defect in BBS8- and BBS6-deficient cells (Fig. 6d, e). To test whether HEF1 activity also underlies the AurA and HDAC6 mediated ciliary disassembly in BBS KD cells, we performed double KD experiments. We observed an appreciable rescue in cilia length in the BBS and HEF1 double KD cells compared to single KD (Fig. 6f, g). We confirmed that double KD did not lead to loss of individual gene KD efficiency (Supp. Figure 4e–g). Recovery of cilia length after inhibition or KD of HEF1, AurA and HDAC6 supports our hypothesis that BBS proteins are involved in the control of ciliary disassembly (Fig. 6h).
Inhibition of ciliary disassembly components rescues BBS mediated ciliary disassembly. Representative immunofluorescence images of cilia stained with antibodies against acetylated α-tubulin (Ac.tubulin, red) and Arl13b (green), from non-targeting control (NTC), BBS8 and BBS6 knockdown (KD) hTERT-RPE1 cells treated with and without HDAC6 inhibitor, tubacin (a). Quantification of ciliary length show that tubacin treatment did not affect ciliary length in control, yet was able to increase ciliary length in BBS8 and BBS6 KD cells (b). Quantification of ciliated cells showed that treatment with tubacin had no effect on ciliation (c). Treatment with AurA inhibitor I was also able to significantly increase ciliary length in BBS8 and BBS6 KD cells. BBS8 and BBS6 KD tubacin-treated cells show a greater increase in cilia length in comparison to their DMSO mock-treated counterparts, while control cells showed minimal increase (d, e). KD of HEF1 increased ciliary length in control hTERT-RPE1 cells. Double KD of HEF1 and BBS8 or BBS6 was able to reverse the ciliary disassembly phenotype observed in BBS8 or BBS6 KD (f, g). A model showing the inhibition or KD of ciliary disassembly components causes rescue of cilia length in BBS KD hTERT-RPE1 cells (h). Statistical analyses in b, c, e, g were done using the Sidak’s multiple comparison test from two independent experiments. The length of at least 1000 cilia from the six different treatment groups (NTC treated, untreated, BBS8/6 KD treated, untreated) in each experiment (Tubacin treatment, AurA treatment and Hef1 KD) were measured. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns not significant. KD Knockdown, NTC non-targeting control, SS serum-starved
To address off-target effects of siRNA we knocked down BBS8 from hTERT-RPE1 cells using single (siRNA 1 or siRNA2) and double siRNAs (siRNA 1 + 2) and looked for levels of HDAC6 via Western blotting. We observed a similar increase in HDAC6 levels in single vs double KD cells, as quantified using Western blotting analysis (Supp. Figure 5c). Furthermore, using different combinations of siRNA against BBS8 from different companies had the same effect on AurA expression levels (Supp. Figure 5a). Therefore, we believe that the phenotype we observe is specific. To rule out differences in cell cycle stages, which could affect changes in ciliary disassembly components, we performed cell cycle analysis via fluorescence-activated cell sorting (FACS) using propidium iodide (PI). This corroborated that the majority of cells remained in G0/G1 phase in both NTC and KD cells (Supp. Figure 5d), confirming that our results were cell cycle independent.
Loss of BBS proteins regulate HDAC6, thereby influencing post-translational modification of β-catenin
Previous studies have shown that ciliogenesis and ciliary disassembly modulate the switch from β-catenin-dependent canonical to non-canonical Wnt signaling pathways [36, 51]. Therefore, we examined the impact of BBS-regulated ciliary disassembly components on Wnt signaling. β-catenin is a direct substrate of HDAC6, which deacetylates β-catenin at lysine residue (K49), inhibiting downstream phosphorylation [52]. This results in β-catenin stabilization and nuclear accumulation. Although we and others have shown that suppression of BBS genes results in stabilization of β-catenin and altered regulation of downstream Wnt targets [18, 38], the mechanisms that cause this stabilization are not yet known. Loss of BBS is not thought to directly affect the core activity of the proteasome [38], which suggests a defect in phosphorylation and subsequent targeting of β-catenin for degradation. Since the stability of β-catenin is mediated by various specific phosphorylation and acetylation events at different sites, we hypothesized that upon loss of BBS, β-catenin becomes differentially modified. Consistent with this, the level of acetylated β-catenin (Lys49) was reduced after BBS8 or BBS6 KD in hTERT-RPE1 cells as quantified by Western blotting (Fig. 7a). We also sought to identify further downstream phosphorylation events of β-catenin that lead to degradation. Using phospho-specific antibodies, we detected a decrease in phosphorylation at T41/S45 and S33/37/T41 as a result of BBS8 and BBS6 KD (Fig. 7b, c).
