Abstract
When temperatures plummet below 0 °C, wood frogs (Rana sylvatica) can endure the freezing of up to ~ 65% of their body water in extracellular ice masses, displaying no measurable brain activity, no breathing, no movement, and a flat-lined heart. To aid survival, frogs retreat into a state of suspended animation characterized by global suppression of metabolic functions and reprioritization of energy usage to essential survival processes that is elicited, in part, by the regulatory controls of microRNAs. The present study is the first to investigate miRNA biogenesis and regulation in the brain of a freeze tolerant vertebrate. Indeed, proper brain function and adaptations to environmental stimuli play a crucial role in coordinating stress responses. Immunoblotting of miRNA biogenesis factors illustrated an overall reduction in the majority of these processing proteins suggesting a potential suppression of miRNA maturation over the freeze–thaw cycle. This was coupled with a large-scale RT-qPCR analysis of relative expression levels of 113 microRNA species in the brains of control, 24 h frozen, and 8 h thawed R. sylvatica. Of the 41 microRNAs differentially regulated during freezing and thawing, only two were significantly upregulated. Bioinformatic target enrichment of the downregulated miRNAs, performed at the low temperatures experienced during freezing and thawing, predicted their involvement in the potential activation of various neuroprotective processes such as synaptic signaling, intracellular signal transduction, and anoxia/ischemia injury protection. The predominantly downregulated microRNA fingerprint identified herein suggests a microRNA-mediated cryoprotective mechanism responsible for maintaining neuronal functions and facilitating successful whole brain freezing and thawing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Freezing is deadly for most organisms. Yet despite the challenges it poses, freeze tolerance has arisen multiple times across the eukaryotic kingdom ranging from microorganisms, plants, insects, invertebrates, reptiles, and amphibians [1,2,3,4,5]. One of the most well-studied and extreme vertebrate models of natural freeze tolerance is the wood frog, Rana sylvatica (also known as Lithobates sylvaticus), capable of freezing up to 65% of total body water in extracellular ice masses. When the wood frog retreats into its frozen state of suspended animation, that can last months at a time, it exhibits no measurable brain activity, no breathing, no movement, and a flat-lined heart [6]. To freeze and thaw months later, unscathed, wood frogs have developed various key adaptations that include the following: (1) initiation of freezing above the freezing point of body fluids at (− 0.5 °C), to minimize damaging instantaneous ice surges that occur if animals are extensively supercooled prior to freezing (2) synthesis and distribution of cryoprotectant glucose to minimize cell volume reduction and limit extracellular ice formation, and (3) global metabolic rate depression coupled with the activation of select “survival” pathways that protect and stabilize cellular macromolecules [5].
Wood frogs are subject to numerous freeze-associated challenges such as anoxia/ischemia, dehydration, hyperglycemia, osmotic shock, mechanical damage from ice crystallization, and even thawing brings with it the dangers of rapid oxygen reperfusion [5]. To survive such extreme environmental stress, these frogs have evolved various protective mechanisms, such as the upregulation of antioxidant defenses [7], coupled with the global reprioritization of energy consumption [8]. While freezing itself is a prolonged dormant state that is characterized by global metabolic rate depression, entrance into the frozen state does not involve extensive gene expression changes [9]. This is likely because the energy-limited frozen state does not facilitate major reorganizations of the cellular environment. To survive, it appears that the frog has adapted molecular mechanisms that are; (1) modulated by environmental stimuli, (2) applicable to nearly all biological pathways, (3) easily inducible, (4) not energetically costly, and (5) rapidly reversible to allow for smooth transitions between active and dormant states [10]. Indeed, studies on various animal species that use hypometabolism have found that this complex metabolic state is orchestrated by a multitude of molecular mechanisms that fit these criteria including; epigenetic modifications of DNA (e.g. methylation) and histones, transcription factors, changes in transcription factor activity and signal transduction, in addition to post-transcriptional regulation of mRNA transcripts by RNA-binding proteins and the inhibitory action of microRNAs (miRNAs) [10].
MicroRNAs are short (~ 22 nt) non-coding RNA transcripts that have been predicted to target more than 60% of protein-coding genes in humans [11, 12]. This large and highly conserved group is proving to be master regulators of virtually all cell processes with broad controls stretching into cell cycle, signal transduction, and energy metabolism pathways, among others [13, 14]. MicroRNAs rely on sequence-specificity to mediate post-transcriptional gene suppression via either translational inhibition or mRNA degradation, where a single miRNA can target multiple mRNAs and each mRNA can have multiple miRNA binding sites [12]. Added to this, the thermodynamic nature of miRNA–mRNA interactions is sensitive to changes in the cellular environment, such as temperature [15]. This leaves the miRNA–mRNA duplex vulnerable to large fluctuations in body temperature such as those experienced by ectothermic species, where drops in body temperature result in a decrease of the free energy required for the formation of the RNA duplex which in turn favors the binding of miRNA–mRNA [15, 16]. Indeed, events such as freezing have been shown to result in significant changes in miRNA functionality and the individual mRNA molecules they target [17].
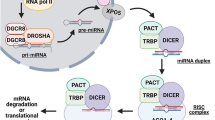
The biogenesis and processing of microRNAs is under tight spatial and temporal control [18]. Canonical microRNA biogenesis is a stepwise process that progresses from long imperfect dsRNA-like hairpins known as primary-miRNAs (pri-miRNAs). Maturation ensues with the cleavage of the 5′ and 3′ ends by DROSHA, a Class 2 RNase III endonuclease, and its binding partner DiGeorge’s syndrome critical region gene 8 (DGCR8), a dsRNA binding protein, to release the ~ 60–70 nt precursor-miRNA (pre-miRNA) [19]. Pre-miRNA then complexes with the nucleoplasmic transporter factor EXPORTIN-5 (XPO5) and RAN–GTP to protect against nuclear degradation and facilitate nuclear export into the cytoplasm for further processing [20]. Once in the cytoplasm, the RNAse III endonuclease DICER cleaves the loop structures to yield ∼ 21 nt miRNA duplexes [21, 22]. This is facilitated by transactivator response RNA binding protein (TRBP) and protein kinase RNA activator (PACT) that complex with DICER to modulate efficient pre-miRNA processing [23, 24]. While one of the miRNA strands is usually degraded, the mature guide strand is incorporated into the effector (RISC) complex along with a member of the Argonaute family of endonucleases (AGO1-4), where miRNAs then use base-pairing to negatively regulate the expression of target mRNAs [12].
Proper brain development and function rely on the gene-regulatory capacity of miRNA networks to coordinate between the transcriptomes and proteomes of the diverse neural cell types and their functional specializations [25, 26]. The brain’s persistent flow of information and adaptations to environmental stimuli plays a pivotal role in the facilitation of stress responses and prolonged hypometabolic periods [27]. Despite the lack of any measurable brain activity or nerve conductance [5], biochemical studies on the brains of frozen R. sylvatica have revealed various neuroprotective aspects such as the increase in li16 and fr10 transcripts that encode novel protein that have been linked with the mitigation of oxygen restriction damage and freeze protection, increased levels of c-FOS, and the elevation of phosphorylated Smad3 levels that has been implicated in the promotion of tissue survival [28,29,30,31]. Other neuronal “activations” found to occur in frozen wood frog brains include the following: increased protein kinase C phosphorylation status, the upregulation of the acidic ribosomal phosphoprotein P0, and increased levels of the large ribosomal subunit protein 7 [32,33,34]. Our initial investigations into the regulatory response of miRNAs in wood frog heart, skeletal muscle, and liver have demonstrated tissue-specific differential expression of miRNAs involved in the maintenance of muscle contraction and reversible protein phosphorylation kinases over the freeze–thaw cycle [35, 36].
