Abstract
Research over the last 25 years on the cell adhesion molecule L1 has revealed its pivotal role in nervous system function. Mutations of the human L1CAM gene have been shown to cause neurodevelopmental disorders such as X-linked hydrocephalus, spastic paraplegia and mental retardation. Impaired L1 function has been also implicated in the aetiology of fetal alcohol spectrum disorders, defective enteric nervous system development and malformations of the renal system. Importantly, aberrant expression of L1 has emerged as a critical factor in the development of human carcinomas, where it enhances cell proliferation, motility and chemoresistance. This discovery promoted collaborative work between tumour biologists and neurobiologists, which has led to a substantial expansion of the basic knowledge about L1 function and regulation. Here we provide an overview of the pathological conditions caused by L1 malfunction. We further discuss how the available data on gene regulation, molecular interactions and posttranslational processing of L1 may contribute to a better understanding of associated neurological and cancerous diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since its discovery in 1984 [1], the cell adhesion molecule L1 has been established as a key player throughout the development of the nervous system. L1 is the founding member of a subgroup of neuronal immunoglobulin (Ig) superfamily cell adhesion molecules (IgSF-CAMs) that includes both vertebrate and invertebrate members. In mammals, this subgroup comprises the close homologue of L1 (CHL1), the neuronal cell adhesion molecule (NrCAM) and neurofascin, all of which share similar protein domain structures [2]. L1 itself consists of a large extracellular part possessing six Ig-like and five fibronectin-repeat III (FNIII-like) domains, that is linked via a single transmembrane sequence to a short intracellular cytoplasmic domain (ICD) (Fig. 1a). The importance of L1 is reflected in pathological mutations in the human L1CAM gene, which underlie a variety of neurological conditions collectively referred to as CRASH syndrome or L1 syndrome [3–6]. The clinical characterization of patients with L1 syndrome, together with phenotypes observed in L1-deficient mouse mutants [7–9], have revealed that the most distinctive molecular actions of L1 relate to neuronal migration, axon growth and synapse formation in the developing and adult brain [2, 10, 11].
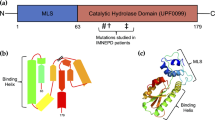
L1 domain structure and proteolytic cleavage maps. a Schematic protein domain model of L1. L1 has a relative molecular mass of about 220 kDa and consists of an large extracellular part, possessing six Ig-like (Ig 1–6) and five FNIII 1–5 domains. The extracellular part is linked via a single transmembrane sequence to a short ICD. b The serine proteases PC5A and plasmin constitutively cleave full-length L1 (L1–220) at dibasic residues in the third FNIII-like domain, thereby generating an ectodomain fragment of about 140 kDa (L1–140) and a membrane-retained fragment of about 80 kDa (L1–80). c The serine protease neuropsin cleaves L1 in response to neural activity. Cleavage occurs at a site near the membrane, leading to the release of an ectodomain fragment of about 200 kDa (L1–200). d The metalloprotease ADAM10 cleaves L1 at a site near the membrane, leading to the release of an ectodomain fragment of about 200 kDa (L1–200) and a membrane-retained stub of 32 kDa (L1–32). Cleavage occurs constitutively in cancer cells and is activity-dependent in neuronal cells. e The metalloprotease ADAM17 cleaves L1 at a site near the membrane, leading to the release of an ectodomain fragment of about 200 kDa (L1–200) and a membrane-retained stub of 32 kDa (L1–32). Cleavage can be induced by stimulation by phorbol esters or pervanadate. f Constitutive sequential cleavage of L1. Constitutive L1 cleavage by ADAM10 or ADAM17 is followed by cleavage of the membrane-retained 32 kDa fragment mediated by γ-secretase, leading to the release of an intracellular fragment of about 28 kDa (L1–28)
The various functions of L1 involve complex homo- and heterophilic interactions of its six Ig-like and five FNIII-like extracellular domains. Similarly, the cytoplasmic domain interacts with a variety of intracellular proteins such as kinases (e.g. casein kinase II, focal adhesion kinase) and adaptor molecules (e.g. ankyrin, AP-2), which modulate the association between L1 and the cytoskeleton [12, 13] and endosomal membrane systems [14–16], respectively. Several lines of evidence indicate that the interactions between L1 and extracellular and intracellular binding partners not only mediate cell–cell adhesion, but also potentiate intracellular signalling cascades. One example is the L1-dependent activation of MAPK-ERK signalling [17] via interaction with β1-integrin [18].