BBS-mediated regulation of ciliary disassembly components alters post-translational modification of β-catenin. Western blot analysis and quantification show reduced acetylation of β-catenin at K49 (a) and consequently reduced phosphorylation at p41/44 (b) and p33/37/41 (c) upon BBS8 and BBS6 knockdown (KD) in hTERT-RPE1 cells, suggesting an increase in stable and active β-catenin. Immunocytochemistry and quantification using an antibody against acetylated β-catenin K49 (green) show reduced expression in the nucleus upon BBS8 and BBS6 KD compared to non-targeting control (NTC) in hTERT-RPE1 cells (d). Cells were co-labeled with Arl13b (red) to confirm reduction in ciliary length. Reduced β-catenin K49 expression could be rescued by treatment with HDAC6 inhibitor tubacin (n ≥ 100 for each group) (d, e). Immunocytochemistry using an antibody against β-catenin pS33/37/T41 (green), a target of β-catenin acetylation, shows expression at the base of the cilium and in the nucleus in NTC hTERT-RPE1 cells (f, g). Ciliary axoneme is marked by acetylated α-tubulin (Ac. tubulin, red), and the basal body by Pericentrin 2 (PCNT2, magenta). Quantification confirms reduced localization of β-catenin pS33/37/T41 at the basal body upon BBS8 and BBS6 KD (g). BBS8 and BBS6 KD cause less β-catenin degradation, resulting in increased levels of total β-catenin in hTERT-RPE1 cells, as quantified from Western blot (h, i). This was confirmed by a TCF/LEF luciferase activity assay that measures the transcriptional activity of β-catenin enzymatically. Luciferase activity in Wnt3a-treated non-targeting control HEK cells is upregulated compared to the untreated control. The Wnt response (luciferase activity) is significantly enhanced upon the suppression of BBS8 and BBS6 (j). Immunofluorescence analysis and quantification show increased stability and nuclear translocation of active β-catenin pS552 (green) in non-ciliated hTERT-RPE1 cells after BBS8 KD (n = 263 for NTC and 100 for BBS8 KD) (k, l). Cells were co-labeled with Ac. tubulin (green) to confirm reduction in ciliary length. Quantification of Western blot data was normalized to GAPDH levels. Bar charts in a–c show relative protein expression in arbitrary units (A.U.). Data in a, b, c, i, j are expressed as mean ± SD, n = 3 separate experiments. Data in g show mean ± SD, two independent experiments. Statistical analyses in a, b, c, g, i were done using the Dunnett’s multiple comparison test. Data analyses in e, j, k were performed using Sidak’s multiple comparison test. p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns not significant. Scale bars: a, e 10 µm; b 2 µm; h 5 µm
To strengthen our findings on Western blot, we performed immunocytochemistry using an antibody against acetylated β-catenin (Lys49) and observed a reduction in nuclear fluorescence intensity upon BBS KD. Treatment with HDAC6 inhibitor tubacin rescued the levels of acetylated β-catenin K49 in BBS-deficient cells (Fig. 7d, e). Previous studies showed that phospho-(S33/37/T41)-β-catenin localizes to the base of the cilium [51, 53]. We observed localization of phospho-(S33/37/T41)-β-catenin not only to the basal body, but also in the nucleus (Fig. 7f). Upon KD of BBS8 and BBS6, this nuclear localization is diminished while localization at the basal body is often absent (Fig. 7f, g).
Reduction of phosphorylation at S33/37/T41 ultimately results in reduced degradation and consequent increase in levels of total β-catenin as observed in Western blot (Fig. 7h, i). This increased level of total β-catenin translated to increased levels of β-catenin activity as measured via luciferase assays (Fig. 7j). Stimulation with Wnt3a conditioned medium significantly increased β-catenin driven TCF/LEF transcription after BBS8 or BBS6 KD. In contrast to phosphorylation at S33/37/T41, phosphorylation at S552 causes stabilization and nuclear accumulation of β-catenin [54]. Consistent with this, and the increased activity of β-catenin, we detected elevated expression and nuclear accumulation of pS552 β-catenin in nonciliated BBS8 KD cells (Fig. 7k, l). Together, these experiments show that loss of BBS molecules influences post-translational modification of β-catenin, which ultimately regulates its signaling activity.
Discussion
In the present work, we demonstrate that ciliary trafficking proteins are required for homeostasis of primary ciliary disassembly components, specific regulation of which is required for ciliary retraction and regulation of signaling pathways. Precise regulation of ciliation is an absolute requirement for tissue differentiation [8, 55]. We focused our attention on BBS proteins known to be required for ciliary trafficking, in particular on a component of the Bbsome, BBS8, and a component of the chaperonin complex, BBS6, thought to be required for Bbsome assembly [55,56,57,58].
The RPE is a ciliated epithelial monolayer essential for visual function. Insights into RPE maturation can be extrapolated to other epithelial tissues for example lung epithelial cells [19]. In mice we showed that as the RPE matures, the primary cilium retracts with only a few stunted cilia being retained after birth. Cilia dysfunction in knockout mice resulted in abnormal RPE patterning in part due to defective RPE maturation (this and previous study) [19]. Retraction of the primary cilium is accompanied by expression of ciliary disassembly components suggesting that loss of the primary cilium is a tightly controlled cellular event. Interestingly, such mechanisms of ciliary disassembly are also observed in other tissues, such as in the auditory hair cells of the inner ear [6]. Here, the kinocilium plays an important role during development of the Organ of Corti but is reabsorbed by post-natal day 10 [59]. Because cilia are not retained, we suggest that they are less likely to be essential for cellular function in adult RPE, but more likely required for regulating signaling processes during development as we have recently demonstrated [19]. Since retraction of the primary cilium in the RPE is accompanied by the expression of ciliary disassembly components in vivo, we focused our attention on the molecular control of these processes.