A growing number of studies by our lab, and others, have implicated miRNAs in various stress responses and adaptations in diverse animals ranging from; marsupial hibernation [37], sea cucumber estivation [38], invertebrate anoxia tolerance [39], insect freeze avoidance [40], and turtle, frog, and insect freeze tolerance [5, 41]. In the present study, which to our knowledge is the first investigation of miRNAs in the brain of a freeze tolerant species, immunoblotting of the miRNA biogenesis proteins revealed a potential suppression of miRNA processing over the freeze–thaw cycle in wood frog brains. Large-scale analysis of the expression levels of 113 miRNAs, using RT-qPCR, found that the 41 differentially regulated miRNAs displayed a pattern of downregulation during both freezing and thawing. Subsequent bioinformatic miRNA target prediction of the downregulated miRNAs was then carried out at physiologically relevant low temperatures of − 2 and 5 °C, for 24 h frozen and 8 h thawed conditions, respectively. Functional enrichment and clustering revealed that the mRNA targets of the downregulated miRNA networks mediate neuroprotective processes and hypoxia/anoxia protection. Our findings suggest a miRNA-mediated mechanism for tuning of neuronal functions and brain cryoprotection over the course of the freeze–thaw cycle. Understanding the role that miRNAs play in facilitating successful freeze tolerance and protecting living brain tissue and neuronal networks from freeze-associated damages sheds light on new approaches on the basic processes that must be maintained for successful brain function.
Materials and methods
Animals
Mature male R. sylvatica were collected from spring meltwater ponds in the Ottawa region and transported on ice to an animal facility at Carleton University. Frogs were washed in a tetracycline bath and acclimated at 5 °C for 2 weeks in containers lined with sphagnum moss. Three experimental treatments were run in parallel with individuals randomly selected for control, frozen, and thawed conditions. Control (non-stressed) frogs were sampled from this condition. For freezing experiments, frogs were placed in an incubator at − 4 °C in plastic containers lined with damp paper towels. An initial (45 min) cooling period was used to initiate freezing during which the body temperature cooled to below − 0.5 °C, at which point ice nucleation was triggered due to skin contact with ice crystals formed on the paper towels, as previously described [73]. Following the initial 45 min, the incubator temperature was raised to − 2.5 °C and a 24-h freezing exposure was timed from this point (maximal ice formation is usually achieved within 12–15 h) [6]. Frogs were randomly assigned to either the freezing group that was sampled after the 24-h freezing exposure, or to the recovery group and were subsequently transferred to 5 °C and sampled after 8 h of thawing. Animals were euthanized by spinal pithing and brains were rapidly dissected, immediately flash frozen in liquid nitrogen, and then stored at − 80 °C until use. Animal care protocols, experimentation, and euthanasia were performed in accordance with the Carleton University Animal Care Committee and followed the guidelines set forth by the Canadian Council on Animal Care.
Total protein extraction
Total protein extracts were prepared as previously described [30]. Briefly, ~ 100 mg of brain tissue (n = 4 individual animals) was homogenized in 1:2 w:v chilled homogenization buffer (20 mM Hepes, 200 mM NaCl, 0.1 mM EDTA, 10 mM NaF, 1 mM Na3VO4, 10 mM β-glycerophosphate, pH 7.5) with a few crystals of phenylmethylsulfonyl fluoride (Bioshop) and 1 μL/mL of protease inhibitor (Cat# PIC002; Bioshop Canada Inc., Burlington, ON, Canada) using a Polytron PT10 homogenizer. Samples were then centrifuged at 4 °C for 15 min at 10,000g, and soluble protein-containing supernatants were collected. Protein concentrations were measured using the BioRad reagent (Cat# 5000002; Hercules, CA) as per the manufacturer’s instructions. Soluble brain protein concentrations were standardized to 10 µg/µL using homogenization buffer and mixed with 1:1 v:v SDS buffer (100 mM Tris–HCl, 4% w:v SDS, 20% glycerol, 0.2% w:v bromophenol blue, 10% v:v β-mercaptoethanol, pH 6.8) to yield a final concentration of 5 µg/µL. Samples were then boiled for 10 min and subsequently stored at − 20 °C until use.
Immunoblotting
Equal amounts of total protein homogenates (20–40 µg depending on target being probed) from each sample were loaded onto 6–15% polyacrylamide gels and prepared with 5% upper stacking gels. Proteins were then separated in SDS-PAGE running buffer (190 mM glycine, 0.1% w/v SDS, 25 mM Tris-base [pH 6.8]) at 180 V for 60–120 min at 4 °C with a BioRad Mini Protean III system. Proteins were then electroblotted by wet transfer onto a 0.45-µm polyvinylidene difluoride membrane in a pre-chilled transfer solution (25 mM Tris [pH 8.8], 192 mM glycine, and 10% v/v methanol) at 4 °C for 1–16 h at 160 mA. Transferred membranes were air-dried for 15 min, reactivated in methanol for 5 min, and incubated in 2–10% v/v skim milk in TBST (20 mM Tris base [pH 7.6], 140 mM NaCl, 0.05% v:v Tween-20, and 90% v:v dd H2O), and blocked for 10–45 min in 2–10% w:v milk in TBST. This was followed by 3 × 5 min washes with TBST and overnight incubations with the appropriate primary antibodies (1:1000 v:v dilution in TBST) at 4 °C. Primary antibodies were purchased for DROSHA (NeoBiolab; Cat# A8336), DGCR8 (GeneTex; Cat# GTX130061), DICER (SantaCruz; Cat# SC-30226), AGO1 (GeneTex; Cat# GTX47799), AGO2 (ECM BioSciences; Cat# AP5281), p-AGO2Tyr393 (ECM BioSciences; Cat# AP5311), RAN (GeneTex; Cat# GTX114139), EXPORTIN-5 (GeneTex; Cat# 130727), TRBP (GeneTex; Cat# GTX485546), and PACT (GeneTex; Cat# GTX114215). After probing with primary antibody, membranes were washed for 3 × 5 min with TBST and incubated with HRP-linked anti-rabbit secondary antibody (BioShop; 1:8000 v:v dilution in TBST) for 30 min at room temperature. Membranes were then washed in TBST and bands were visualized with enhanced chemiluminescence (H2O2 and Luminol) on a ChemiGenius Bio-Imaging System (Syngene, Frederick, MD). Membranes were then stained with Coomassie blue (0.25% w/v Coomassie brilliant blue, 7.5% v:v acetic acid, and 50% v:v methanol) to visualize total protein levels.
Total RNA extraction
Isolation of RNA was conducted as previously described [37]. Approximately 50 mg of brain tissue (n = 4 individual animals) was briefly homogenized using a Polytron PT1200 homogenizer in 1 mL of Trizol (Invitrogen; Cat# 15596-018), as per the manufacturer’s instructions. RNA quality was assessed by measuring the 260/280 nm ratio (> 1.8) using a Take3 micro-volume quantification plate (BioTek) and a PowerWave HT spectrophotometer (BioTek). Total RNA integrity was determined by running RNA isolates on a 1% agarose gel stained with SYBR Green and verifying the presence of sharp bands for 28S and 18S ribosomal RNA. All RNA samples were standardized to a final concentration of 1 μg/μL with RNase-free water. Samples were frozen at − 80 °C until use.