Outside the nervous system, aberrant expression of L1 has been shown to promote proliferation, motility and chemoresistance of cancer cells [19–21]. The critical role of L1 in these processes and the possibility of perturbing aberrant L1 function in carcinomas by administration of specific therapeutic antibodies [22–24] has directed the attention of researchers towards the pathological mechanisms underlying the action of L1 in cancer.
Previous reviews have focused on many facets of L1 function, including its complex binding properties [25, 26], signal transduction [2, 10] and the relationship between the clinical presentation and the genotype in patients with L1 syndrome [5]. The role of L1 in cancer progression with a focus on colon cancer has recently been reviewed [27]. Here we provide an overview of the various disorders that have been associated with loss of function or aberrant function of L1. We discuss the underlying mechanisms and highlight the role of proteolytic processing of L1 in the nervous system and in cancer.
Phenotypic spectrum of L1 syndrome
Mutations in the L1CAM gene cause X-linked disorders of a broad phenotypic spectrum including X-linked hydrocephalus (HSAS, hydrocephalus due to stenosis of the aqueduct of Sylvius), MASA syndrome (mental retardation, aphasia, shuffling gait, adducted thumbs), agenesis of the corpus callosum, and spastic paraplegia (X-linked hereditary spastic paraplegia type 1). The manifestation of these disorders is highly variable and even identical gene mutations may cause distinct intra- and interfamily phenotypes [3, 28]. Most consistent features in affected males are lower limb spasticity, mental retardation, hydrocephalus and flexion deformity of the thumbs [3, 5, 29]. A few reports have also described female carriers who develop minor symptoms comprising adducted thumbs and mild mental retardation [30] as well as subtle abnormalities in corticospinal function [31]. Owing to the fact that all of these phenotypic features represent variable manifestations of mutations in the L1CAM gene, we follow the consensus of naming this group of disorders “L1 syndrome” [6].
To date, about 200 human pathological mutations in the L1CAM gene have been identified, including frameshift, nonsense and, the most frequent, missense mutations [32]. The pathological mechanisms leading to L1 syndrome are not well understood. However, it turns out that both nonsense mutations, which lead to the production of truncated proteins, and missense mutations, which affect structurally important key residues in the extracellular part of L1, result in severe hydrocephalus and early mortality more often than mutations affecting the ICD [29, 33]. These findings, together with the observation that the cytoplasmic part of L1 is not required for homophilic cell–cell adhesion [34], suggested that the adhesive function of the extracellular part of L1 plays a more important role during nervous system development than intracellular signalling mediated by its cytoplasmic part [29]. However, this view might be an oversimplification because cell–cell adhesion mediated by extracellular domains of L1 and the subsequent activation of downstream signalling are closely associated [2, 10]. Indeed, two L1 syndrome-associated mutations that substitute amino acids of the extracellular part of L1 have been shown to promote cell–cell adhesion but fail to stimulate adhesion-dependent activation of EGFR [35]. Recently, an extended view on the possible mechanisms underlying L1 syndrome has been obtained by genetic rescue studies in Drosophila. The authors found evidence that a mutation in the second Ig-like domain of L1 (H210Q) may cause neurological features that are due to synaptic dysfunction rather than axonal pathfinding defects [36]. The same mutation was previously shown to differentially affect homo- and heterophilic binding of L1 [37], which is compatible with the hypothesis that alterations in ligand-specific L1 binding properties represent one important pathological mechanism involved in L1 syndrome.
Table 1 summarizes some interaction partners of L1, which have been associated with L1 malfunction. It is conceivable that such interaction partners might play a role in the variable phenotypic manifestations observed in patients with L1 syndrome. In addition, the intra- and interfamilial variability of L1 syndrome’s phenotypic features, such as X-linked hydrocephalus, suggests the existence of genetic modifiers. Of note, the development of X-linked hydrocephalus in transgenic L1-6D mice that lack the sixth Ig-like domain of L1 [38] has been reported to be strongly dependent on the genetic background [39]. The authors screened for single nucleotide polymorphisms that are present in different L1-6D mice strains and identified possible genetic modifiers of X-linked hydrocephalus such as the polycomb-like protein transcription factor Mtf2 [39]. Interestingly, Mtf2-mutant mice have also been reported to develop hydrocephalus [40]. It will therefore be important to validate whether and how Mtf2 or other candidate genes contribute to the pathogenesis of L1 syndrome.