The most common pathways that govern ciliary disassembly are through AurA, HEF1 and HDAC6 [10, 22]. Previous reports have shown that BBS proteins regulate cilia length [27] and that BBIP10 (BBS18), an additional subunit of the BBSome, interacts directly with HDAC6 [37], yet the molecular mechanisms underlying this phenomenon have not been elucidated. Here, we observed that the loss of BBS proteins resulted in increased levels of HDAC6 and that inhibition of HDAC6 resulted in a rescue of cilia length in BBS KD cells. Similarly, inhibition of AurA and HEF1 also resulted in rescue of cilia length. Therefore, we propose that BBS proteins maintain cilia length by suppressing HEF1-AurA-HDAC6-mediated disassembly (Fig. 8). Although the total levels of pAurA were not changed upon BBS KD, since inhibition of AurA activity rescued the ciliary length defect in these cells, this suggests a difference in activity.
Model of BBS-mediated regulation of ciliary disassembly. BBS proteins interact with Inversin and regulate its expression at the base of the cilium. Inversin inhibits HEF1/AurA, inactivating histone deacetylase HDAC6 thus preventing ciliary disassembly. As a consequence of dormant HDAC6, β-catenin remains acetylated and phosphorylated, thereby undergoing proteasomal degradation. Upon BBS suppression, Inversin expression decreases at the base of the cilium. This leads to phosphorylation and activation of AurA via HEF1, resulting in upregulation and activation of HDAC6. HDAC6 deacetylates β-catenin, hence preventing further phosphorylation and degradation. Consequently, β-catenin is stabilized and translocates to the nucleus activating canonical Wnt signaling. This also regulates ciliary length
Because Inversin (NPHP2), a key ciliary protein, influences ciliary disassembly via inhibiting AurA [35, 36] and is also a key mediator of Wnt signaling, we postulated that the role of BBS proteins in ciliary disassembly may in part be mediated via Inversin. We show that BBS proteins are required for Inversin protein expression at the base of the cilium, thereby regulating AurA phosphorylation and subsequent ciliary disassembly. AurA can also be activated by other proteins such as Pitchfork, calmodulin, trichoplein, HIF1α (Hypoxia-inducible factor 1-alpha) or Plk1(Polo-like-Kinase 1) [60]. Moreover, some kinesins such as Kif2a (Kinesin family member 2a) might also be involved as they are direct targets of AurA [61]. Therefore, BBS proteins could also be acting upon these regulators. In support of this, some of these proteins have already been shown to interact with other ciliary proteins [62, 63]. BBS proteins may also influence other ciliary length regulating components such as CPAP (Centrobin-mediated Regulation of the Centrosomal Protein 4.1-associated Protein), Nde1 (Nuclear distribution protein nudE homolog 1) and OFD1 (oral–facial–digital syndrome 1) proteins [64], which remain to be addressed.
Previous studies showed that BBS proteins interact with proteasomal subunits regulating the composition of the centrosomal proteasome [38, 65]. Consequently, loss of BBS proteins results in altered signal transduction due to defects in proteasomal clearance of key signaling proteins including β-catenin [38, 65]. Here, we show that the loss of BBS proteins decreases the stability of HEF1 and AurA proteins. A partial recovery of these proteins is observed upon inhibiting proteasomal function using MG132, suggesting that BBS proteins protect Hef1 and AurA from proteasomal degradation and thus inhibit ciliary dissasembly. Increased stability of β-catenin (discussed below) and subsequent upregulation of canonical Wnt signaling upon BBS KD may also in part be ascribable to defects in proteasomal clearance. Similarly, the cilia protein RPGRIP1L (RPGR-Interacting Protein 1-Like) has been shown to protect canonical Wnt components (dishevelled) from proteasomal degradation at the basal body [53, 66]. Because HEF1 stabilization differed in BBS8 compared to BBS6 KD cells, it suggests differences in the functional role of the BBSome vs. the BBS chaperonins. One explanation for reduced protein levels of HEF1 in serum-starved BBS8 KD cells could be a feedback loop mechanism. Since it is clear from our experiments that BBS8 KD cells experience increased cilia disassembly, these cells may be trying to maintain homeostasis by mediating increased degradation of HEF1, in an effort to maintain cilia length and limit the extent of cilia disassembly. This phenomenon is intriguing, especially considering that we only see this after BBS8 KD and not after BBS6 KD, which warrants more in depth examination in the future.