MicroRNA polyadenylation and stem-loop reverse transcription
RNA samples were prepared for miRNA analysis as previously described [54]. Polyadenylation was carried out using the PolyA tailing kit from Epi-Bio (Cat# PAP5104H; Epicentre, Madison, WI, USA). Each 10 µL reaction contained; 3 µg RNA, 1 mM ATP, and 0.5 µL (2 U) of E. coli poly (A) polymerase in buffered solution (0.1 M Tris–HCl [pH 8.0], 0.25 M NaCl, and 10 mM MgCl2). Reactions were incubated for 30 min at 37 °C to adenylate, for 5 min at 95 °C to arrest, and immediately chilled on ice. Stem-loop adapter ligation was carried out by combining the polyadenylated products (10 µL samples) with 5 µL of 250 pM stem-loop adapter primers (Supp. Table S1) and heating the mixture for 5 min at 95 °C to denature RNA, incubating it for 5 min at 60 °C to allow annealing, and then immediately chilling on ice. For reverse transcription, each polyadenylated and stem-loop ligated RNA sample (15 µL) was then combined with the following: 1 µL mouse Maloney leukemia virus (M-MLV) reverse transcriptase (2 U) (Cat# 18080044; ThermoFisher Scientific), 1 µL deoxynucleotide triphosphate (dNTP) mixture containing 25 mM of each nucleotide (Cat# R1121; ThermoFisher Scientific), 2 µL 0.1 M dithiothreitol (DTT), and 4 µL 5× first-strand buffer (Cat# 18080044; ThermoFisher Scientific). Samples were then incubated at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min. Polyadenylated, stem-loop ligated, and reverse transcribed cDNA was serially diluted and stored at − 20 °C.
Relative microRNA quantification
All RT-qPCR assays were performed using a BioRad MyIQ2 Detection System (BioRad, Hercules, CA, USA), as previously described [37]. The miRNA-specific forward primers were designed based on annotated miRNA sequences of Xenopus obtained from miRBase (Release 21). All miRNA-specific forward primers and the universal reverse primer were designed using the previously described method to amplify conserved miRNAs [54] (Supp. Table S1). Primers were synthesized by Integrated DNA Technologies. To ensure primer specificity and the amplification of a single PCR product, all RT-qPCR assays were subjected to a post-run melt-curve analysis; reactions that amplified non-specific products were rejected.
Bioinformatic microRNA target identification and pathway enrichment
To characterize the potential miRNA targets, mature miRNA sequences were searched against the 3′UTR sequences from the Xenopus tropicalis reference genome available on the UCSC table browser (JGI 7.0/xenTro7) using FINDTAR3 (v.3.11.12) [74]. FINDTAR3 was used with the following parameters: AT and GC weight of 5, GT weight of 2, a gap opening penalty of − 8, a gap extension penalty of − 2, target duplex with maximum threshold free energy − 20 kcal/mol, and demand strict 5′ seed pairing. Target predictions were carried out at a temperature of − 2 °C for miRNAs differentially expressed during freezing and at 5 °C for miRNAs that changed during thawing. FINDTAR3 generated a list of miRNA-targeted genes that were then functionally enriched and analyzed. To characterize the functions and potential interplay between miRNA-targeted mRNA, protein–protein interactions were mapped using only the high-confidence STRING X. tropicalis interaction database [75]. This was subsequently modeled using CYTOSCAPE software [76] and clustered using Markov clustering (MCL) default clustering parameters on the STRING combined scores. Two groups were queried separately: [1] the 23 miRNAs downregulated in frozen brains, and [2] the 33 miRNAs downregulated in thawed brains. Pathway enrichment analysis for miRNA-targeted clusters was performed on miRNAs found to significantly change in response to freezing and thawing. This analysis was performed using the Gene Ontology (GO) Enrichment Analysis available for X. tropicalis using the PANTHER classification system (v.11.1). The enriched biological processes discussed herein are statistically significant clusters analyzed with Bonferroni tests (p < 0.05).
Data quantification and statistics
For relative protein level quantification, chemiluminescent bands were quantified by densitometry using a ChemiGenious BioImaging Sytem and GeneTools Software (Syngene, MD, USA). To correct for minor variations in sample loading, the band intensity in each lane was standardized against a group of Coomassie blue-stained protein bands. Immunoblot data are expressed as mean ± SEM, relative to control values (n = 3–4 from different animals). Data were statistically analyzed using one-way ANOVA and a Dunnett’s post hoc test and results were considered significant when a Dunnett’s test resulted in p < 0.05. Statistical analyses were performed using the RBIOPLOT statistics and graphing R package [77].
For RT-qPCR analyses, the comparative ΔΔCq method was used to calculate the relative miRNA expression levels. Raw Cq values were transformed to the 2−Cq form, each individual miRNA was then normalized to the reference gene, snord68. Snord68 RNA was experimentally deemed a suitable reference gene as it exhibited stable expression under control, frozen, and thawed conditions, as previously described [78]. Data are reported as mean expression levels, relative to control values (± SEM, where n = 3–4 independent biological replicates of tissue from different animals). Data were statistically analyzed using a one-way ANOVA and a Dunnett’s post hoc test, and miRNAs were considered significantly changing when p < 0.05. Statistical analysis and histogram generation was performed using RBIOPLOT [77].
Results
Protein expression of miRNA biogenesis and processing machinery
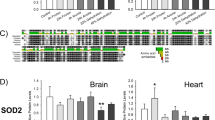
Using immunoblotting, we analyzed protein abundance relative to control for key proteins involved in the microRNA biogenesis pathway in wood frog brains from 24 h frozen and 8 h thawed frogs. Levels of DROSHA and DGCR8, the two sole members of the microprocessor complex, significantly decreased during 24 h freezing to 0.67 ± 0.04 and 0.67 ± 0.06 of control levels, respectively (Fig. 1). This decrease was sustained through the 8-h thawing with DROSHA and DGCR8 levels at 0.63 ± 0.05 and 0.43 ± 0.01 of control, respectively (Fig. 1). EXPORTIN-5 protein levels significantly decreased in the brain during freezing and thawing to 0.75 ± 0.04 and 0.77 ± 0.04 of control, respectively (Fig. 1). DICER and RAN protein levels remained constant over the freeze–thaw cycle (Fig. 1). TRBP protein levels also remained constant during freezing but significantly decreased to 0.58 ± 0.04 of control during thawing (Fig. 1). Protein levels of the dsRNA binding protein PACT decreased to 0.55 ± 0.03 of control during freezing and returned to control levels during thawing (Fig. 1). AGO1 protein levels were found to remain constant over the freeze–thaw cycle (Fig. 1). Total protein levels of the predominant AGO2 significantly decreased during both freezing and thawing to 0.75 ± 0.05 and 0.56 ± 0.01 of control, respectively (Fig. 1). Levels of p-AGO2Tyr393 remained unchanged during freezing but significantly decreased to 0.60 ± 0.03 of control during 8 h thawing.