L1 malfunction in fetal alcohol spectrum disorders
Fetal alcohol spectrum disorders (FASD) are another pathological condition that has been associated with loss of L1 function. FASD comprise neuropathological abnormalities such as mental retardation, hydrocephalus, and agenesis of the corpus callosum, which are similar to those observed in L1 syndrome [41]. Several in vitro studies have shown that ethanol inhibits L1-mediated cell–cell adhesion [42], signalling [43, 44] and neurite outgrowth [45]. Furthermore, specific peptide antagonists prevent ethanol-dependent inhibition of L1 as well as ethanol teratogenesis in mouse embryos [46, 47]. However, the inhibitory effects of ethanol on the adhesive or neurite growth-promoting functions of L1 are not observed in S2 cells [48] or cortical neurons [49], suggesting that these effects are dependent on the cellular context.
Recently, the first insights into the molecular mechanisms underlying ethanol-dependent inhibition of L1 have been obtained through the identification of an ethanol-binding site at the interface between the first and fourth Ig-like domains of L1 [50]. Previous studies have revealed that the four N-terminal Ig-like domains of L1 [51], as well as those of related cell adhesion molecules such as axonin-1/TAG-1 [52] or Dscam [53], adopt a horseshoe-shaped conformation in which the first and the second domains fold back to interact with the fourth and third domains. Binding studies using purified L1 proteins, where single domains or the hinge region between Ig-like domains II and III were deleted, have shown that integrity of the horseshoe conformation is required for homo- and heterophilic interactions of L1 [37, 54].
The location of the ethanol binding site has been found to be close to amino acid residues L120 and G121 that are mutated in L1 syndrome patients [37], suggesting that similar mechanisms underlie the pathology of these mutations and FASD [50]. The collected data support the hypothesis that the pathophysiology of FASD depends, at least in part, on the deleterious effects of ethanol on protein domain integrity of L1, which in turn leads to impaired ligand interactions during nervous system development. It remains an open question as to whether ethanol also binds to and thereby inhibits the function of the L1 family members CHL1, NrCAM and neurofascin, which also might have the potential to form a horseshoe conformation. Therefore, further research is required to address this issue.
L1 syndrome in combination with Hirschsprung’s disease
Hirschsprung’s disease (HSCR) is a complex multigenic disorder in which there is massive distension of the bowel and functional intestinal obstruction due to the absence of ganglion cells. Based on several investigations these defects have been attributed to defective migration of neural crest cells [55]. About ten genes, including the most frequently implicated RET receptor tyrosine kinase gene, are associated with HSCR. In addition, several susceptibility loci have been identified as possible modifier genes [56–58]. Comparative immunohistological analyses of ganglionic and aganglionic segments from patients with HSCR have revealed that the protein expression of L1, but not that of the cell adhesion molecules Thy-1 and alpha5 integrin, is impaired in hypertrophied nerve bundles in aganglionic segments, suggesting a role of L1 in HSCR pathogenesis [59]. In the meantime, HSCR has been diagnosed in several patients with L1 syndrome further suggesting a link between L1 malfunction and HSCR [60–64]. However, it should be noted that, since the incidence of HSCR is low in L1 syndrome patients (about 3%) but probably higher in patients with X-linked hydrocephalus [60], L1 mutations alone are probably not sufficient to cause HSCR. Recently, a role for L1 in HSCR onset has been further supported by histological studies in L1-deficient mice, which showed that changes in the migration and differentiation of neural crest-derived cells within the developing enteric nervous system indeed depend on L1 [65, 66]. Taken together, the available data suggest that L1 is a genetic modifier of HSCR. Further studies are required to investigate the role of L1 malfunction in HSCR and whether L1CAM gene mutations interfere with the function of RET or other HSCR-associated genes.