In an effort to elucidate the underlying mechanisms affecting β-catenin stability upon loss of cilia function, we focused our attention on post-translational modifications (PTMs) of proteins associated with proteasomal degradation [41]. HDAC6 physically interacts and acetylates β-catenin [52] resulting in altered Wnt signaling. Several studies have demonstrated the importance of PTMs such as ubiquitination, phosphorylation, acetylation and glutamylation in various signaling processes [67]. PTMs such as phosphorylation also actively influence the process of ciliogenesis and maintenance [68]. Targeting β-catenin for degradation is a complex process involving PTMs at various sites [41]. HDAC6 deacetylation of β-catenin at K49 inhibits downstream phosphorylation. Because HDAC6 was increased in the absence of BBS proteins, we observed less acetylation at K49, which results in less β-catenin phosphorylation at T41/S45 and S33/37/T41. Thus, less β-catenin is targeted for degradation. Moreover, upon loss of BBS proteins, β-catenin is actively phosphorylated at S552, a modification that increases its stabilization and nuclear localization (our data and previous data [19]). Together, these dysregulated PTMs result in aberrant canonical-Wnt hyperactivation upon loss of BBS protein function. Although we and others [52] have observed that HDAC6 and AurA can modify β-catenin levels, very little is known about the nature of this regulation which needs further elucidation.
In conclusion, we observed transient expression of the primary cilium in the developing mouse RPE. As the RPE matures, primary cilia retract, which are accompanied by altered expression of Hef1, AurorA and Hdac6, key mediators of ciliary disassembly. In control cells, BBS proteins protect against ciliary disassembly whereas the loss of BBS proteins results in altered ciliary disassembly components, including HDAC6, likely via interaction and trafficking of Inversin. This results in HDAC6-mediated downstream cross-talk between ciliary disassembly signaling and canonical-Wnt signaling, leading to PTMs resulting in canonical-Wnt hyperactivation. Taken together, we furthered our understanding into how ciliary proteins modulate cellular signaling pathways and contribute to maturation of epithelial tissues.
Materials and methods
Animals
All mouse experiments had ethical approval from appropriate governing bodies. Experiments were performed in accordance with guidelines provided by ARVO (Association for Research in Vision and Ophthalmology). Animals were housed under a 12 h light–dark cycle. The morning after mating was considered E0.5 and up to 24 h after birth was considered P0. C57BL/6 mice were used for control at embryonic (E14.5, E16.5 and E18.5), and post-natal stages (P1, P7 and adult). Bbs6 and Bbs8 knockout mice have been previously described [1, 69].
Cell culture
hTERT-RPE1 and HEK293T cells were obtained from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM)-F12 (Thermo Fisher) or DMEM (Thermo Fisher) supplemented with heat-inactivated 10% fetal bovine serum (FBS) (LONZA), and 1% penicillin/streptomycin (P/S) (Thermo Fisher) (Referred to as complete media). Immortalized kidney medullary (KM) cells were obtained from P. Beales and cultured as previously described [27].
Antibodies and siRNAs
For immunofluorescence, antibodies used were anti-Arl13b (Rb, 1:1000; Proteintech, 17711-1-AP), anti-acetylated α-tubulin (Mm, 1:800; Sigma, T6793), anti-GT335 (Mm, 1:200; Adipogen, AG-20B-0020), anti-β-catenin (Rb, 1:150; CST, D10A8), anti- acetylated α-tubulin (Rb, 1:1000; Abcam, ab11317), anti-pericentrin2 (Gt, 1:200; SantaCruz, SC28145), and anti-ZO-1 (Mm, 1:150; Thermo Fisher, ZO1-1A12, 339188), anti-GFP (Rb, 1:5000; Clontech, 632592), anti-Aurora A (D3E4Q) (Rb, 1:100; CST, 14475), anti-phospho-Aurora A (Thr288) (C39D8) (Rb, 1:50; CST, 3079), anti-phospho-β-Catenin (Ser33/37/Thr41) (Rb, 1: 200; CST, 9561), anti-Acetyl-β-Catenin (Lys49) (D7C2) (Rb, 1: 150; CST, 9030), and anti-NPHP2 (Rb, 1:150; Abcam, ab65187). The primary antibodies were detected using Alexa Fluor 488, 555 and 568 (1:400; Molecular Probes) and CF™640R (1:400; Biotium) conjugated secondary antibodies.
For western blot, antibodies used were anti-myc-Tag (9B11) (Mm, 1:1000, CST, 2276), anti-GFP (Rb, 1:1000; Clontech, 632592), anti-Aurora A (D3E4Q) (Rb, 1:1000; CST, 14475), anti-phospho-Aurora A (Thr288) (C39D8) (Rb, 1:1000; CST, 3079), anti-β-Catenin (D10A8) (Rb, 1: 1000; CST, 8480), anti-phospho-β-Catenin (Ser33/37/Thr41) (Rb, 1: 1000; CST, 9561), anti-phospho-β-Catenin (Thr41/Ser45) (Rb, 1: 1000; CST, 9565), anti-Acetyl-β-Catenin (Lys49) (D7C2) (Rb, 1: 1000; CST, 9030), anti-HEF1/NEDD9 (2G9) (Mm, 1: 1000; CST, 4044), anti-HDAC6 (D2E5) (Rb, 1: 1000; CST 7558). Secondary antibodies used were IRDye 800 and IRDye 680 (Rb, Mm or Gt; 1:10000; Li-cor Bioscience).