Analysis of miRNA biogenesis pathway protein levels in wood frog brain over a freeze–thaw cycle using immunoblotting. The histogram shows protein levels, relative to control, of DROSHA, DGCR8, XPO5, RAN, DICER, TRBP, PACT, AGO1, AGO2, and p-AGO2Tyr393 under control, 24 h frozen, and 8 h thawed conditions. Data are mean ± SEM of 3–4 independent biological replicates, relative to control values. Data are analyzed using a one-way ANOVA with a Dunnett’s post hoc test, *p < 0.05
Differential miRNA expression over the freeze–thaw cycle
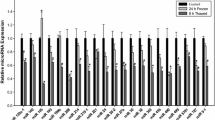
The primary goal of this study was to identify freeze and thaw-specific patterns of miRNA expression in the brains of R. sylvatica. Of the 113 miRNAs successfully quantified by RT-qPCR, 41 miRNAs were differentially regulated over the freeze–thaw cycle, relative to control levels (Fig. 2). During freezing, expression of 24 miRNAs significantly changed (Fig. 2). Nearly all of the miRNAs differentially expressed during freezing were downregulated, with 23 miRNAs significantly decreasing to 0.48–0.84 of control levels. The subset of miRNAs significantly downregulated during freezing comprised: rsy-miR-103/107-3p, rsy-miR-10b-5p, rsy-miR-125a-5p, rsy-miR-126-3p, rsy-miR-140-5p, rsy-miR-145-5p, rsy-miR-155-5p, rsy-miR-183-5p, rsy-miR-184-3p, rsy-miR-18-5p, rsy-miR-193-3p, rsy-miR-196b-5p, rsy-miR-204-5p, rsy-miR-210-3p, rsy-miR-214-3p, rsy-miR-24a-5p, rsy-miR-2970-5p, rsy-miR-30b-5p, rsy-miR-30d-5p, rsy-miR-31b-5p, rsy-miR-367-3p, rsy-miR-449b-5p, and rsy-miR-9407-5p (Fig. 2 and Supp. Table S2). Only rsy-miR-451-5p expression levels significantly increased during freezing to 2.11 ± 0.32 of control (Fig. 2 and Supp. Table S2).
Heatmap of RT-qPCR measured expression levels of 113 miRNA species examined in 24 h frozen and 8 h thawed wood frog brain, relative to control. All miRNAs were standardized against Snord68 reference gene expression. Data are mean ± SEM of 3–4 independent biological replicates. Statistical testing used a one-way ANOVA with a Dunnett’s post hoc test, *p < 0.05, **p < 0.01, and ***p <0.001. The legend provides a visual reference for the color gradient used. Different shades of red represent significant downregulation of miRNA in the frozen and thawed states versus control; increasing redness signifies greater relative downregulation during stress. Black represents no significant changes. Increasing greenness represents greater upregulation of miRNA during stress versus control. For the relative expression ± SEM values of all 113 miRNA species examined, refer to Supplementary Table S2
During thawing, 34 miRNAs were differentially expressed (Fig. 2). Similar to the pattern observed during freezing, the majority of the differentially expressed miRNAs were found to significantly decrease during thawing, with 33 miRNAs significantly decreasing to 0.42–0.85 of control (Fig. 2 and Supp. Table S2). The subset of miRNAs downregulated during thawing were as follows: rsy-miR-let-7f-3p, rsy-miR-103/107-3p, rsy-miR-10b-5p, rsy-miR-125a-5p, rsy-miR-126-3p, rsy-miR-129-5p, rsy-miR-130a-5p, rsy-miR-140-5p, rsy-miR-145-5p, rsy-miR-155-5p, rsy-miR-181a-3p, rsy-miR-183-5p, rsy-miR-184-3p, rsy-miR-193-3p, rsy-miR-196b-5p, rsy-miR-204-5p, rsy-miR-208-3p, rsy-miR-210-3p, rsy-miR-212-3p, rsy-miR-221-3p, rsy-miR-222-3p, rsy-miR-22-5p, rsy-miR-24a-5p, rsy-miR-26-3p, rsy-miR-26-5p, rsy-miR-30b-5p, rsy-miR-30d-5p, rsy-miR-31b-5p, rsy-miR-365-3p, rsy-miR-425-5p, rsy-miR-449c-3p, rsy-miR-96-5p, and rsy-miR-98-5p (Fig. 2 and Supp. Table S2). Despite a backdrop of unchanging and downregulated miRNAs, rsy-miR-192-5p was upregulated during thawing to 2.19 ± 0.39 of control.
A subset of miRNAs were shown to be downregulated during both freezing and thawing: rsy-miR-103/107-3p, rsy-miR-10b-5p, rsy-miR-125a-5p, rsy-miR-126-3p, rsy-miR-140-5p, rsy-miR-145-5p, rsy-miR-155-5p, rsy-miR-183-5p, rsy-miR-184-3p, rsy-miR-193-3p, rsy-miR-196b-5p, rsy-miR-204-5p, rsy-miR-210-3p, rsy-miR-24a-5p, rsy-miR-30b-5p, rsy-miR-30d-5p, and rsy-miR-31b-5p (Fig. 2 and Supp. Table S2).
Bioinformatic analyses of miRNA-targeted pathways
Using miRNA–mRNA target predictions at physiologically relevant temperatures, protein–protein interaction networks, and biological process enrichment, the following targeted gene networks and processes were identified. The functionally enriched clusters predicted that the key pathways targeted by the miRNAs downregulated during the 24-h frozen condition consisted of the following: (1) intracellular signal transduction pathways, (2) RNA processing and mRNA splicing, (3) synaptic signaling, (4) lipid phosphorylation, (5) microtubule nucleation, (6) DNA replication, (7) generation of precursor metabolites and energy, (8) developmental process, and (9) proton transport and ATP hydrolysis (Fig. 3, Supp. Table S3, Supp. Figure S1). The key networks targeted by the miRNAs that were downregulated after 8 h thawing were comprised of (1) intracellular signal transduction, (2) DNA replication, (3) proton transport, (4) RNA processing and mRNA splicing, (5) protein folding and chaperones, (6) synaptic signaling, (7) ATP hydrolysis, (8) microtubule nucleation, (9) generation of precursor metabolites and energy, (10) lipid phosphorylation, (11) oligosaccharide-lipid intermediate biosynthesis, and (12) developmental process (Fig. 4, Supp. Table S4, and Supp. Figure S2). The majority of remaining clusters in both freezing and thawing remained unclassified, likely due to the limited nature of annotated X. tropicalis network interactions.