L1 syndrome in combination with renal malformations
L1 has also been implicated as a possible candidate gene in familial cases of renal malformations [67, 68]. It has been shown that L1 is expressed in a spatiotemporally restricted pattern in human epithelial kidney cells [69] and is required for branching morphogenesis of the ureteric bud in the rabbit [70]. Consistent with an important role of L1 in kidney development, L1-deficient mice exhibit diverse renal abnormalities [68]. Similar malformations of the renal system such as bilateral duplex kidneys and ureters have been found in boys with L1 syndrome suggesting that loss of L1 function impairs kidney morphogenesis during development [67]. The pathological mechanism is still unclear but might involve dysregulated proliferation of collecting duct cells [68]. It is worth noting that these defects must be distinguished from renal dysfunction in other patients who carry a large gene deletion encompassing the L1CAM and AVPR2 genes which leads to the development of L1 syndrome in combination with X-linked nephrogenic diabetes insipidus [71].
Aberrant L1 function in human carcinomas
Besides its broad expression in the nervous system, L1 has also been observed in a limited number of nonneuronal tissues including leucocytes [72] as well as ovarian and urogenital epithelial cells [70, 73]. However, many studies have identified elevated or aberrant expression of L1 as a critical factor in the development of different types of human carcinomas. As a common feature, tumour cells expressing L1 show increased proliferation, motility and metastatic invasion into surrounding tissues (Table 2). In addition, L1 expression has been shown to act as an antiapoptotic factor and to confer chemoresistance on pancreatic ductal adenocarcinomas [21] and colorectal carcinomas associated with a poor prognosis [74, 75].
It is likely that L1-mediated changes in cell–cell adhesion and signalling are involved in several steps during the transition from normal cells to invasive tumour cells. Cell–cell adhesion of neighbouring tumour cells could, for example, favour bidirectional signalling via L1 and integrins, since the same cell types often express both binding partners [76]. Recent studies further support a role for L1-integrin interactions in cancer and have demonstrated that both functional L1 and alpha5-integrin are required to promote chemoresistance in pancreatic ductal adenocarcinoma cell lines through inhibition of nitric oxide-dependent caspase [21, 77] (see also Table 1).
Neuropilin-1 (NP-1), originally described as a transmembrane receptor of semaphorins [78, 79] and vascular endothelial growth factor [80], has been identified as a ligand for L1 both in the nervous system [81] and in cancer cells [82]. In the nervous system, the interaction between L1 and NP-1 is well established and has been recognized as an important signalling mechanism for axon guidance [81, 83]. More recent evidence also suggests that L1 expressed on ovarian carcinoma cells interacts in trans mode with NP-1 on mesothelial cells, thereby mediating cell–cell adhesion and eventually bidirectional signalling [82]. Other observations suggest that L1 and NP-1 in endothelial tumour cells could also interact in cis favouring transendothelial migration and angiogenesis during tumour progression, as evidenced by antibody perturbation experiments [84]. However, it remains unclear whether the interactions between L1 and NP-1 play a role in other types of cancer cells such as ovarian carcinomas [73] and melanoma cells [85], in which L1 has been found to promote transendothelial migration.
Although several lines of evidence suggest a cooperative mode of action for L1, integrins and NP-1, that may also involve vascular endothelial growth factor receptor signalling [86–88], the molecular hierarchy of these processes in cancer cells is not well understood. Further investigations are required to elucidate the interactions of these molecules and their possible association in protein complexes. This would also be important for the development of therapeutic strategies, since the targeting of these molecules by specific therapeutic antibodies or inhibitory peptides could be instrumental in interfering with cancer progression.
Pathological L1CAM gene regulation in neurons and cancer
The L1CAM gene is located at chromosome Xq28 and spans about 16 kb with 28 coding exons. The full length open reading frame consists of 3,825 bp encoding for a 1,275 amino acid polypeptide [89]. Two alternatively spliced exons have been identified comprising exons 2 and 27 that are encoded in the neuronal full-length form of L1 but are lacking in the short isoform. Exon 2 is required for optimal ligand binding and promotion of neurite growth [90, 91], whereas exon 27 facilitates clathrin-mediated endocytosis of L1 [14]. The short isoform is expressed in nonneuronal cells such as Schwann cells, kidney cells, and blood lymphocytes [90, 92, 93], whereas full-length L1 is predominantly expressed in neurons. However, oligodendrocytes have been reported to express both L1 isoforms in a maturation-dependent manner [94].