For flow cytometry, a primary antibody against Aurora A (D3E4Q) (Rb, 1:100; CST, 14475) and a secondary Alexa Fluor 488-labelled anti-rabbit antibody (1: 250) were used. Propidium iodide (PI, Thermo Fisher, P3566)) at 50 μg/ml was used for cell cycle analysis.
siRNA was used in our study to knockdown (KD) BBS6 and BBS8 in hTERT-RPE1 cells. BBS8 siRNA (HSC.RNAI.N198309.12; IDT), BBS8 siRNA (L-021417-02-0005; Dharmacon), BBS6 siRNA (L-013300-00-0005), HEF1 siRNA (hs.Ri.NEDD9.13; IDT) and non-targeting siRNA (D-001810-10-05) were used.
Transfections and treatments: Plasmid transfections were performed using GeneTrap transfection reagent (made at NEI, NIH, Bethesda, MD, USA). In brief, 6 μl of GeneTrap transfection reagent was diluted in 90 μl of DMEM, incubated for 5 min followed by addition of 2 μg plasmid and then incubated for 20 min at room temperature (RT). The transfection mix was added dropwise to cells in a 6-well plate containing 2 ml complete media. siRNA transfections were performed in 6-well plates with Lipofectamine RNAiMax transfection reagent (Thermo Fisher; 13778150) using a reverse transfection protocol according to the manufacturer’s instructions. To induce cilia formation, 24 h post-transfection cells were serum-starved with Opti-MEM (Thermo Scientific) for up to 48 h. For proteasome, AurA or HDAC6 inhibition experiments, 48 h post-transfections cells were serum-starved for 24 h followed by treatment with 10 µM MG132 (Calbiochem), 1 µM AurA Inhibitor I (Selleckchem) and 2 µM tubacin (Sigma), respectively, for the indicated time (5 h) followed by western blot analysis and immunofluorescence. DMSO was taken as vehicle control for treatments.
Tissue preparation and Immunohistochemistry
Mouse eyes were enucleated and immersed in cold phosphate-buffered saline (PBS) and kept on ice for 20 min. Eyes were then placed in 1X PBS and the cornea, lens and retina were removed. The resulting eyecups were then fixed with 4% paraformaldehyde (PFA) in PBS for 1 h at RT, then washed three times with 1X PBS and incubated with 50 mM NH4Cl for 10 min, followed by incubation with β-Mercaptoethanol for 10 min. Eyecups were permeabilized with PBSTX (0.3% Triton-X) and blocked with blocking buffer (0.1% Ovalbumin, 0.5% Fish gelatine in PBS) at RT for 2 h followed by overnight incubation in primary antibody at 4 °C. Samples were washed three times with 1X PBS followed by incubation in secondary antibody and DAPI for 2 h at RT. Finally, samples were washed with PBS for three times before mounting on glass slides. Cells close to the optic nerve were considered near the center; cells closer to the edge of the eye cup were considered as peripheral. For isolation of pure RPE cells, eyecups were incubated with trypsin at 37 °C for 30–90 min depending on the age of tissue. RPE cells were mechanically removed from underlying choroid and isolated by hand. Choroidal and retinal contamination was checked via qPCR. Contaminated samples were removed from the analysis.
hTERT-RPE1 cells were fixed with 4% PFA for 10 min and permeabilized for 15 min with PBSTX. Immunostaining was performed as described above. KM cells were fixed with 100% methanol for 10 min on ice. Immunostaining was performed as described above.
Specimens were imaged on a Leica DM6000B microscope (Leica, Bensheim, Germany). Images were deconvoluted and co-localization profiles were generated with Leica imaging software (BlindDeblur Algorithm, one iteration step). Images were processed and cilia length measurements were performed using Fiji/ImageJ software (NIH, Bethesda, USA).
PLA assay
Direct in situ protein–protein interactions were investigated by means of a proximity ligation assay (PLA) using Duolink In Situ FarRed Kit Mouse/Rabbit (Sigma) according to the manufacturer’s instruction. Cells were incubated with anti-myc and anti-Inversin primary antibodies followed by anti-rabbit PLUS and anti-mouse MINUS secondary PLA probes. The two complementary oligonucleotides were then hybridized, ligated and rolling circle amplified by the provided polymerase, resulting in fluorescence signals when the targeted proteins were closer than 40 nm. TRITC–phalloidin (Sigma) was used for visualization of cells and DAPI for nuclear staining.