Representative functional target enrichment and network clusters of the subset of miRNAs downregulated in the brains of 24 h frozen wood frogs. Downstream miRNA target prediction was performed at − 2 °C using FINDTAR3. Protein–protein interactions of the downstream networks was performed using the STRING high-confidence filter on the X. tropicalis database. MCL clustering and visualization was performed on CYTOSCAPE and coupled with functional biological enrichment using GO ANALYSIS. Refer to Supplementary Table S3 for more information on individual clusters, proteins, and the targeting of individual miRNA species. Refer to Supplementary Figure S1 for full 24 h frozen cluster map
Representative functional target enrichment and network clusters of the subset of miRNAs downregulated in the brains of 8 h thawed wood frogs. Downstream miRNA target prediction was performed at 5 °C using FINDTAR3. Protein–protein interactions of the downstream networks was performed using the STRING high-confidence filter on the X. tropicalis database. MCL clustering and visualization was performed on CYTOSCAPE and coupled with functional biological enrichment using GO ANALYSIS. Refer to Supplementary Table S4 for more information on individual clusters, proteins, and the targeting of individual miRNA species. Refer to Supplementary Figure S2 for full 8 h thawed cluster map
Discussion
The rapid and reversible nature of miRNA-mediated gene silencing renders these small molecules as excellent candidates for fine-tuning the gene expression profile required to protect brain networks and processes during prolonged periods of severe environmental stress. Studies on diverse animal models have demonstrated that the brain is critical for coordinating environmental stimuli and for mounting molecular stress responses. While global metabolic rate depression is one of the main characteristics of freeze tolerance, neuroprotective processes, such as the promotion of tissue survival, have been shown to be active and upregulated in frozen wood frog brains, emphasizing the importance of maintaining brain functionality [30]. One of the main questions we sought to investigate was—what happens to the brains of freeze tolerant animals? Indeed, while it has been reported that brains from frozen wood frogs display no measurable brain activity and both visual observations and magnetic resonance imaging have shown ice accumulation in brain ventricles, little other physiological work has been done on freeze tolerant brains [5]. In the present study, we investigated the regulation of microRNA biogenesis and the expression levels of 113 miRNAs over the course of a freeze–thaw cycle in wood frog brain. This lead to the identification of a global pattern of downregulation of both miRNA biogenesis and relative expression levels. This freeze–thaw specific miRNA fingerprint of downregulation suggests that wood frog brains may utilize miRNAs in a cryoprotective manner to maintain neural networks and circumvent extensive transcriptional and translational repression.
Recent studies have demonstrated that the activity, function, and abundance of miRNA–protein complexes can alter miRNA biogenesis via the regulation of environmental and stress-responsive signaling pathways [14]. An examination of the miRNA biogenesis and processing proteins revealed an overall pattern of downregulation over the freeze–thaw cycle (Fig. 1). As the efficiency of DROSHA-mediated processing is crucial for determining miRNA abundance, the observed reduction of microprocessor protein levels suggests a freeze-induced reduction in initial miRNA maturation and pre-miRNA synthesis that could be a result of the energy limitations imposed during freezing and thawing [42]. Furthermore, reduced levels of EXPORTIN-5 have been shown to lead not only to reductions in cytoplasmic miRNA levels, but also to pre-miRNA nucleolytic degradation in the nucleus (Fig. 1) [43]. However, this warrants further investigation as protein levels of the RAN GTP-binding protein, that complexes with EXPORTIN-5 to transport pre-miRNAs into the cytoplasm, remained constant over the freeze–thaw cycle (Fig. 1) [20, 44]. Following nuclear export, nascent pre-miRNAs are cleaved into mature miRNA duplexes by DICER’s endonuclease activity and while DICER levels remained constant during both freezing and thawing, protein levels of two other components of the RISC complex, TRBP and PACT, were downregulated during thawing and freezing, respectively (Fig. 1). Indeed, reduced levels of TRBP and PACT lower the efficiency of post-transcriptional gene silencing and the depletion of TRBP could play a role in DICER destabilization and miRNA downregulation [18, 45]. However, it should be noted that TRBP and PACT are non-essential facilitators of DICER-mediated pre-miRNA processing [46].
After processing, the miRNA guide strand is then loaded onto an Argonaute protein effector complex. While levels of AGO1, that is known to interact with only ~ 30% of miRNAs, did not change in wood frog brains, protein levels of AGO2, that has been shown to interact with 60% of all miRNAs, were downregulated over the freeze–thaw cycle [47] (Fig. 1). AGO2 is the sole Argonaute with slicer activity and the loss of AGO2 has been demonstrated to result in a reduction of mature miRNA expression and activity [45]. However, levels of p-AGO2Tyr393 were downregulated during thawing which contradicted the overall observed suppression of miRNA biogenesis since this phosphorylation, commonly associated with hypoxic conditions, has been shown to result in the inhibition of miRNA processing and DICER binding (Fig. 1) [48]. Indeed, AGO2 is a prime candidate for the regulation of miRNA biogenesis and function and our findings suggest that there is likely diminished mature mRNA translational inhibition, destabilization, and/or target degradation. Whereas our data suggest an overall reduction in miRNA biogenesis and processing, especially during the nuclear maturation steps, future studies should explore the regulation of various non-canonical miRNA biogenesis mechanisms such as the DROSHA and DGCR8-independent pathway, the DICER-independent pathway, and the terminal uridylyl transferase dependent pathway that might be used to circumvent the reduced levels of the miRNA biogenesis factors analyzed in the current study [18].
Initial studies on frozen wood frog miRNAs examined the regulation of miR-16 and miR-21 in liver and skeletal muscle and 53 miRNA species in cardiac and skeletal muscle over the course of a freeze–thaw cycle [35, 36]. The present large-scale study provides us with a better understanding of the cellular mechanisms that contribute to successful brain freeze tolerance. During freezing, 23 of the 113 miRNA species analyzed in brain were shown to be significantly downregulated whereas rsy-miR-451-5p was the only miRNA that was freeze-upregulated (Fig. 2). Previous work on miR-451 has shown that this miRNA acts as a conditional glucose-sensing switch, whereby high glucose levels result in its overexpression, which in turn leads to the suppression of downstream protein kinases and the PI3K/AKT pathway, resulting in unrestrained mTOR activation [49, 50]. Indeed, during freezing, not just the brain, but all wood frog organs, are flooded with large amounts of cryoprotectant glucose that protect against mechanical ice damage and excessive cellular dehydration [5]. The upregulation of rsy-miR-451-5p could be a result of abundant glucose levels but could also be involved in maintaining neural functions during freezing. In addition, miR-451 has also been shown to protect against anoxia/reoxygenation injury, a stress that frozen wood frog brains are vulnerable to, through the inhibition of 14-3-3ζ and high mobility group box 1 expression, respectively [51, 52].
While the majority of miRNA species measured exhibited constant expression levels over the freeze–thaw cycle, 23 of the differentially expressed miRNAs were significantly downregulated during freezing (Fig. 2). Predicted targets of the freeze-downregulated miRNAs, generated from annotated Xenopus sequences, were collectively analyzed and found to potentially be involved in maintaining cellular processes such as intracellular signal transduction, RNA processing, and synaptic signaling, among others (Fig. 3). The reduced levels of these differentially expressed miRNAs implies that the gene transcripts under their control are more likely to be translated, which in turn implies that the protein product that the gene encodes will be expressed and may play a role in facilitating freeze tolerance. The various biological processes targeted by the downregulated miRNAs appear to be involved in the maintenance of basic neuronal cell function and survival. Targeting of proteins essential for neurotransmitter release and pre-synaptic functions such as synaptosomal-associated protein of 25 kDa (SNAP25) suggests an enhancement of their roles during freezing, possibly in a protective manner. Downregulated miRNAs 103/107-5p, 10b-5p, 183-5p, 184-3p, 18-5p, 193-3p, 196b-5p, 210-3p, 214-3p, 2970-5p, 449b-5p (Supp. Table S3) were predicted to target SNAP25, a known pre-synaptic reactive oxygen species (ROS) sensor that exhibits ROS-induced functional impairment. This suggests that its predicted upregulation during freezing and thawing could be in an effort to minimize impairment and promote synaptic vesicle priming and signaling [53].
