Abstract
Polycystin-1 (PC1) has been proposed as a chief mechanosensing molecule implicated in skeletogenesis and bone remodeling. Mechanotransduction via PC1 involves proteolytic cleavage of its cytoplasmic tail (CT) and interaction with intracellular pathways and transcription factors to regulate cell function. Here we demonstrate the interaction of PC1-CT with JAK2/STAT3 signaling axis in mechanically stimulated human osteoblastic cells, leading to transcriptional induction of Runx2 gene, a master regulator of osteoblastic differentiation. Primary osteoblast-like PC1-expressing cells subjected to mechanical-stretching exhibited a PC1-dependent increase of the phosphorylated(p)/active form of JAK2. Specific interaction of PC1-CT with pJAK2 was observed after stretching while pre-treatment of cells with PC1 (anti-IgPKD1) and JAK2 inhibitors abolished JAK2 activation. Consistently, mechanostimulation triggered PC1-mediated phosphorylation and nuclear translocation of STAT3. The nuclear phosphorylated(p)/DNA-binding competent pSTAT3 levels were augmented after stretching followed by elevated DNA-binding activity. Pre-treatment with a STAT3 inhibitor either alone or in combination with anti-IgPKD1 abrogated this effect. Moreover, PC1-mediated mechanostimulation induced elevation of Runx2 mRNA levels. ChIP assays revealed direct regulation of Runx2 promoter activity by STAT3/Runx2 after mechanical-stretching that was PC1-dependent. Our findings show that mechanical load upregulates expression of Runx2 gene via potentiation of PC1–JAK2/STAT3 signaling axis, culminating to possibly control osteoblastic differentiation and ultimately bone formation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mechanical forces have a decisive impact on bone remodeling, a vital process for preservation of the skeleton’s structural integrity, regeneration of damaged bone and maintenance of metabolic homeostasis, holding great promise for bone-tissue engineering and orthopedic treatments [1]. Mechanical competence of bone tissue relies on its adaptive ability to reform its mass, three dimensional structure and mechanical properties to meet the prevailing dynamic loading environment. Resident bone cells including osteoblasts, osteocytes and osteoclasts regulate bone’s adaptive response by perceiving and translating mechanical energy into a cascade of structural and biochemical changes within the cells [2]. Previous studies from our group have shown that mechanical stimuli applied on osteoblastic cells induce activation of tissue-specific transcription factors such as Runt-related transcription factor 2 (Runx2) and activator protein-1 (AP-1) to integrate upstream signaling pathways eventually modulating anabolic cell responses [3–7].
Significant advances have been made over the last years towards identification of mechanosensing proteins, intracellular pathways and nuclear factors that cooperate to control osteoblast response to mechanical signals [1, 5, 6, 8, 9]. Animal models have revealed polycystin-1 (PC1; encoded by Pkd1 gene) as a key mechanosensing molecule in osteoblasts with a primary role in bone formation and regulation of skeletogenesis [8, 9]. PC1 is proposed to ‘sense’ mechanical forces either directly or indirectly through interaction of its long extracellular domain with most mechanosensing proteins [10–12]. Upon mechanical stimulation, the C-terminal cytoplasmic tail (CT) of PC1 is proteolytically cleaved and released from the membrane, eliciting a mechanoresponse. Nuclear translocated CT has been shown to interact with several transcription factors such as AP-1 and signal transducer and activator of transcription 3 (STAT3) affecting transcriptional gene regulation and cell function [10–14].
Studies of Pkd1-deficient mice demonstrate the essential role of PC1 in the induction of osteoblast mechanoresponse to mechanical loading [15]. Pkd1m1Bei homozygous mutant mice (bearing inactivating missense mutations of Pkd1 gene) exhibit decreased expression of the master regulator of osteoblast differentiation, Runx2 and abnormal bone development, whereas heterozygous mice present osteopenia [16]. Moreover, conditional deletion of Pkd1 in osteoblasts causes a gene dosage-dependent reduction in bone development and a proportional reduction of osteoblastic gene expression including Runx2-II isoform [17].
Although there is evidence for the critical role of PC1 in proliferation and differentiation of mechanically stimulated bone cells, the underpinning intracellular pathways mediating this response in osteoblasts remain elusive.
In different cell types, PC1 CT cleavages have been associated with modulation of the mammalian target of rapamycin (mTOR), cAMP and Wnt signaling pathways [18, 19]. Studies in pre-osteoblastic cell line MC3T3-E1 have shown that PC1 mediates proliferation and differentiation induced by mechanical tensile strain via the Akt/β-catenin pathway and Ca2+ signaling [20], while overexpression of PC1 CT in the same cell line enhances the Runx2 distal promoter activity [16]. Furthermore, in an in vitro model of mechanically stimulated human osteoblast-like cells we have shown that PC1 regulates osteoblastic gene expression via the calcineurin/nuclear factor of activated T-cells (NFAT) signaling pathway with potential control on bone formation [21].
Recent evidence indicates the capacity of PC1 to regulate the activity of the STAT transcription factors in experimental autosomal polycystic kidney disease (APKD) in vitro and in vivo models [22, 23]. PC1 has been reported to directly activate STAT1 and STAT3 via Janus kinase 2 (JAK2) that is associated with PC1-CT in renal HEK293T cells. Additionally, in renal epithelial cells, PC1-CT was shown to function as a nuclear co-activator of STAT1/3/6 along with cytokines and growth factors [22, 23]. Interestingly, studies on JAK and STAT knock-out mice have revealed a crucial role of the JAK/STAT signaling cascade in bone development and metabolism [24]. Compared to other STATs, profound effects of STAT3 have been shown on bone homeostasis mainly involving transduction of anabolic signals in osteoblasts and promotion of differentiation. Inactivation of STAT3 resulted in unresponsive osteoblasts to mechanical loading and limited bone formation in knock-out mice [25].
Runx2 is tightly related to osteoblastic differentiation during bone development and remodeling. Importantly, previous work from our group demonstrates that Runx2 gene harbors functional STAT3-binding sites on its promoter and can physically interact with STAT3 upon stimulation of osteoblasts with growth hormone (GH) [26, 27]. Additional studies in pre-osteoblast MC3T3-E1 cells reinforce this interaction by showing that Runx2 is regulated by STAT3 during the differentiation process in the presence of heparin sulfate [28] and interleukin (IL)-6 [24]. Therefore, a direct link of PC1–JAK2/STAT3 signaling and Runx2 induction in mechanically stimulated osteoblasts was speculated.
This study investigates the ability of PC1 to deliver mechanical stimuli to human osteoblastic cells through JAK/STAT pathways eventually regulating cell differentiation. Indeed, the results of our experiments suggest an essential role of PC1 in the activation of JAK2/STAT3 cascade and positive regulation of Runx2 promoter activity upon mechanical loading of osteoblastic cells, posing that PC1–JAK/STAT axis could be a potential novel target for enhancing bone formation.