Fluorescence-activated cell sorting (FACS) analyses
For FACS, RPE1 cells were stained with an antibody directed against intracellular AurA. Recovered cells were fixed using 4% PFA for 10 min and washed three times with PBS. Cells were permeabilized with 90% ice-cold methanol for 30 min and rinsed three times. Cells were blocked using 0.5% bovine serum albumin (BSA) in PBS and incubated with AurA for 1 h at RT, then washed three times with 0.5% BSA/PBS. After incubation with a secondary antibody (Alexa Fluor 488-labelled anti-rabbit) for 45 min at RT, cells were finally washed with 0.5% BSA/PBS. Cells were acquired using the Invitrogen Attune NxT Flow Cytometer (Thermo Fisher Scientific, Inc., USA), and analyzed using FlowJo (Treestar, CA, USA). For cell cycle analysis, RPE1 cells were detached and fixed with 70% ethanol for 30 min, followed by incubation with 100 μg/ml RNase A solution and 50 μg/ml propidium-iodide solution. Samples were acquired and analyzed as described above.
Pulldown and western blotting
For pulldown experiments, HEK cells were transiently co-transfected with Inversin-GFP and pCMV-BBS6/2-myc [27] or empty plasmids. 48 h post-transfections cells were lysed using RIPA buffer supplemented with EDTA-free protease inhibitor cocktail (Halt™ Protease and Phosphatase Inhibitor Cocktail (100X), Thermo Fisher). Pulldown was performed using agarose beads (GFP-Trap-A, ChromoTek) according to the manufacturer’s instructions followed by western blot. Proteins were harvested in Laemmli sample buffer and separated on 8–10% SDS–polyacrylamide gel (PAGE), followed by transfer PVDF membrane (Immobilon®-FL PVDF membrane, Sigma, 05317). The blots were blocked with 5% milk or Applichem blocking buffer (0.2% AppliChem Blocking Reagent, 10 mM TrisHCl, 150 mM NaCl, 0.04% NaN3, in ddH2O; pH 7.4) and probed with antibodies as listed above. The blots were scanned for infrared fluorescence at 680 or 800 nm using the Odyssey Infrared Imaging System (Licor). Densitometry analysis was performed with Fiji/ImageJ software (NIH, Bethesda, MD, USA) and the expression levels were normalized to the input.
Quantitative real-time reverse transcriptase polymerase chain reaction
The total RNA was extracted either from hTERT-RPE1 cells, retina or RPE tissue using TRIzol reagent (Thermo Fisher, 15596026) following manufacturer’s instructions. 1 μg of RNA was reverse transcribed to cDNA using GoScript reverse transcription system (Promega, A5000). cDNA was amplified on a StepOnePlus™ Real-Time PCR System (Applied Biosystems, 4376600) using SYBR Green (Thermo Fisher; Platinum™ SYBR™ Green qPCR SuperMix-UDG, 11733046) according to the manufacturer’s recommendation. The following cycling conditions were used: 95 °C for 10 min followed by 40 cycles of 95 for 15 s, 60 for 1 min. Specificity of the amplified product was determined by melt curve analysis. Relative target gene expression was normalized to GAPDH and analyzed by comparative Ct or 2ΔΔCT method [70, 71]. For a list of primers used, see Table 1.
Electron microscopy
Electron microscopy was performed as previously described [72].
Luciferase assay
To monitor the activity of the Wnt/β-catenin signaling pathway characterized by TCF/LEF-dependent target gene transcription, reporter gene assays were performed with the Dual-Glo® Luciferase Assay System (Promega) in a 96-well plate. BBS6 and BBS8 siRNA-mediated KD in HEK293T cells were performed using Lipofectamine RNAiMax transfection reagent (Thermo Fisher) using a reverse transfection protocol according to the manufacturer’s instructions. After 24 h cells were transiently transfected with plasmids by X-tremeGENE™ 9 DNA Transfection Reagent (Sigma) according to the manufacturer’s protocol. Plasmids used were pRL-TK (Renilla luciferase, 1 ng), TopFlash (5 ng) and the total amount of transfected DNA was equalized to 80 ng by addition of pcDNA3. 24 h post-transfections cells were stimulated with Wnt3a conditioned media and luciferase activity was measured after 24 h in a Tecan Infinite M200 Pro plate reader. Firefly luciferase activity was normalized to Renilla luciferase activity in each well.
Statistical analysis
Statistical analysis was performed using Graphpad Prism 7.0 software (GraphPad Software Inc., San Diego, CA, USA). For multigroup comparisons, ANOVA followed by Dunnett’s multiple comparison test, Turkeys multiple comparison test and Sidak’s multiple comparison test was performed depending of the data to be compared. Differences between two groups were compared using a nonparametric Mann–Whitney U test. p value of 0.05 and below was considered statistically significant. Statistical tests and number of repetitions are described in the legends. Box plots show median (middle line), edge of boxes is top and bottom quartiles (25–75%), and whiskers represent the ranges for the upper 25% and the bottom 25% of data values. Outliers were excluded using the ROUT method (GraphPad Prism). Bar plots show mean ± standard deviation (SD).