Interestingly, a similar activation of long-term potentiation and axon guidance was reported in other miRNA studies of brains utilizing hypometabolic survival strategies including during mammalian hibernation and frog estivation [54, 55]. Interestingly, this pattern of downregulation and the promotion of neuroprotective mechanisms have also been reported in whole brains of another environmentally stressed frog, the estivating African-clawed frog, where 12 of the 43 miRNAs measured exhibited significant downregulation [55]. The downregulation of miRNAs in the brains of environmentally stressed anurans may be an evolutionarily conserved response. Indeed, while freezing is a unique physiological response, it builds on and exaggerates many pre-existing strategies that stem from the general ability of anurans to endure wide variations in body hydration, including severe dehydration under arid conditions [5, 56]. Since cellular dehydration, due to water exit into extracellular ice masses, is one of the main stresses on cells of freezing frogs, it is not surprising that they enlist the ancient defense strategies for dehydration resistance that are shared among all amphibians. The target miRNAs that changed during freezing were largely involved in regulating brain neural plasticity. Despite the fact that wood frogs display no measurable brain activity during freezing, upon thawing they are able to immediately resume normal functions with no apparent neurological damage or deleterious behavioral changes. Therefore, we propose the existence of miRNA-mediated neuroprotective mechanisms to circumvent the degeneration of neural synapses and protect against negative effects of prolonged states of hypometabolic dormancy. Indeed, our data suggest that rather than modulating metabolic rate depression, the differentially expressed miRNAs in brains of frozen wood frogs appear to be responsible for neural network plasticity and maintenance. Our findings point away from a miRNA-mediated mechanism for a freeze-induced state of translational suppression but rather towards a role for miRNAs as tuners of neuronal maintenance and function.
Thawing exhibited a similar but more pronounced response to that of freezing, with the downregulation of 33 miRNAs and the upregulation of only rsy-miR-192-5p (Fig. 2 and Supp. Table S2). Bioinformatic target enrichment, performed using Xenopus sequences, implicated the downregulated miRNAs in the predicted activation of numerous brain-specific cytoprotective processes such as chaperone-mediated protein folding, synaptic signaling, and intracellular signal transduction (Fig. 4 and Supp. Table S4). Protein folding networks are a crucial cell preservation strategy essential for ensuring protein homeostasis via the proper assembly of new proteins, the prevention of their aggregation, and the promotion of their efficient folding [57]. Molecular chaperones have been shown to be cold/freeze upregulated in cold-hardy arthropods and in oxygen-limited states including those experienced by anoxia tolerant turtles [58,59,60,61]. Furthermore, the importance of chaperones in mediating successful freeze–thaw has also been demonstrated in yeast, where the deletion of chaperones resulted in reduced cell viability to freeze–thaw events [62]. As such, the predicted activation of chaperone-mediated protein folding in the brains of thawing wood frogs through the upregulation of heat shock proteins Hsp10 and Hsp60 mitochondrial chaperones, and other proteins in the network (Fig. 4) underscores the involvement of stress-induced chaperone expression in protecting nervous system tissues. Interestingly, a recent study on zebrafish has shown that injection of Hsp60 triggers tissue regeneration and wound healing, another possible role for the protein during thawing [63].
The observed thaw-induced downregulation of rsy-miR-181a-3p could potentially serve a similar function to the role it plays in the brains of ischemic mice, where it was found to promote long-lasting brain recovery via its action as a modulator of apoptosis that is capable of inducing the upregulation of apoptosis-inhibiting and pro-survival proteins, XIAP and BCL2, respectively [64, 65]. Indeed, BCL2 has been shown to play a central role in the regulation of neuronal cell survival and neurodegeneration and is a known target of miR-204 that was found to be downregulated during both freezing and thawing [66, 67] (Fig. 2). In addition, miR-210, that was also downregulated during both freezing and thawing, is also known to provide neuroprotection against hypoxic/ischemic brain injury, as its suppression has been associated with increased levels of glucocorticoid receptors which resulted in improvement to long-term neurological function recovery and reduced brain infarct size [68]. However, despite this pattern of downregulation observed during thawing, rsy-miR-192-5p was identified as the sole thaw-upregulated miRNA (Fig. 2). The enhanced expression of miR-192 has been shown to be tightly associated with Smad3, where Smad3 is able to mediate TGF-β1-induced miR-192 overexpression by binding to, and activating the miR-192 promoter [69]. Interestingly, this corroborates the aforementioned significant upregulation of p-Smad3 protein levels in R. sylvatica brains during both freezing, and to a larger extent during thawing [30, 69]. A known target of miR-192 that could be targeted in thawed brains is Semaphorin 3A; the suggested miR-192 mediated silencing of Semaphorin 3A could be an ischemia protection mechanism. Indeed, elevated Semaphorin 3A has been shown to be a critical facilitator of cerebrovascular permeability and ischemia-induced brain damage and as such its silencing should aid in protecting frogs from ischemic damage associated with thawing [70, 71].
Taken together, the miRNA biogenesis and relative expression findings suggest that a global reduction in miRNA biogenesis proteins during both freezing and thawing may be facilitating an overall downregulation of differentially expressed miRNAs (Figs. 1, 2). It should be noted that since the majority of miRNAs examined in this study displayed no significant changes over the freeze–thaw cycle, this suggests that the prominent downregulation of differentially expressed miRNAs observed is not a result of an indiscriminate or global downregulation of miRNA biogenesis machinery and miRNA relative abundance levels. Indeed, many of the downregulated miRNAs appear to be involved in providing neuroprotection against hypoxic/ischemic brain injury during both freezing and thawing. For example, downregulation of the plasticity-related miR-24 over the freeze–thaw cycle has been linked with the induction and persistence of long-term potentiation through the rapid release of target mRNA transcript inhibition [72]. Furthermore, numerous pathways such as those involved in intracellular signal transduction were found to be targeted during both freezing and thawing, suggesting a consistent enhancement of their regulatory functions over the course of the freeze–thaw cycle (Figs. 3, 4). These processes are involved in neuronal maintenance and could act to better adapt neurons to environmental cues and protect delicate brain tissue from freeze–thaw induced damages. Since miRNA expression in brains is region-specific, neuron type-specific, and cell compartment-specific [25], further dissection of the observed whole-brain miRNA fingerprint may provide us with additional insights into the miRNA involvement of these various factors. It should be noted that miRNAs act in concert with various other cellular regulatory mechanisms to facilitate global metabolic adaptations and as such should be considered in the light of other regulators.
Our findings demonstrate, for the first time, that wood frog brains appear to circumvent the transcriptional and translational repression that is typical of other organs, possibly to sustain brain function through prolonged periods of environmental stress. The correlation between the downregulation of miRNA biogenesis protein levels and the differentially expressed miRNAs over the freeze–thaw cycle suggests that key brain functions are required for the overall adaptation of the frogs to freezing. These results suggest that the 41 differentially expressed miRNAs are not only freeze–thaw responsive but have been bioinformatically predicted to be involved in maintaining brain function by possibly targeting pathways involved in neuronal maintenance, survival, and anoxia/ischemia protective mechanisms. Taken together, this study provides us with insights into the molecular underpinnings of neuronal adaptations to environmental stress and the mechanisms that facilitate natural freeze tolerance in vertebrate central nervous systems.