Materials and methods
Reagents and antibodies
Cell culture media (including fetal bovine serum, FBS) and all tissue culture reagents were obtained from Invitrogen (CA, USA). The anti-CT rabbit polyclonal antibody was kindly provided by Baltimore PKD Core Center (University of Maryland, USA; see “Acknowledgements”) [29]. The inhibitory anti-IgPKD1 antibody was a generous gift from Dr. O. Ibraghimov-Beskrovnaya (Genzyme, MA, USA) [30, 31]. The PC1 antibody (sc-130554, mouse monoclonal) was obtained from Santa Cruz Biotechnology (CA, USA). The STAT3 antibody (G.659.4, mouse monoclonal) was purchased from Thermo Scientific (IL, USA), the human actin antibody (MAB-1501, mouse monoclonal) was purchased from Millipore (MA, USA) and the SP-1 antibody (sc-59) was purchased from Santa Cruz Biotechnology. The JAK2 (ab37226, mouse monoclonal), pJAK2 (ab32101, rabbit monoclonal) and pSTAT3 (ab76315, rabbit monoclonal) antibodies were obtained from Abcam (UK). The secondary antibodies used for western immunoblotting were purchased from Thermo Scientific (Ontario, Canada) and Millipore (goat anti-mouse 31430 and goat anti-rabbit AP132P, respectively) and those used for immunofluorescence (goat anti-rabbit sc-2780 and goat anti-mouse sc-2010) were from Santa Cruz Biotechnology. The JAK2 inhibitor AG490 (658491) and the STAT3 inhibitor VII (ethyl-1-(4-cyano-2,3,5,6-tetrafluorophenyl)-6,7,8-trifluoro-4-oxo-1,4-dihydroquinoline-3-carboxylate) were purchased from Calbiochem (Merck, Germany). DAPI and all other chemical reagents were from Sigma-Aldrich Chemie (Germany). Duolink in situ Ligation Kit (92004, 92006, 92002, 92007) was obtained from Olink Bioscience (Uppsala, Sweden). All FACS reagents were purchased from BD Biosciences (NJ, USA). The STAT3-TransAM Kit and the ChIP-IT High Sensitivity Kit (53040) were purchased from Active Motif (CA, USA). The SimpleChIP Human Runx2 Promoter Primers (12376), the Runx2 antibody (8486, rabbit monoclonal) and the normal rabbit IgG (2729) were obtained from Cell Signaling Technology (MA, USA). The oligonucleotides used were custom ordered from Invitrogen.
Cell cultures
Human osteoblastic cells (i.e. cells with the potential to differentiate into mature osteoblasts) were obtained from explant cultures of periodontal ligament tissues as detailed previously [4–7]. These cells have well-established osteoblast-like properties (hence pre-osteoblasts) as verified by the intense immunohistochemical staining for alkaline phosphatase and vimentin, a highly reliable marker of cells originating from the mesenchyme [3]. The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % FBS and antibiotics. All experiments were carried out with cells from the third to the sixth passage after being checked for their osteoblastic characteristics.
All experiments were carried out in accordance with the approved guidelines and regulations set by the Ethics Committee of the National and Kapodistrian University of Athens Medical School. Furthermore, the entire study protocol was approved by the Ethics Committee of the National and Kapodistrian University of Athens Medical School.
Mechanical stretch application and treatment of cultured human osteoblastic cells
Approximately 4 × 105 cells were seeded onto 50-mm dishes with a flexible, hydrophilic growth surface (Sarstedt, Germany) and cultivated until they reached confluence. The medium was then changed to DMEM supplemented with 0.1 % FBS, to remain quiescent. Twenty-four hours later, the cell cultures were stretched continuously for the indicated periods of time employing an established stretching apparatus, as detailed previously [4–7]. This stretching system allows mechanical stimulation of human osteoblastic cells to be quantitated by calculating the percent expansion of the membrane at the bottom surface of the culture dishes using a specific formula previously described [4, 6, 32, 33]. The percentage of stretch applied to the cells in our study was 2.5 %, an amount that closely resembles the strain forces/mechanical load exerted on osteoblastic cells in vivo [4, 5].
Control (unstretched) cultures were incubated under the same conditions for the minimum and maximum period of stretch application. Cells were treated with the inhibitors AG490 (25 μM) and STAT3 VII (4 μM) for 2 h and inhibitory anti-IgPKD1 antibody (dilution 1:50) for 3 h prior to stretching in starvation medium.
Western immunoblotting
After stretch application, cells were washed with ice-cold phosphate-buffered saline (PBS) and whole-cell, cytoplasmic or nuclear lysates were obtained as follows. Total cell extracts were prepared in SDS sample buffer as described [34–36]. Cytoplasmic and nuclear extracts were prepared according to Schreiber et al. [36], except that the nuclear pellet was extracted with 20 mM HEPES–NaOH, pH 7.5, 20 % glycerol, 0.38 M NaCl, 2 mM MgCl2, 10 mM NaF, 10 mM [Na2]-β-glycerophosphate, 10 mM [Na2]p-nitrophenyl phosphate, 0.1 mM Na3VO4, 2 mM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 2 μg/ml pepstatin, 2 μg/ml leupeptin and 2 mM dithiothreitol. All protein extracts were aliquoted and stored at −80 °C until use.
Protein extracts were resolved in 10 % SDS–polyacrylamide gels followed by electrotransfer onto polyvinylidene fluoride membranes (Amersham Biosciences, Germany). Membranes were blocked for 1 h at room temperature with 5 % milk in Tris-buffered saline with Tween (TBST; 10 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1 % Tween-20) and probed with the primary antibody overnight at 4 °C. After incubation with a horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich Chemie), immunoreactive bands were visualized by the enhanced chemiluminescence (ECL) Kit (Amersham Biosciences, Germany).
The primary antibodies were used at the following dilutions: anti-pJAK2, 1:250, anti-pSTAT3, 1:200 and anti-actin, 1:5000. Both secondary antibodies (goat anti-mouse IgG, goat anti-rabbit IgG) were used at a 1:2500 dilution.
Duolink proximity ligation assay (PLA) and image analysis
Duolink PLA is a method that enables the visualization of protein–protein interactions in cells using fluorescence detection. PLA retains the dependency of proximal binding of antibodies and provides a mean for signal amplification [37]. This technology utilizes DNA oligonucleotides conjugated to antibodies (proximity probes) to provide a template for ligation of subsequently added circularization DNA oligonucleotides (http://www.sigmaaldrich.com/life-science/molecular-biology/molecular-biology-products.html?TablePage=112232138). The ligation will only generate circular products if the proximity probes bind in close proximity. The length and orientation of the oligonucleotides, as well as the size of the affinity reagents, determine the distance requirement for in situ PLA. Once a circular ligation product is formed, it can be amplified by rolling circle amplification (RCA). The signal amplification provided by RCA facilitates detection of single ligation events and also enumeration and analysis by a specific digital image processing tool. PLA probes, anti-goat PLUS, anti-mouse MINUS and detection reagent Duolink II red were obtained from Olink Bioscience.
Human osteoblastic cells were serum-deprived in medium containing 0.1 % FBS for 24 h and exposed to mechanical stretch for 1 h. The cells were cultured either on coverslips (unstretched) or on the flexible, hydrophilic growth surface plates (stretched). After reaching ~75 % confluency in culture, cells were starved overnight and the next day subjected to mechanical stretch. Immediately afterwards, the cells were fixed with 4 % paraformaldehyde (PFA) in PBS for 10 min at room temperature. Following permeabilization for 10 min at room temperature, PLA was performed according to the Duolink II protocol using different sets of primary antibodies (CT/JAK2 and CT/pJAK2). For controls, one of the primary antibodies of each set was omitted and the protocol was carried out without any further changes. Coverslips and membranes were mounted with DII mounting medium containing DAPI (Olink Bioscience). Cells were then incubated with the PLA probes diluted 1:5 in antibody diluent (Olink Bioscience) in a humidified chamber for 1 h at 37 °C. Subsequent hybridization, ligation, amplification and detection were performed according to manufacturer’s instructions, as detailed before [37].
Fluorescence images were acquired using a Zeiss Axiovert microscope (Carl Zeiss Microscopy, Thornwood, NY, USA). For each coverslip, images were taken from at least ten randomly selected fields of view. Data analysis was performed using Duolink ImageTool Software that had been developed for quantification of PLA signals. For statistical analysis, the resulting ‘PLA signals per cell’ in all groups were normalized to the mean value of the control group. Representative images for each condition were prepared for figure presentation by applying brightness and contrast adjustments uniformly using Adobe Photoshop CS5 software.