References
Ross AJ et al (2005) Disruption of Bardet–Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet 37:1135–1140
Schneider L et al (2005) PDGFRαα signaling is regulated through the primary cilium in fibroblasts. Curr Biol 15:1861–1866
Goetz SC, Anderson KV (2010) The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet 11:331–344
May-Simera HL, Kelley MW (2012) Cilia, Wnt signaling, and the cytoskeleton. Cilia 1:7
Wheatley DN, Wang AM, Strugnell GE (1996) Expression of primary cilia in mammalian cells. Cell Biol Int 20:73–81
Denman-Johnson K, Forge A (1999) Establishment of hair bundle polarity and orientation in the developing vestibular system of the mouse. J Neurocytol 28:821–835
McDermott KM, Liu BY, Tlsty TD, Pazour GJ (2010) Primary cilia regulate branching morphogenesis during mammary gland development. Curr Biol 20:731–737
Blitzer AL et al (2011) Primary cilia dynamics instruct tissue patterning and repair of corneal endothelium. Proc Natl Acad Sci 108:2819–2824
Iomini C, Tejada K, Mo W, Vaananen H, Piperno G (2004) Primary cilia of human endothelial cells disassemble under laminar shear stress. J Cell Biol 164:811–817
Liang Y, Meng D, Zhu B, Pan J (2016) Mechanism of ciliary disassembly. Cell Mol Life Sci 73:1787–1802
Keeling J, Tsiokas L, Maskey D (2016) Cellular mechanisms of ciliary length control. Cells 5:6
Caspary T, Larkins CE, Anderson KV (2007) The graded response to sonic hedgehog depends on cilia architecture. Dev Cell 12:767–778
Tran PV et al (2008) THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet 40:403–410
Besschetnova TY et al (2010) Identification of signaling pathways regulating primary cilium length and flow-mediated adaptation. Curr Biol 20:182–187
Thompson CL, Chapple JP, Knight MM (2014) Primary cilia disassembly down-regulates mechanosensitive hedgehog signalling: a feedback mechanism controlling ADAMTS-5 expression in chondrocytes. Osteoarthr Cartil 22:490–498
Sánchez I, Dynlacht BD (2016) Cilium assembly and disassembly. Nat Cell Biol 18:711–717
May-Simera H, Nagel-Wolfrum K, Wolfrum U (2016) Cilia—the sensory antennae in the eye. Prog Retin. Eye Res. https://doi.org/10.1016/j.preteyeres.2017.05.001
Wheway G, Parry DA, Johnson CA (2014) The role of primary cilia in the development and disease of the retina. Organogenesis 10:69–85
May-simera HL et al (2018) Primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells article primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells. Cell Rep. 22:189–205
Plotnikova OV, Pugacheva EN, Golemis EA (2009) Primary cilia and the cell cycle. Methods Cell Biol 94:137–160
Kukic I, Rivera-Molina F, Toomre D (2016) The IN/OUT assay: a new tool to study ciliogenesis. Cilia 5:23
Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis E (2007) a. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129:1351–1363
Kim S, Lee K, Choi J-H, Ringstad N, Dynlacht BD (2015) Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nat Commun 6:8087
Fisher SK, Steinberg RH (1982) Origin and organization of pigment epithelial apical projections to cones in cat retina. J Compd Neurol 206:131–145
Nishiyama K, Sakaguchi H, Hu JG, Bok D, Hollyfield JG (2002) Claudin localization in cilia of the retinal pigment epithelium. Anat Rec 267:196–203
Williams CL et al (2011) MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol 192:1023–1041
Hernandez-Hernandez V et al (2013) Bardet-Biedl syndrome proteins control cilia length through regulation of actin polymerisation. Hum Mol Genet 22:3858–3868
Patzke S et al (2010) CSPP is a ciliary protein interacting with nephrocystin 8 and required for cilia formation. Mol Biol Cell 21:2555–2567
Wei Q et al (2012) The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol 14:950–957
Goetz SC, Bangs F, Barrington CL, Katsanis N, Anderson KV (2017) The Meckel syndrome- associated protein MKS1 functionally interacts with components of the BBSome and IFT complexes to mediate ciliary trafficking and hedgehog signaling. PLoS One 12:e0173399
Nachury MV et al (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129:1201–1213
Jin H, Nachury MV (2007) The BBSome. Curr Biol 19:472–473
Otto EA et al (2003) Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet 34:413–420
Plotnikova OV, Golemis EA, Pugacheva EN, Ciliogenesis Cell Cycle-Dependent (2008) Cell cycle–dependent ciliogenesis and cancer. Cancer Res 68:2058–2061
Mergen M et al (2013) The nephronophthisis gene product NPHP2/Inversin interacts with Aurora A and interferes with HDAC6-mediated cilia disassembly. Nephrol Dial Transpl 28:2744–2753
Simons M et al (2005) Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet 37:537–543
Loktev AV et al (2008) A BBSome Subunit Links Ciliogenesis, Microtubule Stability, and Acetylation. Dev Cell 15:854–865
Gerdes JM et al (2007) Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat Genet 39:1350–1360
Oh EC, Katsanis N (2013) Context-dependent regulation of Wnt signaling through the primary cilium. J Am Soc Nephrol 24:10–18
Liu C et al (2002) Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108:837–847