References
Costanzo JP, Lee RE, Ultsch GR (2008) Physiological ecology of overwintering in hatchling turtles. J Exp Zool Part A Ecol Genet Physiol 309A:297–379
Denlinger DL, Lee RE (2010) Low temperature biology of insects. Cambridge University Press, Cambridge
Holmstrup M (2014) The ins and outs of water dynamics in cold tolerant soil invertebrates. J Therm Biol 45:117–123
Strimbeck GR, Schaberg PG, Fossdal CG et al (2015) Extreme low temperature tolerance in woody plants. Front Plant Sci 6:884
Storey KB, Storey JM (2017) Molecular physiology of freeze tolerance in vertebrates. Physiol Rev 97:623–665
Storey KB (1990) Life in a frozen state: adaptive strategies for natural freeze tolerance in amphibians and reptiles. Am J Physiol 258:R559–R568
Dawson NJ, Katzenback BA, Storey KB (2015) Free-radical first responders: the characterization of CuZnSOD and MnSOD regulation during freezing of the freeze-tolerant North American wood frog, Rana sylvatica. Biochim Biophys Acta Gen Subj 1850:97–106
Storey KB, Storey JM (2004) Metabolic rate depression in animals: transcriptional and translational controls. Biol Rev Camb Philos Soc 79:207–233
Storey KB (2006) Reptile freeze tolerance: metabolism and gene expression. Cryobiology 52:1–16
Storey KB (2015) Regulation of hypometabolism: insights into epigenetic controls. J Exp Biol 218:150–159
Ebert MS, Sharp PA (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149:515–524
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Biggar KK, Storey KB (2011) The emerging roles of microRNAs in the molecular responses of metabolic rate depression. J Mol Cell Biol 3:167–175
Leung AKL, Sharp PA (2010) MicroRNA functions in stress responses. Mol Cell 40:205–215
Wu P, Nakano S, Sugimoto N (2002) Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur J Biochem 269:2821–2830
Biggar KK, Storey KB (2014) Insight into temperature-dependent microRNA function in mammalian hibernators. Temperature 1:84–86
Biggar KK, Storey KB (2015) Low-temperature microRNA expression in the painted turtle, Chrysemys picta during freezing stress. FEBS Lett 589:3665–3670
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524
Huang Y, Shen XJ, Zou Q et al (2011) Biological functions of microRNAs: a review. J Physiol Biochem 67:129–139
Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10:185–191
Jaskiewicz L, Filipowicz W (2008) Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol 320:77–97
MacFarlane L-A, Murphy PR (2010) MicroRNA: biogenesis, function and role in cancer. Curr Genom 11:537–561
Lee HY, Doudna JA (2012) TRBP alters human precursor microRNA processing in vitro. RNA 18:2012–2019
Fukunaga R, Han BW, Hung J-H et al (2012) Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 151:533–546
O’Carroll D, Schaefer A (2013) General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology 38:39–54
Abe M, Bonini NM (2013) MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol 23:30–36
Storey KB, Storey JM (2007) Tribute to PL Lutz: putting life on ‘pause’—molecular regulation of hypometabolism. J Exp Biol 210:1700–1714
Greenway SC, Storey KB (2000) Activation of mitogen-activated protein kinases during natural freezing and thawing in the wood frog. Mol Cell Biochem 209:29–37
Cai Q, Storey KB (1997) Upregulation of a novel gene by freezing exposure in the freeze-tolerant wood frog (Rana sylvatica). Gene 198:305–312
Aguilar OA, Hadj-Moussa H, Storey KB (2016) Regulation of SMAD transcription factors during freezing in the freeze tolerant wood frog, Rana sylvatica. Comp Biochem Physiol B Biochem Mol Biol 201:64–71
Sullivan KJ, Storey KB (2012) Environmental stress responsive expression of the gene li16 in Rana sylvatica, the freeze tolerant wood frog. Cryobiology 64:192–200
Wu S, De Croos JNA, Storey KB (2008) Cold acclimation-induced up-regulation of the ribosomal protein L7 gene in the freeze tolerant wood frog, Rana sylvatica. Gene 424:48–55
Wu S, Storey KB (2005) Up-regulation of acidic ribosomal phosphoprotein P0 in response to freezing or anoxia in the freeze tolerant wood frog, Rana sylvatica. Cryobiology 50:71–82
Dieni CA, Storey KB (2014) Protein kinase C in the wood frog, Rana sylvatica: reassessing the tissue-specific regulation of PKC isozymes during freezing. PeerJ 2:e558
Bansal S, Luu BE, Storey KB (2016) MicroRNA regulation in heart and skeletal muscle over the freeze–thaw cycle in the freeze tolerant wood frog. J Comp Physiol B 186:229–241
Biggar KK, Dubuc A, Storey K (2009) MicroRNA regulation below zero: differential expression of miRNA-21 and miRNA-16 during freezing in wood frogs. Cryobiology 59:317–321
Hadj-Moussa H, Moggridge JA, Luu BE et al (2016) The hibernating South American marsupial, Dromiciops gliroides, displays torpor-sensitive microRNA expression patterns. Sci Rep 6:24627
Chen M, Storey KB (2014) Large-scale identification and comparative analysis of miRNA expression profile in the respiratory tree of the sea cucumber Apostichopus japonicus during aestivation. Mar Genom 13:39–44
Biggar KK, Kornfeld SF, Maistrovski Y, Storey KB (2012) MicroRNA regulation in extreme environments: differential expression of microRNAs in the intertidal snail Littorina littorea during extended periods of freezing and anoxia. Genom Proteom Bioinform 10:302–309
Lyons PJ, Crapoulet N, Storey KB, Morin PJ (2015) Identification and profiling of miRNAs in the freeze-avoiding gall moth Epiblema scudderiana via next-generation sequencing. Mol Cell Biochem 410:155–163
Lyons PJ, Lang-Ouellette D, Morin PJ (2013) CryomiRs: towards the identification of a cold-associated family of microRNAs. Comp Biochem Physiol Part D Genom Proteom 8:358–364
Tang X, Wen S, Zheng D et al (2013) Acetylation of Drosha on the N-terminus inhibits its degradation by ubiquitination. PLoS One 8:e72503
Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17:3011–3016
Okada C, Yamashita E, Lee SJ et al (2009) A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326:1275–1279
Winter J, Jung S, Keller S et al (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234
Lee Y, Hur I, Park S-Y et al (2006) The role of PACT in the RNA silencing pathway. EMBO J 25:522–532
Wang D, Zhang Z, O’Loughlin E et al (2012) Quantitative functions of Argonaute proteins in mammalian development. Genes Dev 26:693–704
Shen J, Xia W, Khotskaya YB et al (2013) EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497:383–387
Godlewski J, Bronisz A, Nowicki MO et al (2010) microRNA-451: a conditional switch controlling glioma cell proliferation and migration. Cell Cycle 9:2742–2748
Tian Y, Nan Y, Han L et al (2012) MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol 40:1105–1112
Yu D, dos Santos CO, Zhao G et al (2010) miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev 24:1620–1633
Xie J, Hu X, Yi C et al (2016) MicroRNA-451 protects against cardiomyocyte anoxia/reoxygenation injury by inhibiting high mobility group box 1 expression. Mol Med Rep 13:5335–5341
Giniatullin AR, Darios F, Shakirzyanova A et al (2006) SNAP25 is a pre-synaptic target for the depressant action of reactive oxygen species on transmitter release. J Neurochem 98:1789–1797