Fluorescence-activated cell sorting (FACS) analysis
Human osteoblastic cells were treated as described in previous sections and exposed to mechanical stretch. After the set time points, cells were harvested and fixed with 1 % PFA for 15 min at room temperature. Cells were permeabilized with 0.01 % glycine in PBST (PBS with 0.01 % Triton X-100) for 20 min and blocked for 20 min with 5 % BSA in PBS with 0.01 % Tween-20. Following washing with PBS, samples were further incubated with the secondary antibody (1:250) for another hour at room temperature. Finally, cells were washed twice, resuspended in PBS and subjected to analysis. A total of 10,000 events were analyzed per sample. All antibody incubations were performed at room temperature. Flow cytometry analyses were performed in a FACSCalibur flow cytometer using Cell Quest software (BD Biosciences). The antibodies used were: PC1 at 1:500 dilution; JAK2, pJAK2, STAT3 and pSTAT3 at 1:200 dilution.
TransAM DNA-binding assay
Nuclear protein extraction from cells was performed as described previously [36]. Five micrograms of nuclear extract were tested for STAT3 DNA-binding activity by employing STAT3-TransAM kit, using the STAT consensus binding site (5′-TTCCCGGAA-3′) and a specific pSTAT3 antibody, according to manufacturer’s instructions (Active Motif). The results (optical density measured at 450 nm) were expressed as percentage increase over basal (untreated cells). All samples were run in triplicates.
RNA isolation
Human osteoblastic cells were serum-deprived in medium containing 0.1 % FBS for 24 h and exposed to mechanical stretch for various time points (cells were pre-treated with specific inhibitors in certain experiments). Total RNA was isolated using the RNeasy Mini Kit (Qiagen, CA, USA) according to manufacturer’s instructions and cDNA was synthesized using the Fermentas Reverse Transcriptase (Maxima RT, EP0741, Fermentas, Ontario, Canada).
Semi-quantitative PCR analysis
End-point PCR analysis was performed for the human Runx2 gene using the Maxima Hot Start Green PCR Master Mix (K1069, Fermentas) according to manufacturer’s instructions. The human Runx2 promoter (−621 to −587) specific primers containing an OSE2 motif (ACCACA) and an adjacent putative STAT-binding site (TGccaGGAA) have been previously described [26] and were used for end-point PCR of chromatin preparations (see below). The semi-quantitative PCR approach was performed in parallel by assaying human actin mRNA levels as a reference gene, as described previously [38].
End-point PCR for each gene was performed as follows: for Runx2, 32 cycles and 59 °C annealing; for actin, 28 cycles and 57 °C annealing. Reaction products were verified on a 2 % agarose gel with size markers (New England Biolabs, MA, USA) and stained with ethidium bromide. The intensity of density area was analyzed using the Quantity-One Program (Bio-Rad Laboratories, CA, USA). The final PCR product was expressed as the ratio to actin used for scanning analysis.
Real time RT-PCR analysis
Expression of Runx2 and gapdh genes was determined by real time RT-PCR using the iCycler MyiQ detection system (Bio-Rad Laboratories), SYBRGreen (Qiagen) and specific primers for Runx2 (Hs_RUNX2_1_SG QuantiTect Primer Assay (200) QT00020517 (NM_004348, NM_001015051, NM_001024630) (https://www.qiagen.com/fi/shop/pcr/real-time-pcr-enzymes-and-kits/two-step-qrt-pcr/quantitect-primer-assays/?catno=QT00020517&autoSuggest=true#geneglobe) and for gapdh (Hs_GAPDH_2_SG QuantiTect Primer Assay QT01192646 (NM_002046, NM_001289745) (https://www.qiagen.com/fi/shop/pcr/real-time-pcr-enzymes-and-kits/two-step-qrt-pcr/quantitect-primer-assays/?catno=QT01192646#geneglobe) as described previously [39, 40]. One microliter of cDNA was amplified as a template in a 25-μl reaction mixture containing 12.5 μl 2 × QuantiFast SYBRGreen PCR Master Mix (Qiagen), 2.5 μl of primers and 9 μl deionized water. The mixture was heated initially at 95 °C for 5 min and then followed by 40 cycles with denaturation at 95 °C for 10 s and combined annealing/extension at 60 °C for 30 s. Data were analyzed by the comparative threshold cycle method.
Chromatin immunoprecipitation (ChIP) and real time PCR
For the ChIP experiments, the ChIP-IT High Sensitivity Kit that enables identification of transcription factor-binding to specific DNA loci was used according to manufacturer’s instructions (Active Motif). Osteoblastic cells chromatin was prepared according to the manual using between 10,000 and 100,000 cells per chromatin preparation. The immunoprecipitation reaction was performed using previously tested high quality antibodies against: Runx2 (rabbit monoclonal, 5 μl per sample; Cell Signaling Technology), pSTAT3 (rabbit monoclonal, 5 μl per sample; Abcam), or normal rabbit IgG as negative control (2 μl per sample; Cell Signaling Technology).
The enriched DNA was initially quantified by real time PCR using the SimpleChIP Human Runx2 Promoter Primers (Cell Signaling Technology) according to manufacturer’s instructions. Real time PCR was run using the iCycler MyiQ detection system (Bio-Rad Laboratories), SsoAdvancedTM Universal SYBR® Green Supermix (Bio-Rad Laboratories) and specific primers (Cell Signaling Technology). Five microliters of cDNA was amplified as a template in a 25-μl reaction mixture containing 12 μl 2 × qPCR mix (dNTPs, Sso7d fusion polymerase, MgCl2, SYBR® Green I, ROX normalization dyes; Bio-Rad Laboratories), 2 μl of primers and 6 μl deionized water. The mixture was heated initially at 95 °C for 3 min and then followed by 40 cycles with denaturation at 95 °C for 15 s and combined annealing/extension at 65 °C for 60 s.
Furthermore, the same enriched DNA was quantified by end-point PCR employing specific primers for a previously described human Runx2 promoter sequence [26]. The forward oligonucleotide sequence used is: OSE2–STAT, 5′-CCTTCTGGATGCCAGGAAAGGCCTTACCACAAGCC-3′ (derived from the human Runx2 promoter (−621 to −587) and containing an OSE2 motif (ACCACA) and an adjacent putative STAT-binding site (TGccaGGAA) [26]. Cycle information for Runx2: 32 cycles and 59 °C annealing. As negative control, a commercially available set of primers were used to amplify a non-coding region of the promoter (Active Motif). Reaction products were verified on a 3 % agarose gel with size markers (New England Biolabs) and stained with ethidium bromide. The intensity of density area was analyzed using the Quantity-One Program (Bio-Rad Laboratories). For data analysis subtraction of the irrelevant IgG background was performed and the results were plotted as ‘Fold enrichment or enrichment relative to the input’ where input refers to untreated cells without any antibody.
Image and data analysis
For densitometry quantification analysis, three independent Western blot and end-point PCR results were analyzed using the Quantity-One Program (Bio-Rad Laboratories). RT-PCR data were analyzed by the comparative threshold cycle method.
Statistical analysis was performed using the SPSS V16.0 program. Differences between two groups were assessed using analysis of variances followed by a Student’s t test and one-way ANOVA (*P < 0.05, n.s. = not significant).
Results
Mechanical stretching activates JAK2 in human osteoblastic cells via PC1
To explore the effect of mechanical stress on JAK2 activation in human osteoblastic cells, we mechanically stimulated (stretched) primary osteoblast-like cells (pre-osteoblasts) for 1/2, 1 and 6 h as previously described [3, 21]. Western immunoblot analysis was performed in whole-cell extracts obtained from unstretched and mechanically stretched cells at all time points. Phosphorylated(p)/active JAK2 (pJAK2) protein levels increased at 1 h of mechanical stretching compared to unstretched controls (P < 0.001). Total JAK2 levels remained unchanged throughout these time points (Fig. 1a, b).