Taking advantage of different transgenic L1-lacZ reporter mouse lines, previous work has identified a neural restrictive silencer element (NRSE) in the first intron of L1 [89]. The authors demonstrated that activation of NRSE by the repressor element 1 silencing factor/neuron-restrictive silencing factor (REST/NRSF) facilitates neuron-restricted L1 expression in the embryonic nervous system [89]. However, expression of REST/NRSF is not exclusively observed in nonneuronal cells and proceeds, albeit at relative low levels, in postmitotic neurons from several brain regions [95]. In addition, increased neuronal REST/NRSF expression has been observed in response to kainate-induced seizures [95] and ischaemia, where REST/NRSF contributes to ischaemia-induced neuronal death [96]. Interestingly, L1 protein expression decreases in the same hippocampal regions following sustained ischaemia [97], suggesting that REST/NRSF may not only repress nonneuronal L1 expression but also acts as a repressor of neuronal L1 expression under pathological conditions.
Recently, more than 1,800 genes have been predicted to contain NRSE sites and to represent putative target genes of REST/NRSF [98]. Moreover, dysregulation of REST/NRSF has been implicated in neurological disorders such as Huntington’s disease [99, 100] and Down syndrome (DS) [101, 102]. In the R6/2 transgenic mouse model of Huntington’s disease aberrant silencing activity of REST/NRSF has been shown to pathologically reduce expression of selected neuronal genes [99]. It is currently not clear whether expression of L1 is affected in this model. However, in DS model mice, dosage imbalance of the dual specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (Dyrk1A), which plays an important role in DS pathogenesis [103], causes reduced expression levels of REST/NRSF in embryonic neurons and increased expression levels in adult neurons, thereby affecting expression of L1 and other neuronal genes [101]. As a consequence, impaired neurite growth reminiscent of the morphological defects in neurons of DS model mice and patients has been observed [101].
These studies indicate that alterations in REST/NRSF expression levels are critically involved in several neurological disorders. Not unexpectedly, dysregulation of L1 expression levels is observed in most of these disorders, possibly attracting additional research attention on the role of L1 gene expression regulation in the pathophysiology of the nervous system. On the other hand, REST/NRSF has also emerged as a tumour suppressor, and diminished expression of REST/NRSF has been associated with colon cancer and transformation of human mammary epithelial cells [104, 105]. It appears conceivable that dysregulation of REST/NRSF may also play a role in aberrant L1 expression in human carcinomas. For example, REST/NRSF has been found to be inactivated in small-cell lung cancer [106], and increased expression levels of the neuronal isoform of L1 have been found in cells of the small-cell lung carcinoma cell line NCI-H69 [107].
In most other types of cancer cells with aberrant L1 expression, the nonneuronal isoform predominates [107–111]. In H6c7 pancreatic cancer cells, the gene expression regulation of L1 involves binding of the transcription factor slug to the L1 promoter, which induces aberrant L1 expression upon administration of TGF-β [108]. In colon cancer cells, the L1 promoter contains a LEF/TCF binding site which drives L1 expression by β-catenin/TCF signalling. Based on these observations, the authors proposed that aberrant expression of L1 in colon cancer may occur as a consequence of mutations in components of the Wnt signalling pathway [20]. More recently, analyses of the DNA methylation patterning in the core promoter and TCF-binding sites in the L1 promoter have indicated that DNA hypomethylation at L1 CpG islands may contribute to aberrant L1 expression in colorectal cancer [112]. The combined data suggest that both genetic and epigenetic mechanisms may lead to the dysregulation of transcription factors and/or alterations in the DNA methylation pattern of the L1 promoter, respectively, thereby inducing aberrant L1 expression in cancer.
Proteolytic processing of L1 in neurons and cancer cells
Previous investigations have revealed that L1 is proteolytically cleaved both in cultured cells and neuronal tissues in vivo (Fig. 1). The cleavage of full-length L1 (L1–220) occurs in the third FNIII-like domain and gives rise to an N-terminal ectodomain fragment of 150–180 kDa (L1–150) and a membrane-retained fragment of 80–90 kDa (L1–85) [113–118]. Subsequent studies have shown that this cleavage is most likely mediated by the serine protease proprotein convertase PC5A (Fig. 1b) during protein biosynthesis and intracellular transport [119]. Interestingly, the N-terminal L1–150 fragment is not released and remains in a complex with uncleaved, membrane-bound L1 [119]. Another serine protease, plasmin, has also been shown to cleave L1 at a dibasic motif in the third FNIII-like domain (Fig. 1b), which led to the release of soluble L1 from the cells of a variety of cultured cell lines, as well as the disruption of cell–cell adhesion mediated by homophilic L1 interactions [120].