Biggar KK, Wu C-W, Storey KB (2014) High-throughput amplification of mature microRNAs in uncharacterized animal models using polyadenylated RNA and stem–loop reverse transcription polymerase chain reaction. Anal Biochem 462:32–34
Luu BE, Storey KB (2015) Dehydration triggers differential microRNA expression in Xenopus laevis brain. Gene 573:64–69
Costanzo JP, Lee RE, Lortz PH (1993) Physiological responses of freeze-tolerant and -intolerant frogs: clues to evolution of anuran freeze tolerance. Am J Physiol 265:R721–R725
Hartl FU, Bracher A, Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475:324–332
Storey KB, Storey JM (2012) Insect cold hardiness: metabolic, gene, and protein adaptation. Can J Zool 90:456–475
Jiang J-J, Xia E-H, Gao C-W, Gao L-Z (2016) The complete mitochondrial genome of western painted turtle, Chrysemys picta bellii (Chrysemys, Emydidae). Mitochondrial DNA 27:787–788
Ramaglia V, Buck LT (2004) Time-dependent expression of heat shock proteins 70 and 90 in tissues of the anoxic western painted turtle. J Exp Biol 207:3775–3784
Storey K, Storey JM (2011) Heat shock proteins and hypometabolism: adaptive strategy for proteome preservation. Res Rep Biol 2:57–68
Naicker MC, Jo IS, Im H (2012) Identification of chaperones in freeze tolerance in Saccharomyces cerevisiae. J Microbiol 50:882–887
Pei W, Tanaka K, Huang SC et al (2016) Extracellular HSP60 triggers tissue regeneration and wound healing by regulating inflammation and cell proliferation. NPJ Regen Med 1:16013
Moon J, Xu L, Giffard RG (2013) Inhibition of microRNA-181 reduces forebrain ischemia-induced neuronal loss. J Cereb Blood Flow Metab 33:1976–1982
Xu L-J, Ouyang Y-B, Xiong X et al (2015) Post-stroke treatment with miR-181 antagomir reduces injury and improves long-term behavioral recovery in mice after focal cerebral ischemia. Exp Neurol 264:1–7
Sacconi A, Biagioni F, Canu V et al (2012) miR-204 targets Bcl-2 expression and enhances responsiveness of gastric cancer. Cell Death Dis 3:e423
Akhtar RS, Ness JM, Roth KA (2004) Bcl-2 family regulation of neuronal development and neurodegeneration. Biochim Biophys Acta Mol Cell Res 1644:189–203
Ma Q, Dasgupta C, Li Y et al (2016) Inhibition of microRNA-210 provides neuroprotection in hypoxic–ischemic brain injury in neonatal rats. Neurobiol Dis 89:202–212
Chung ACK, Huang XR, Meng X, Lan HY (2010) miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 21:1317–1325
Hou ST, Nilchi L, Li X et al (2015) Semaphorin3A elevates vascular permeability and contributes to cerebral ischemia-induced brain damage. Sci Rep 5:7890
Yan-Chun L, Hong-Mei Y, Zhi-Hong C et al (2017) MicroRNA-192-5p promote the proliferation and metastasis of hepatocellular carcinoma cell by targeting SEMA3A. Appl Immunohistochem Mol Morphol 25:251–260
Ryan MM, Ryan B, Kyrke-Smith M et al (2012) Temporal profiling of gene networks associated with the late phase of long-term potentiation in vivo. PLoS One 7:e40538
Aguilar OA, Hadj-Moussa H, Storey KB (2017) Freeze-responsive regulation of MEF2 proteins and downstream gene networks in muscles of the wood frog, Rana sylvatica. J Therm Biol 67:1–8
Ye W, Lv Q, Wong C-KA et al (2008) The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One 3:e1719
Szklarczyk D, Franceschini A, Wyder S et al (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Zhang J, Storey KB (2016) RBioplot: an easy-to-use R pipeline for automated statistical analysis and data visualization in molecular biology and biochemistry. PeerJ 4:e2436
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Acknowledgements
This work was supported by a Discovery Grant (Grant #6793) from the Natural Sciences and Engineering Research Council (NSERC) of Canada. KBS holds the Canada Research Chair in Molecular Physiology and HH holds a NSERC Postgraduate scholarship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2018_2821_MOESM1_ESM.xlsx
Primers used for analysis of miRNA expression in the brain of R. sylvatica, including miRNA-specific forward primers, universal reverse primer, and the stem-loop adapter for reverse-transcription. (XLSX 13 kb)
18_2018_2821_MOESM2_ESM.xlsx
Relative expression levels of 113 miRNA species examined in the brain of R. sylvatica. MicroRNA relative expression was evaluated by RT-qPCR of reverse-transcribed, polyadenylated transcripts. Data represent means of n = 3–4 biological replicates from different animals ± SEM. Relative expression of genes was calculated by standardizing against snord68 expression. Control values were adjusted to 1 and the 24 h frozen and 8 h thawed values were expressed relative to the control. Statistical testing used a one-wat ANOVA with a Dunnett's post hoc test, *p < 0.05, **p < 0.01, and ***p <0.001. (XLSX 18 kb)
18_2018_2821_MOESM3_ESM.xlsx
Identity of the significantly enriched biological processes, protein members, and miRNA species identified using MCL clustered protein networks and functional GO ANALYSIS of the miRNAs downregulated in 24 h frozen frog brain. (XLSX 17 kb)
18_2018_2821_MOESM4_ESM.xlsx
Identity of the significantly enriched biological processes, protein members, and miRNA species identified using MCL clustered protein networks and functional GO ANALYSIS of the miRNAs downregulated in 8 h thawed frog brain. (XLSX 22 kb)
18_2018_2821_MOESM5_ESM.jpg
Full cluster map of functional target enrichment and network clustering of the subset of miRNAs downregulated in the brains of 24 h frozen wood frogs. Downstream miRNA target prediction was performed at -2 °C using FINDTAR3. Protein–protein interactions of the downstream networks was performed using the STRING high-confidence filter on the X. tropicalis database. MCL clustering and visualization was performed on CYTOSCAPE and coupled with functional biological enrichment using GO ANALYSIS. Refer to Supplementary Table S3 for more information on individual clusters, proteins, and the targeting of individual miRNA species. (JPEG 351 kb)
18_2018_2821_MOESM6_ESM.jpg
Full cluster map of functional target enrichment and network clustering of the subset of miRNAs downregulated in the brains of 8 h thawed wood frogs. Downstream miRNA target prediction was performed at 5 °C using FINDTAR3. Protein–protein interactions of the downstream networks was performed using the STRING high-confidence filter on the X. tropicalis database. MCL clustering and visualization was performed on CYTOSCAPE and coupled with functional biological enrichment using GO ANALYSIS. Refer to Supplementary Table S4 for more information on individual clusters, proteins, and the targeting of individual miRNA species. (JPEG 302 kb)
Rights and permissions
About this article
Cite this article
Hadj-Moussa, H., Storey, K.B. Micromanaging freeze tolerance: the biogenesis and regulation of neuroprotective microRNAs in frozen brains. Cell. Mol. Life Sci. 75, 3635–3647 (2018). https://doi.org/10.1007/s00018-018-2821-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2821-0