Mechanical stretching activates JAK2 in human osteoblastic cells. a Western immunoblotting showing pJAK2 and total JAK2 protein levels in osteoblastic cells following mechanical stimulation for 0–6 h and b corresponding densitometric analysis (***P < 0.001 vs. unstretched cells). Whole-cell protein extracts were isolated and JAK2, pJAK2 protein detection (~130 kDa) was performed using specific primary anti-JAK2, anti-pJAK2 antibodies and distinct secondary antibodies (anti rabbit and anti-mouse antibodies, accordingly). c–g Cells were grown to over-confluency followed by overnight starvation, treatment with specific inhibitors (anti-IgPKD1, AG490 or their combination) where indicated, application of mechanical stretch and collection for FACS analysis. FACS analysis data of JAK2 and pJAK2 levels after mechanical stretching of d untreated cells, e cells pre-treated with a combination of anti-IgPKD1 and AG490, f cells pre-treated with anti-IgPKD1 only and g cells pre-treated with AG490 only. Ten thousand events were analyzed per sample. The graphs depict quantitative estimation of JAK2 (red line) and pJAK2 (blue line) levels. Data show average values of at least three independent experiments and * P < 0.05 vs. unstretched cells
To verify the specificity of JAK2 activation upon mechanical stimulation and the involvement of PC1 in this process, we treated osteoblastic cells with a potent JAK2 inhibitor (AG490) or/and an antibody that blocks the mechanosensing extracellular N-terminal part of PC1 (anti-IgPKD1) prior to mechanical strain application for the aforementioned time points. PC1, pJAK2 and total JAK2 levels were further evaluated by fluorescence-activated cell sorting (FACS) analysis of whole-cell suspensions (Fig. 1c–g). PC1 levels were not significantly altered after exposure of cells to mechanical load with or without pre-treatment with the inhibitors for the indicated time points, as previously observed [21] (data not shown). However, after 1/2 h of mechanical stretching there was a notable rise of pJAK2 that continued to increase significantly up to 6 h (P < 0.05, Fig. 1c, d). By contrast, the total JAK2 species did not show any considerable changes throughout the experimental time period (Fig. 1c, d). Pre-treatment of osteoblastic cells with the above inhibitors, either alone or in combination, effectively reversed the activation of JAK2 upon mechanical stimulation (Fig. 1c–g). Specifically, treatment of cells with both anti-IgPKD1 and AG490 prior to mechanical stimulation led to complete loss of the observed increase in pJAK2 levels (Fig. 1e). Treatment of cells with either anti-IgPKD1 (Fig. 1f) or AG490 alone (Fig. 1g) prior to mechanical strain application resulted in marked reduction of pJAK2 levels, suggesting that JAK2 activation in osteoblastic cells after mechanical stretching is PC1-dependent.
Mechanical stretching induces a strong interaction of PC1-CT and pJAK2 in human osteoblastic cells
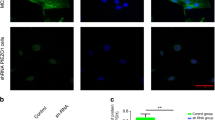
To detect possible protein–protein interaction (complex formation) between PC1-CT and pJAK2 or JAK2 in human osteoblastic cells upon mechanical stretching, we employed the Duolink antibody-based proximity ligation assay (PLA) as previously described [41]. Using specific antibodies directed against the PC1-CT and either JAK2 or pJAK2, a specific interaction was observed between PC1-CT/JAK2 and PC1-CT/pJAK2 in osteoblastic cells (Fig. 2a, b). Specifically, a PC1-CT/JAK2 complex was observed in unstretched cells (Fig. 2a, top left) that was significantly reduced after 1 h of stretching (P < 0.05, Fig. 2a, top middle). This finding suggests that the CT of PC1 is in close proximity to and interacts physically with JAK2 in quiescent osteoblastic cells, whereas mechanical stimulation significantly lessens this interaction. Furthermore, strong complexes (dimers) were observed between PC1-CT and pJAK2 upon mechanical stretching of cells for 1 h (Fig. 2a, bottom middle). The ratio of interactions (complex formation) of PC1-CT/JAK2 and PC1-CT/pJAK2 in unstretched cells is increased by three-fold in mechanically stimulated cells (0.53 vs. 1.74, respectively, Fig. 2b). Control experiments performed with rabbit pre-immune serum or AG490 or without primary antibody against PC1-CT, JAK2 or pJAK2 prior to stretching yielded no fluorescent signal, verifying the specificity of the detected PC1-CT/JAK2 and PC1-CT/pJAK2 protein–protein interactions (Fig. 2a). These results indicate the specific interaction of PC1-CT with pJAK2 upon mechanical stimulation of osteoblastic cells.
Mechanical stretching induces a strong interaction of PC1-CT with pJAK2 in human osteoblastic cells. a Duolink in situ PLA immunofluorescence images showing protein–protein interaction and specific signal generation of PC1-CT and JAK2 in unstretched cells (top left) and cells stretched for 1 h (top middle) as well as of PC1-CT and pJAK2 in unstretched cells (bottom left) and cells stretched for 1 h (bottom middle). Antibodies against PC1-CT, JAK2, pJAK2 and DAPI stain were employed in the Duolink assay according to manufacturer’s instructions (scale bar 20 μm). b The graph depicts data analyses for PLA signal quantification using the Duolink ImageTool Software. For statistical analysis, the resulting ‘PLA signals per cell’ in all groups were normalized to the mean value of the control group (dark blue PC1-CT and JAK2 in unstretched cells, light blue PC1-CT and JAK2 in cells stretched for 1 h, red PC1-CT and pJAK2 in unstretched cells, and pink PC1-CT and pJAK2 in cells stretched for 1 h). Data show average values of at least three independent experiments and *P < 0.05 of PC1-CT/JAK2 complex formation between unstretched and 1 h-stretched cells
Mechanical stretching augments pSTAT3 translocation to the nucleus by activating JAK2/STAT3 signaling pathway in human osteoblastic cells
Since STAT3 is the substrate of JAK2, we proceeded to investigate the effect of mechanical stimulation on phosphorylated(p)/active STAT3 (pSTAT3) levels in human osteoblastic cells. To this end, we first examined the nuclear translocation of pSTAT3 after mechanical stretching by western immunoblotting (Fig. 3a). At 1/2 h of stretch application, the cytoplasmic pSTAT3 species decreases while its nuclear counterpart increases with a nuclear to cytoplasmic ratio of 2.1 compared to unstretched cells. This ratio is markedly decreased at 1 and 6 h of stretching (Fig. 3b), indicating that mechanical load induces activation of the JAK2/STAT3 signaling pathway.
Mechanical stretching increases pSTAT3 translocation to the nucleus and enhances its DNA-binding capacity by activating JAK2/STAT3 signaling pathway in human osteoblastic cells. a Western immunoblotting showing cytoplasmic and nuclear protein levels of pSTAT3, actin and SP-1 in osteoblastic cells after mechanical stimulation for 0–6 h and b corresponding densitometric analysis. Cytoplasmic and nuclear protein extracts were isolated and pSTAT3 protein detection (~86–91 kDa) was performed using specific anti-pSTAT3 antibodies. Actin (~42 kDa) was probed with an anti-actin antibody as loading control. SP-1 (~106 kDa) was probed with an anti-SP-1 antibody as loading control of nuclear extracts. Data show the average values of at least three independent experiments. c Quantitative results from the TransAM cell-based assay used for detection of STAT3-binding activity in mechanostimulated osteoblastic cells. Cells were grown to over-confluency, treated with anti-IgPKD1 or STAT3 VII inhibitors or their combination where indicated, subjected to mechanical stress for various time points (0–6 h) and nuclear protein extracts were isolated. The graph depicts quantified colorimetric measurements of STAT3-binding activity in unstretched cells or cells stretched for the indicated time-points following pre-treatment with the indicated inhibitors (blue untreated cells, red cells pre-treated with anti-IgPKD1 only, green cells pre-treated with STAT3 VII only, and purple cells pre-treated with anti-IgPKD1 and STAT3 VII). Data show average values of at least three independent experiments. *P < 0.05 of anti-IgPKD1- or STAT3 VII-treated vs. untreated cells. **P < 0.05 of anti-IgPKD1 + STAT3 VII-treated vs. untreated cells
Mechanical stretching enhances the DNA-binding potential of STAT3 in human osteoblastic cells
To assess the DNA-binding activity of STAT3 in mechanostimulated human osteoblastic cells, we employed the TransAM STAT3 cell-based assay which allows sensitive, non-radioactive determination of specific transcription factor activation in mammalian tissues and cell extracts, as previously described [21]. For the TransAM STAT3 assay, the STAT3 consensus binding site oligonucleotide sequence is being used as a template for the isolated nuclear extracts.