More recently, the serine protease neuropsin, also known as kallikrein 8, has been shown to cleave L1 enriched in synaptosomal fractions from brain tissue. Neuropsin-mediated cleavage takes place at a site near the membrane which results in an N-terminal fragment of about 180 kDa comprising the extracellular part of L1 (Fig. 1c) [121]. Proteolytic processing of L1 occurs in an activity-dependent manner and promotes the removal of L1 from so-called orphan boutons, an immature form of presynaptic terminals. Importantly, cleavage of L1 has been observed to be markedly decreased in neuropsin-deficient mice, suggesting that L1 indeed represents a specific substrate for neuropsin. As a consequence of neuropsin-mediated cleavage of L1, orphan boutons devoid of surface L1 exhibit altered morphology and form functional synapses [122]. Together, these studies indicate that proteolytic processing of L1 is facilitated by different serine proteases and is of functional significance for both cell–cell adhesion in cultured cells as well as activity-dependent synapse formation in the nervous system.
While earlier work had already indicated that a larger soluble L1-fragment of 180–200 kDa could be isolated from the supernatant of cultured PC12 rat phaeochromocytoma cells, sympathetic neurons and mouse cerebellar cells [118, 123], more recent studies have revealed that a large soluble form of L1 is secreted by cultured tumour cell lines. This release occurs spontaneously but is strongly enhanced by phorbol ester and pervanadate treatment [72, 124, 125]. It turned out that soluble L1 in the supernatant comprises the whole ectodomain that was cleaved off the membrane by the activity of metalloprotease(s) (Fig. 1d, e) [119, 124, 126]. The metalloproteases responsible for L1 cleavage were identified as ADAM10 [127] and also, under certain conditions, ADAM17 [82, 128]. Thus, different types of serine proteases and metalloproteases have the capacity to generate large soluble L1 ectodomains.
Soluble L1, when immobilized as a substrate, supports cell adhesion and the migration of tumour cells, lymphocytes, and cultured neurons [129], which also display enhance neurite growth in response to this substrate [130, 131]. In addition, there is evidence that soluble L1 generated in vivo can be bound by the extracellular matrix and could become immobilized as a putative substrate for migrating cells [126]. However, whether L1 cleavage serves only to render the extracellular fragments a permissive substrate remains to be investigated.
A novel functional link was born out of the observation that metalloprotease-cleaved L1 triggers cell migration by autocrine binding to integrins (Fig. 2c) [127]. This effect requires the Arg-Gly-Asp integrin binding site in the sixth domain of L1 and is blocked by metalloprotease inhibitors that prevent L1 shedding [127]. In the light of these findings it has become conceivable that expression of L1 also augments cell migration in various cell lines cultured on extracellular matrix proteins such as fibronectin, laminin and vitronectin [18, 132]. However, an alternative explanation for the enhanced cell migration by L1 was put forward by Panicker et al. [133]. The authors did not study L1 cleavage but noted that in the presence of L1 the cell surface recycling of β1-integrin is much faster and thereby can promote cell migration. Also, in this setting the Arg-Gly-Asp integrin binding site in the sixth domain of L1 is required [133].
L1 interaction and processing. L1 is schematically shown in a horseshoe-shaped conformation in which Ig-like domains one and two fold back to interact with the Ig-like domains three and four. a, b Full-length L1 (L1–220) can bind in cis mode (a) or in trans mode (b) to itself (homophilic) or to integrin binding partners (heterophilic) on the same cells (a) or neighbouring cells (b). c L1 can be cleaved from the cell surface by ectodomain shedding involving ADAM metalloproteases. Soluble L1 of about 200 kDa (L1–200) can bind back to integrins in an autocrine (same cell) or paracrine (another cell) fashion, thereby promoting cell migration, metastasis and angiogenesis. d Constitutive sequential L1 cleavage. Following ectodomain shedding by ADAM, the membrane-retained stub of about 32 kDa (L1–32) can be further cleaved by γ-secretase resulting in the release of a soluble L1-intracellular domain of about 28 kDa (L1–28) into the cytoplasm. The L1–28 fragment undergoes nuclear translocation and is involved in gene regulation
Proteolytic processing of L1 by metalloproteases is also of functional significance for neuronal cells. ADAM10 is able to cleave L1 in mouse brain and this process regulates L1-dependent haptotactic migration of cerebellar neurons in vitro [128]. Both ADAM10 and ADAM17 have been found to be involved in neurite outgrowth of cerebellar neurons [128]. Interestingly, abnormally elevated levels of the soluble L1 ectodomain has also been found in the cerebrospinal fluid of patients with Alzheimer’s disease and other dementia syndromes [134]. On the other hand, soluble L1 comprising the whole ectodomain is present in the nanogram range in the serum and ascites of patients with ovarian or endometrial carcinoma [19]. Additional analysis showed that the tumours indeed express ADAM10. These results suggest that cleavage of L1 from the membrane is associated not only with cancer but also with neurodegenerative diseases. However, while detection of soluble L1 in cancerous diseases may represent a valuable prognostic tool [19], the mechanisms leading to elevated levels of soluble L1 in neurodegenerative diseases are completely unclear. Future studies are required to test whether altered activity or expression levels of serine proteases and/or metalloproteases may account for this observation.