Valenta T, Hausmann G, Basler K (2012) The many faces and functions of β-catenin. EMBO J 31:2714–2736
Boatright JH et al (2015) Methodologies for analysis of patterning in the mouse RPE sheet. Mol Vis 21:40–60
Pugacheva EN, Golemis EA (2005) The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol 7:937–946
Moore FE et al (2010) The WW-HECT protein Smurf2 interacts with the Docking Protein NEDD9/HEF1 for Aurora A activation. Cell Div 5:22
Konari K et al (1995) Development of the blood-retinal barrier in vitro “ formation of tight junctions as revealed by occludin and ZO-1 Correlates with the barrier function of chick retinal pigment epithelial. Exp Eye Res 61:99–108
Burke JM, Cao F, Irving PE, Skumatz CMB (1999) Expression of E-Cadherin by human retinal pigment epithelium: delayed expression in vitro. Invest Ophthalmol Vis Sci 40:2963–2970
Gibbs D et al (2004) Role of myosin VIIa and Rab27a in the motility and localization of RPE melanosomes. J Cell Sci 26:6473–6483
Hubbert C et al (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417:455–458
Hideshima T et al (2005) Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci USA 102:8567–8572
Yuan H et al (2012) Overcoming CML acquired resistance by specific inhibition of Aurora A kinase in the KCL-22 cell model. Carcinogenesis 33:285–293
Corbit KC et al (2008) Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol 10:70–76
Li Y, Zhang X, Polakiewicz RD, Yao TP, Comb MJ (2008) HDAC6 is required for epidermal growth factor-induced β-catenin nuclear localization. J Biol Chem 283:12686–12690
Gerhardt C, Leu T, Lier JM, Rüther U (2016) The cilia-regulated proteasome and its role in the development of ciliopathies and cancer. Cilia 5:14
Verheyen EM, Gottardi CJ (2010) Regulation of Wnt/β-catenin signaling by protein kinases. Dev Dyn 239:34–44
Forcioli-Conti N, Lacas-Gervais S, Dani C, Peraldi P (2015) The primary cilium undergoes dynamic size modifications during adipocyte differentiation of human adipose stem cells. Biochem Biophys Res Commun 458:117–122
Nachury MV et al (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129:1201–1213
Jin H, Nachury MV (2009) The BBSome. Curr Biol 19:R472–R473
Seo S et al (2010) BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci USA 107:1488–1493
Sobkowicz HM, Slapnick SM, August BK (1995) The kinocilium of auditory hair cells and evidence for its morphogenetic role during the regeneration of stereocilia and cuticular plates. J Neurocytol 24:633–653
Korobeynikov V, Deneka AY, Golemis EA (2017) Mechanisms for nonmitotic activation of Aurora-A at cilia. Biochem Soc Trans 45:37–49
Jang C, Coppinger JA, Seki A, Yates JR, Fang G (2009) Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J Cell Sci 122:1334–1341
Otto EA et al (2005) Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet 37:282–288
Kinzel D et al (2010) Pitchfork regulates primary cilia disassembly and left-right asymmetry. Dev Cell 19:66–77
Gabriel E et al (2016) CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J 35:699–791
Liu YP et al (2014) Ciliopathy proteins regulate paracrine signaling by modulating proteasomal degradation of mediators. J Clin Invest 124:2059–2070
Mahuzier A et al (2012) Dishevelled stabilization by the ciliopathy protein Rpgrip1 l is essential for planar cell polarity. J Cell Biol 198:927–940
Long H, Wang Q, Huang K (2015) Ciliary/flagellar protein ubiquitination. Cells 4:474–482
Cao M, Li G, Pan J (2009) Regulation of cilia assembly, disassembly, and length by protein phosphorylation. Methods Cell Biol 94:333–346
Tadenev ALD et al (2011) Loss of Bardet-Biedl syndrome protein-8 (BBS8) perturbs olfactory function, protein localization, and axon targeting. Proc Natl Acad Sci USA 108:10320–10325
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and. Methods 25:402–408
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:45e–45
Sedmak T, Wolfrum U (2010) Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J Cell Biol 189:171–186
Acknowledgments
This work was supported by the Alexander Von Humboldt Foundation and the Johannes Gutenberg University. The authors thank Uwe Wolfrum, Kerstin Nagel-Wolfrum and Anne Régnier-Vigouroux for abundant discussion and proofreading. We especially thank Elisabeth Sehn and Gabriele Stern-Schneider for expert technical assistance and Tina Sedmark for providing TEM specimens for analysis.
Author information
Authors and Affiliations
Contributions
SRP and HLM-S were responsible for conception and experimental design. SRP, VK, LB, A-KV, SS, LRO-C and HLM-S performed experiments. SRP and VK generated figures. SRP and HLM-S co-wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declares that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patnaik, S.R., Kretschmer, V., Brücker, L. et al. Bardet–Biedl Syndrome proteins regulate cilia disassembly during tissue maturation. Cell. Mol. Life Sci. 76, 757–775 (2019). https://doi.org/10.1007/s00018-018-2966-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2966-x