Cells untreated or pre-treated with the STAT3 inhibitor VII or anti-IgPKD1 alone, as well as with their combination were subjected to mechanical stretch for 0–6 h. STAT3 inhibitor VII is a cell-permeable dihydroquinoline compound that potently and specifically blocks cellular JAK1/JAK2/TYK2/STAT3 activation as well as the transcription of STAT3 target genes [42]. Following mechanical stimulation, nuclear extracts were isolated for assaying STAT3 binding potential. A significant increase of STAT3 DNA-binding activity after 1/2 h of mechanical stimulation was observed that was gradually decreased at 1 and 6 h time points (Fig. 3c). Pre-treatment of cells with the two inhibitors either alone (P < 0.05) or in combination reduced drastically this effect (P < 0.01, Fig. 3c).
Upregulation of Runx2 mRNA via PC1–JAK2/STAT3 signaling axis in mechanically-stimulated human osteoblastic cells
Previous work in our laboratory has shown that exposure of osteoblastic cells to short-term mechanical stretch affects osteoblastic gene transcription and specifically Runx2 mRNA and protein expression levels [7, 27]. In this vein, untreated human osteoblastic cells or cells pre-treated with anti-IgPKD1, AG490, their combination or STAT3 VII inhibitor were subjected to mechanical stretching for 0–24 h and total RNA was isolated and assayed. As depicted in Fig. 4a, unstretched cells displayed low levels of Runx2 mRNA in end-point PCR analysis. However, Runx2 mRNA levels were increased by more than two-fold after 6 h of continuously applied mechanical load and remained elevated until 24 h. This upregulation of Runx2 mRNA was reduced drastically upon pre-treatment of cells with anti-IgPKD1, AG490 or STAT3 VII inhibitors alone, whilst a statistically significant drop of Runx2 mRNA levels was observed upon cell pre-treatment with the combination of anti-IgPKD1 and AG490 compared to untreated cells (P < 0.05).
Mechanical stretching upregulates Runx2 mRNA via PC1-JAK2/STAT3 signaling axis in human osteoblastic cells. a Effect of mechanical stimulation on Runx2 gene expression levels as monitored by end-point PCR. Quiescent cells were subjected to mechanical stretch for 0–24 h following pre-treatment with specific inhibitors (or their combination) where indicated. cDNA was synthesized from total RNA and Runx2 expression levels were assessed by end-point PCR (grey untreated cells, blue cells pre-treated with anti-IgPKD1 only, purple cells pre-treated with AG490 only, light violet cells pre-treated with anti-IgPKD1 and AG490, and green cells pre-treated with STAT3 VII). Human actin was used as the reference gene for normalization. Data show average values of at least three independent experiments and *P < 0.05 of anti-IgPKD1 + AG490-treated vs. untreated cells. b Effect of mechanical stimulation on Runx2 gene expression levels as monitored by Real time-PCR. Quiescent cells were subjected to mechanical stretch for 0–24 h following pre-treatment with specific inhibitors (or their combination) where indicated. cDNA was synthesized from total RNA and Runx2 expression levels were assessed by Real time-PCR (grey untreated cells, and light violet cells pre-treated with anti-IgPKD1 and AG490). Human gapdh was used as the reference gene for normalization. Data (fold change) show average values of at least three independent experiments and *P < 0.05 of anti-IgPKD1 + AG490-treated vs. untreated cells
The above result was further supported by Real time-PCR, a more sensitive and precise method to measure changes in mRNA levels compared to end-point PCR. Data analysis from the Real time-PCR experiments confirmed the previously observed induction of Runx2 mRNA levels upon mechanical stimulation which, however, were markedly reduced when cells were pre-treated with the combination of anti-IgPKD1 and AG490 inhibitors (P < 0.05, Fig. 4b).
Taking all the above data into account, PC1-mediated mechanostimulation of osteoblastic cells was shown to raise Runx2 mRNA expression levels through potentiation of the JAK2/STAT3 signaling cascade.
PC1-dependent STAT3 and Runx2 interaction with Runx2 promoter in mechanically stimulated human osteoblastic cells
Based on computational transcription factor-binding analysis and previous work from our group [26, 27], we identified functional STAT3- and Runx2-binding sites on the promoter sequence of the human Runx2 gene (Fig. 5a). In light of our previous data showing that Runx2 gene expression levels can be modulated by the PC1–JAK2/STAT3 axis, we run chromatin immunoprecipitation (ChIP) experiments to investigate the direct involvement of activated STAT3 (pSTAT3) and Runx2 transcription factors in the upregulation of Runx2 promoter activity in mechanically stretched human osteoblastic cells.
PC1-dependent pSTAT3 and Runx2 interaction with Runx2 promoter in mechanically-stretched human osteoblastic cells. a The human Runx2 promoter proximal to the transcription start site with designated STAT3- and Runx2-binding sites. b Real time-qPCR results (fold enrichment relative to the input) using the SimpleChIP Human Runx2 Promoter Primers after immunoprecipitation of chromatin preparations derived from unstretched cells and cells stretched for 6 h with pSTAT3 antibody or Runx2 antibody, or an IgG negative control (data not shown). Input refers to no treatment without any antibody. Pre-treatment with a combination of specific inhibitors was performed where indicated. Purple chromatin preparations from unstretched or 6 h-stretched cells pre-treated with anti-IgPKD1 and AG490, orange chromatin preparations from unstretched or 6 h-stretched cells pre-treated with anti-IgPKD1 and AG490 and immunoprecipitated with pSTAT3 antibody, and light orange chromatin preparations from unstretched or 6 h-stretched cells pre-treated with anti-IgPKD1 and AG490 and immunoprecipitated with Runx2 antibody. Data are presented as fold enrichment of specific promoter region sequence relative to the input (untreated cells) of at least three independent experiments
Chromatin preparations from cells either untreated or treated with both anti-IgPKD1 and AG490 (a combination of inhibitors that had most robustly reversed the effects seen in our previous experiments) and then subjected to 6 h of mechanical stretching, were immunoprecipitated with a highly specific pSTAT3 antibody (Y705). The previously observed increase of STAT3-binding activity to Runx2 promoter at 6 h of stretching was reduced when cells were treated with both inhibitors (anti-IgPKD1 and AG490) and pSTAT3 antibody prior to mechanical stretch application compared to untreated controls (input, Fig. 5b). These results demonstrate the direct participation of PC1 in STAT3-mediated upregulation of Runx2 promoter activity. Similar results were obtained when the same chromatin preparations where immunoprecipitated with a highly specific Runx2 antibody. Again, the elevated Runx2-binding activity to Runx2 promoter after 6 h of stretching was reduced after pre-treatment of cells with anti-IgPKD1, AG490 and Runx2 antibody compared to input (Fig. 5b). A normal rabbit IgG antibody was employed as negative control in all ChIP-qPCR experiments (data not shown).
The above data demonstrate the PC1-dependent direct regulation of Runx2 promoter activity by pSTAT3 and Runx2 transcription factors in mechanostimulated osteoblastic cells.
PC1-dependent direct binding of STAT3 and Runx2 transcription factors to Runx2 promoter in mechanically stretched human osteoblastic cells
Given that pSTAT3 and Runx2 can associate with Runx2 promoter, we went on to identify the exact promoter sequences engaged in this interaction. Chromatin preparations were used to run end-point PCR experiments with a set of primers that contain the specific sequence of Runx2 promoter from 3′ −587 to −621 5′ encompassing binding motifs for both STAT3 and Runx2 (Fig. 6a). In accordance with the Real time-PCR results, the increased binding of STAT3 to Runx2 promoter at 6 h of stretching was reduced upon pre-treatment of cells with both inhibitors (anti-IgPKD1 and AG490) and pSTAT3 antibody compared to untreated controls (input, Fig. 6b). In a similar manner, when the same chromatin preparations where immunoprecipitated with Runx2 antibody from cells treated with both inhibitors (anti-IgPKD1 and AG490) a marked decrease of Runx2 binding to Runx2 promoter was also observed at 6 h of stretch application compared to input (Fig. 6b).
PC1-dependent direct binding of pSTAT3 and Runx2 to Runx2 promoter in mechanically-stretched human osteoblastic cells. a The human Runx2 promoter from 3′ −587 to −621 5′ with designated functional STAT3- (−607 to −604, GGAA) and Runx2- (−596 to −591, ACCACA) binding sites. b The graphs depict end-point PCR results (enrichment relative to input) using the relevant oligonucleotide sequence (see “Materials and methods”) after immunoprecipitation with pSTAT3 antibody or Runx2 antibody upon mechanical stimulation (6 h) following pre-treatment with a combination of specific inhibitors where indicated. Purple chromatin preparations from unstretched or 6 h-stretched cells pre-treated with anti-IgPKD1 and AG490, orange chromatin preparations from unstretched or 6 h-stretched cells pre-treated with anti-IgPKD1 and AG490 and immunoprecipitated with pSTAT3 antibody, and light orange chromatin preparations from unstretched or 6 h-stretched cells pre-treated with anti-IgPKD1 and AG490 and immunoprecipitated with Runx2 antibody. Data are presented as enrichment of specific promoter region sequence relative to the input (untreated cells) from at least three independent experiments
These data provide strong evidence that in mechanically stimulated osteoblastic cells PC1 mediates direct binding of pSTAT3 and Runx2 transcription factors to adjacent sites within Runx2 promoter, thus upregulating its activity towards osteoblast differentiation.
Discussion
This study identifies PC1–JAK2/STAT3 signaling axis as a sensitive intracellular pathway that ‘funnels’ osteoblastic cell response to mechanical loading. Runx2 was uncovered as a direct target gene of PC1–JAK2/STAT3 mechanotransduction cascade coupling extracellular mechanical forces to differentiation process in human osteoblast-like cells (pre-osteoblasts).
Mechanical loading presents an effective stimulus capable to alter gene expression and induce bone remodeling, comprising an essential component of regenerative medicine with clinical applications in skeletal bone defects, degenerative bone diseases and bone-tissue engineering. Elucidation of mechanotransduction pathways and molecular targets that regulate bone remodeling and bone repair in human osteoblastic cells constitutes an ongoing research interest of our laboratory [3–7, 21, 26, 27].
In the present study, mechanical stress (stretching) sensed by PC1-expressing osteoblastic cells was shown to trigger a tight interaction between PC1-CT and JAK2 proteins. Human osteoblastic cells exhibited an increase in the phosphorylated/active form of JAK2 (pJAK2) at 1 h of mechanical stretching (immediate-early response), which remained elevated up to 6 h of mechanical stimulation. Pre-treatment of cells with an antibody against the mechanosensing extracellular N-terminal part of PC1 (anti-IgPKD1) and/or a specific JAK2 inhibitor (AG490) abrogated this effect, demonstrating the requirement of PC1 for JAK2 activation as well as the specificity of this interaction. Furthermore, fluorescence confocal microscopy experiments verified the physical interaction of PC1 and JAK/pJAK2 proteins. Complex formation was detected between PC1-CT and JAK2 in unstretched osteoblastic cells that was reduced upon mechanical stimulation for 1 h. Moreover, PC1-CT interacted with pJAK2 after 1 h of mechanical stretching while the ratio of interactions (complex formation) of PC-CT/JAK2 and PC-CT-pJAK2 in unstretched cells was increased by three-fold in 1 h-stretched cells.
This is the first time that a direct interaction of PC1 with JAK2 is demonstrated in osteoblastic cells subjected to mechanical strain. It is likely that mechanical forces induce a conformational change to PC1 that stabilizes JAK2 in an active state, possibly by protecting tyrosine phosphorylated groups from the action of protein phosphatases.
Previous studies in renal epithelium and ADPKD kidneys have shown that PC1–PC2 heterodimer is essential for the interaction and activation of JAK2 by PC1 [30, 43]. In agreement with that, PC1 was recently shown to constitutively bind to JAK2 in HEK293 cells upon proteolytic cleavage of PC1-CT by calpains as a response to changes in intracellular calcium [44]. In the same cells, PC1-CT was found to activate STAT3 in a JAK2-dependent manner leading to tyrosine phosphorylation and ensuing transcriptional activity [22]. Notably, this study identifies JAK2 as the principal kinase involved in STAT3 potentiation by PC1.
The physiological role of JAK2 in osteoblasts and bone development is poorly understood mainly because global JAK2 knock-out mice die of anemia at E12.5 before bone formation starts [45]. Nonetheless, studies of mouse embryonic fibroblasts derived from JAK2 null embryos demonstrate that JAK2 deficiency decouples GH receptor signaling from its downstream effectors, implying that JAK2 may be involved in bone development due to the important role of GH in bone formation [46].
On the other hand, the substrate of activated JAK2, STAT3 has been attributed a critical role in skeletal growth by mediating anabolic signals in osteoblasts and regulating bone formation [47, 48]. Selective inactivation of STAT3 in osteoblasts reduced body and bone mass as well as bone strength and load-driven bone formation in mice [48]. Furthermore, severe spinal deformation owing to delayed endochondral ossification and abnormal growth plate morphology was also detected in about 10 % of those mice [25, 48].
STAT3 is a key signal transduction protein that integrates signaling by numerous growth factors, cytokines and oncoproteins [49]. Upon stimulation, activated JAK phosphorylates the tyrosine residue of YXXQ motif, leading to recruitment through binding to the Src homology 2 (SH2) domain and tyrosine phosphorylation at the cytoplasmic tail of STAT3. Activated STAT3 dimerizes and translocates to the nucleus where it binds to target-gene promoter sequences and induces gene expression [49, 50]. In our study, PC1-expressing osteoblastic cells subjected to mechanical stretching revealed that phosphorylated/DNA-binding competent STAT3 (pSTAT3) increased at 1/2–1 h and returned to normal at 6 h. In accordance with the activation mechanism of STAT3, stretching of cultured cells pre-treated with STAT3 VII inhibitor or anti-IgPKD1 antibody attenuated the observed decrease in the abundance of the cytoplasmic (unphosphorylated/inactive) STAT3 species (data not shown). The specific activation of the STAT3 transcription factor was further explored under conditions of mechanical loading in osteoblastic cells. Indeed, TransAM DNA-binding experiments revealed a statistically significant elevation of pSTAT3 DNA-binding activity in nuclear extracts from osteoblastic cells after 1/2 h of mechanical stimulation that was gradually decreased after 1 h to 6 h. Furthermore, incubation of cells with STAT3 VII and anti-IgPKD1 inhibitors hampered the obtained effect, corroborating that mechanically stimulated PC1 triggers nuclear translocation of STAT3 and augments its DNA-binding activity in osteoblastic cells.
The effect of mechanical stretching on STAT3 expression levels has been previously investigated in murine osteoblast-like MG-63 cells. Upon cell exposure to cyclic tension force, STAT3 expression was upregulated along with the levels of the cytoskeletal (actin-binding) protein, Girdin affecting both cell proliferation and migration [47]. Moreover, mechanically triggered bone formation was found suppressed in STAT3 knock-out mice indicating that STAT3 deficiency hinders osteoblast recruitment and activity [48]. The novelty of our study relies on the fact that mechanical induction of STAT3 activation in osteoblastic cells is mediated through the transmembrane mechanosensor PC1 as blocking of the mechanosensing extracellular N-terminal part of the protein prior to mechanical stimulation abrogated STAT3 translocation to the nucleus. This finding is of importance for bone mechanobiology since it unveils PC1–JAK2/STAT3 as a novel mechanotransduction cascade in osteoblastic cells with potential regulatory role over bone-cell function. Remarkably, among various developmental pathways, the JAK/STAT3 pathway has a prominent role in osteoblastic differentiation, proliferation and survival [25, 48].
Importantly, mechanoresponse in Pkd1-deficient mice has been suggested to engage stimulation of the transcription factor Runx2, a critical regulator of osteoblastic differentiation [51–53]. Additionally, previous studies from our group have shown that mechanical loading (strain/stretching) can modulate both Runx2 expression levels and activity [7, 27]. We, therefore, examined the likelihood that Runx2 can be also a target of PC1–JAK2/STAT3 pathway upon mechanical stimulation of osteoblastic cells. Computational analysis revealed the presence of functional STAT3- and Runx2-binding sites in Runx2 promoter whilst earlier work in our laboratory have demonstrated that STAT3 can physically interact with Runx2 in the human osteoblastic cell line Saos-226. These data prompted us to investigate whether PC1-mediated mechanostimulation can influence the expression of Runx2 mRNA levels by direct control of Runx2 promoter activity through pSTAT3 and Runx2 transcription factors. Indeed, mechanical stretching of osteoblastic cells led to elevation of Runx2 mRNA at 6 h while pre-treatment with anti-IgPKD1 or AG490 inhibitors plus pSTAT3/Runx2 antibodies abrogated this effect. Reduction of Runx2 mRNA expression upon single anti-IgPKD1 treatment was also observed, however, without reaching statistical significance. This may be attributed to the nature of anti-IgPKD1 being a ‘masking’ antibody (rather than a targeted chemical compound) that partially blocks the extracellular N-terminal part of PC1, possibly allowing cellular recovery/compensatory mechanisms to take place.
In addition, ChIP experiments corroborated the previous result by further demonstrating PC1-mediated direct binding of pSTAT3 and Runx2 to neighboring sites within Runx2 promoter in mechanically stretched osteoblastic cells.
All the above can be incorporated into a model where the N-terminal part of PC1 functions as an ‘antenna’ domain in human osteoblastic cells sensing extracellular mechanical cues and eliciting a mechanoresponse through cleavage of its CT. PC1-CT in turn associates specifically with JAK2 triggering its activation and consequent STAT3 phosphorylation. The active form of STAT3 then translocates to the nucleus where it binds to Runx2 promoter and in concert with Runx2 (and perhaps released CT) augments Runx2 transcription (Fig. 7). Runx2 gene expression levels play a crucial and multifaceted role during bone development by triggering osteoblastic differentiation (i.e. progression of immature osteoblasts to the mature osteoblast phenotype) hence the rate of bone formation.
Schematic model of the proposed crosstalk between PC1 and JAK2/STAT3 signaling in human osteoblastic cells under mechanical stimulation. The extracellular N-terminal part of PC1 acts as a mechanosensing domain responding to mechanical stimuli and triggering a mechanotransduction cascade through cleavage of its C-terminal cytoplasmic tail (CT). PC1-CT interacts physically with JAK2 protein inducing its phosphorylation/activation (pJAK2) and, in turn, STAT3 phosphorylation (P). The phosphorylated hence potentiated form of STAT3 translocates to the nucleus where it binds to cognate sequence on Runx2 promoter and in cooperation with adjacently bound Runx2 (autoregulatory loop) and possibly diffusible CT upregulates Runx2 gene transcription (green vertical arrow), a gene that dictates entry to and progression of osteoblastic differentiation
It is, therefore, posed that PC1–JAK2/STAT3 is a new anabolic pathway in osteoblastic cells that functions to couple external mechanical signals to modulation of Runx2 gene expression and control of bone remodeling. Since bone remodeling is an important constituent of regenerative medicine affecting bone growth and repair, PC1–JAK2/STAT3 signaling axis can be regarded as a promising molecular approach to deliver in vivo mechanical stimuli to osteoblasts for bone-tissue engineering and therapeutic manipulation of skeletal bone deformities.
References
Rosa N, Simoes R, Magalhaes FD, Marques AT (2015) From mechanical stimulus to bone formation: a review. Med Eng Phys 37:719–728
Raggatt LJ, Partridge NC (2010) Cellular and molecular mechanisms of bone remodeling. J Biol Chem 285:25103–25108
Basdra EK, Komposch G (1997) Osteoblast-like properties of human periodontal ligament cells: an in vitro analysis. Eur J Orthod 19:615–621
Basdra EK, Papavassiliou AG, Huber LA (1995) Rab and rho GTPases are involved in specific response of periodontal ligament fibroblasts to mechanical stretching. Biochim Biophys Acta 1268:209–213
Kletsas D, Basdra EK, Papavassiliou AG (1998) Mechanical stress induces DNA synthesis in PDL fibroblasts by a mechanism unrelated to autocrine growth factor action. FEBS Lett 430:358–362
Peverali FA, Basdra EK, Papavassiliou AG (2001) Stretch-mediated activation of selective MAPK subtypes and potentiation of AP-1 binding in human osteoblastic cells. Mol Med 7:68–78
Ziros PG, Gil AP, Georgakopoulos T, Habeos I, Kletsas D, Basdra EK, Papavassiliou AG (2002) The bone-specific transcriptional regulator Cbfa1 is a target of mechanical signals in osteoblastic cells. J Biol Chem 277:23934–23941
Papachristou DJ, Papachroni KK, Basdra EK, Papavassiliou AG (2009) Signaling networks and transcription factors regulating mechanotransduction in bone. BioEssays 31:794–804
Papachroni KK, Karatzas DN, Papavassiliou KA, Basdra EK, Papavassiliou AG (2009) Mechanotransduction in osteoblast regulation and bone disease. Trends Mol Med 15:208–216
Dalagiorgou G, Basdra EK, Papavassiliou AG (2010) Polycystin-1: function as a mechanosensor. Int J Biochem Cell Biol 42:1610–1613
Hanaoka K (2000) Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature 408:990–994
Newby LJ, Streets AJ, Zhao Y, Harris PC, Ward CJ, Ong AC (2002) Identification, characterization, and localization of a novel kidney polycystin-1-polycystin-2 complex. J Biol Chem 277:20763–20773
Qian F, Germino FJ, Cai Y, Zhang X, Somlo S, Germino GG (1997) PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat Genet 16:179–183
Tsiokas L, Kim E, Arnould T, Sukhatme VP, Walz G (1997) Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc Natl Acad Sci USA 94:6965–6970
Xiao Z, Dallas M, Qiu N, Nicolella D, Cao L, Johnson M, Bonewald L, Quarles LD (2011) Conditional deletion of Pkd1 in osteocytes disrupts skeletal mechanosensing in mice. FASEB J 25:2418–2432
Xiao Z, Zhang S, Mahlios J, Zhou G, Magenheimer BS, Guo D, Dallas SL, Maser R, Calvet JP, Bonewald L, Quarles LD (2006) Cilia-like structures and polycystin-1 in osteoblasts/osteocytes and associated abnormalities in skeletogenesis and Runx2 expression. J Biol Chem 281:30884–30895
Xiao Z, Zhang S, Cao L, Qiu N, David V, Quarles LD (2010) Conditional disruption of Pkd1 in osteoblasts results in osteopenia due to direct impairment of bone formation. J Biol Chem 285:1177–1187
Bertuccio CA, Caplan MJ (2013) Polycystin-1C terminus cleavage and its relation with polycystin-2, two proteins involved in polycystic kidney disease. Medicina (B Aires) 73:155–162
Merrick D, Bertuccio CA, Chapin HC, Lal M, Chauvet V, Caplan MJ (2014) Polycystin-1 cleavage and the regulation of transcriptional pathways. Pediatr Nephrol 29:505–511
Wang H, Sun W, Ma J, Pan Y, Wang L, Zhang W (2014) Polycystin-1 mediates mechanical strain-induced osteoblastic mechanoresponses via potentiation of intracellular calcium and Akt/beta-catenin pathway. PLoS One 9:e91730
Dalagiorgou G, Piperi C, Georgopoulou U, Adamopoulos C, Basdra EK, Papavassiliou AG (2013) Mechanical stimulation of polycystin-1 induces human osteoblastic gene expression via potentiation of the calcineurin/NFAT signaling axis. Cell Mol Life Sci 70:167–180
Talbot JJ, Shillingford JM, Vasanth S, Doerr N, Mukherjee S, Kinter MT, Watnick T, Weimbs T (2011) Polycystin-1 regulates STAT activity by a dual mechanism. Proc Natl Acad Sci USA 108:7985–7990
Weimbs T, Olsan EE, Talbot JJ (2013) Regulation of STATs by polycystin-1 and their role in polycystic kidney disease. JAKSTAT 2:e23650
Li Y, Backesjo CM, Haldosen LA, Lindgren U (2008) IL-6 receptor expression and IL-6 effects change during osteoblast differentiation. Cytokine 43:165–173
Li J (2013) JAK-STAT and bone metabolism. JAKSTAT 2:e23930
Ziros PG, Georgakopoulos T, Habeos I, Basdra EK, Papavassiliou AG (2004) Growth hormone attenuates the transcriptional activity of Runx2 by facilitating its physical association with Stat3beta. J Bone Miner Res 19:1892–1904
Ziros PG, Basdra EK, Papavassiliou AG (2008) Runx2: of bone and stretch. Int J Biochem Cell Biol 40:1659–1663
Jackson RA, Murali S, van Wijnen AJ, Stein GS, Nurcombe V, Cool SM (2007) Heparan sulfate regulates the anabolic activity of MC3T3-E1 preosteoblast cells by induction of Runx2. J Cell Physiol 210:38–50
Qian F, Boletta A, Bhunia AK, Xu H, Liu L, Ahrabi AK, Watnick TJ, Zhou F, Germino GG (2002) Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc Natl Acad Sci USA 99:16981–16986
Ibraghimov-Beskrovnaya O, Bukanov NO, Donohue LC, Dackowski WR, Klinger KW, Landes GM (2000) Strong homophilic interactions of the Ig-like domains of polycystin-1, the protein product of an autosomal dominant polycystic kidney disease gene, PKD1. Hum Mol Genet 9:1641–1649
Streets AJ, Wagner BE, Harris PC, Ward CJ, Ong AC (2009) Homophilic and heterophilic polycystin 1 interactions regulate E-cadherin recruitment and junction assembly in MDCK cells. J Cell Sci 122:1410–1417
Naruse K, Sokabe M (1993) Involvement of stretch-activated ion channels in Ca2+ mobilization to mechanical stretch in endothelial cells. Am J Physiol 264:C1037–C1044
Yousefian JZNP, Miller B, Shanfeld J, Davidovitch Z (1992) Effect of different types of stress on human periodontal ligament cells in vitro. In: Davidovitch Z (ed) Biological mechanisms of tooth movement and craniofacial adaptation. The Ohio State University, College of Dentistry, Columbus, pp 319–329
Fujii S, Maeda H, Wada N, Kano Y, Akamine A (2006) Establishing and characterizing human periodontal ligament fibroblasts immortalized by SV40T-antigen and hTERT gene transfer. Cell Tissue Res 324:117–125
Marais R, Wynne J, Treisman R (1993) The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73:381–393
Schreiber E, Matthias P, Muller MM, Schaffner W (1989) Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res 17:6419
Karamouzis MV, Dalagiorgou G, Georgopoulou U, Nonni A, Kontos M, Papavassiliou AG (2016) HER-3 targeting alters the dimerization pattern of ErbB protein family members in breast carcinomas. Oncotarget 7:5576–5597
Farmaki E, Mkrtchian S, Papazian I, Papavassiliou AG, Kiaris H (2011) ERp29 regulates response to doxorubicin by a PERK-mediated mechanism. Biochim Biophys Acta 1813:1165–1171
Nokhbehsaim M, Keser S, Nogueira AV, Cirelli JA, Jepsen S, Jäger A, Eick S, Deschner J (2014) Beneficial effects of adiponectin on periodontal ligament cells under normal and regenerative conditions. J Diabetes Res 2014:796565
Nokhbehsaim M, Keser S, Nogueira AV, Jäger A, Jepsen S, Cirelli JA, Bourauel C, Eick S, Deschner J (2014) Leptin effects on the regenerative capacity of human periodontal cells. Int J Endocrinol 2014:180304
Jerman S, Ward HH, Lee R, Lopes CA, Fry AM, MacDougall M, Wandinger-Ness A (2014) OFD1 and flotillins are integral components of a ciliary signaling protein complex organized by polycystins in renal epithelia and odontoblasts. PLoS One 9:e106330
Xu J, Cole DC, Chang CP, Ayyad R, Asselin M, Hao W, Gibbons J, Jelinsky SA, Saraf KA, Park K (2008) Inhibition of the signal transducer and activator of transcription-3 (STAT3) signaling pathway by 4-oxo-1-phenyl-1,4-dihydroquinoline-3-carboxylic acid esters. J Med Chem 51:4115–4121
Bhunia AK, Piontek K, Boletta A, Liu L, Qian F, Xu PN, Germino FJ, Germino GG (2002) PKD1 induces p21(waf1) and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell 109:157–168
Kim H, Kang AY, Ko AR, Park HC, So I, Park JH, Cheong HI, Hwang YH, Ahn C (2014) Calpain-mediated proteolysis of polycystin-1 C-terminus induces JAK2 and ERK signal alterations. Exp Cell Res 320:62–68
Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN (1998) Jak2 is essential for signaling through a variety of cytokine receptors. Cell 93:385–395
Neubauer H, Cumano A, Müller M, Wu H, Huffstadt U, Pfeffer K (1998) Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell 93:397–409
Hu JT, Li Y, Yu B, Gao GJ, Zhou T, Li S (2015) Girdin/GIV is upregulated by cyclic tension, propagates mechanical signal transduction, and is required for the cellular proliferation and migration of MG-63 cells. Biochem Biophys Res Commun 464:493–499
Zhou H, Newnum AB, Martin JR, Li P, Nelson MT, Moh A, Fu XY, Yokota H, Li J (2011) Osteoblast/osteocyte-specific inactivation of Stat3 decreases load-driven bone formation and accumulates reactive oxygen species. Bone 49:404–411
Bromberg J (2002) Stat proteins and oncogenesis. J Clin Invest 109:1139–1142
Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC (2006) Stat3 activation of NF-{kappa}B p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci USA 103:7264–7269
Hou B, Kolpakova-Hart E, Fukai N, Wu K, Olsen BR (2009) The polycystic kidney disease 1 (Pkd1) gene is required for the responses of osteochondroprogenitor cells to midpalatal suture expansion in mice. Bone 44:1121–1133
Qiu N, Cao L, David V, Quarles LD, Xiao Z (2010) Kif3a deficiency reverses the skeletal abnormalities in Pkd1 deficient mice by restoring the balance between osteogenesis and adipogenesis. PLoS One 5:e15240
Xiao Z, Zhang S, Magenheimer BS, Luo J, Quarles LD (2008) Polycystin-1 regulates skeletogenesis through stimulation of the osteoblast-specific transcription factor RUNX2-II. J Biol Chem 283:12624–12634
Acknowledgments
We thank Dr. O. Ibraghimov-Beskrovnaya and H. Husson (Genzyme Co., MA, USA) for their generous gift of the anti-IgPKD1 inhibitory antibody. “Studies utilized resources provided by the NIDDK sponsored Baltimore Polycystic Kidney Disease Research and Clinical Core Center, P30 DK090868”.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest with any financial organization regarding the material discussed in the manuscript.
Additional information
G. Dalagiorgou, C. Piperi and C. Adamopoulos contributed equally to this work.
Rights and permissions
About this article
Cite this article
Dalagiorgou, G., Piperi, C., Adamopoulos, C. et al. Mechanosensor polycystin-1 potentiates differentiation of human osteoblastic cells by upregulating Runx2 expression via induction of JAK2/STAT3 signaling axis. Cell. Mol. Life Sci. 74, 921–936 (2017). https://doi.org/10.1007/s00018-016-2394-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2394-8