There are also many open questions about the mechanisms that regulate L1 cleavage in cancer. For example, the cellular compartment in which ADAM10-mediated cleavage of L1 is accomplished has not yet been identified. Studies have revealed that L1 cleavage mediated by metalloproteases takes place not only at the plasma membrane, but also in released microvesicles, called exosomes, which are derived from late endosomal compartments [82, 111, 135]. Recent work has shown that L1 cleavage fragments are present in late endosomal membranes as well as in lipid raft-like membrane compartments [136]. More detailed analyses of the intracellular trafficking of L1 and metalloproteases with respect to the cellular context and experimental conditions are required to resolve this question.
L1 as a regulator of gene transcription
Following cleavage of L1 by ADAM10 and ADAM17 a 32-kDa stub is retained in the membrane and can be used to track the cleavage process (Fig. 2c) [19, 125]. What happens to the L1–32 membrane-retained stub that is left behind after ADAM ectodomain shedding? Maretzky et al. [128] were the first to demonstrate that L1–32 undergoes further cleavage by the γ-secretase complex to release a soluble L1 intracellular domain (L1-ICD, Fig. 2d). The γ-secretase complex forms a membrane protease that processes a wide variety of integral membrane proteins and clears protein stubs from the lipid bilayer [137]. The process of regulated intramembrane proteolysis is an essential step in a variety of signalling pathways [138]. Nuclear translocation and transcriptional regulation of proteins such as Notch, CD44 and the amyloid precursor protein have been shown to depend on ADAM-mediated cleavage followed by γ-secretase activity [138]. A recent study has demonstrated that EpCAM, an adhesion molecule highly overexpressed in epithelial tumours, signals to the nucleus in the same fashion [139]. For L1, several investigations have shown that expression in tumours causes a variety of effects including altered gene expression (Table 2). It has been shown that the L1-ICD is important for gene regulation as point mutations in this part abolished all its effects [76]. Using an antibody to the C-terminal part of L1, it was recently been found that L1-ICD is present in the nucleus and that the processing of L1 by ADAM10 and γ-secretase is necessary for nuclear translocation and L1-dependent gene regulation (Fig. 2d) [136]. Thus, L1-mediated nuclear signalling by regulated intramembrane proteolysis is reminiscent of the mode of signalling mediated by other cell surface receptors. As cleavage by ADAM10 is a prerequisite for γ-secretase activity and downstream signalling, the production of soluble L1 is not arbitrary and can be viewed as a sign of active L1 signalling. A remaining question deals with how the cleaved L1-ICD reaches the nucleus and how it engages in the regulation of target gene transcription. It should be noted that the genes regulated by L1 in tumour cell lines appear to differ and depend on the cellular background examined. This suggests that L1 does not act on its own, but instead acts in concert with transcription regulating factors. It remains to be investigated whether these pathways identified in tumours are also instrumental in the proper functioning of the developing and adult nervous system.
Conclusions and perspectives
Research over the last 25 years on the role of L1 in the nervous system and tumours has been a cross-border process. The discovery of L1 in 1984 was followed by substantial progress in the understanding of various neuronal functions of this molecule and was accompanied by parallel investigations into the role of aberrant L1 expression in cancer cells. Tumour biologists were prompted to communicate with neurobiologists in order to exchange reagents and ideas, and it soon became clear that hallmarks of nervous system development such as cell proliferation, adhesion and migration involve signalling processes that are also critical in the development of carcinomas. Nevertheless, there is still an important gap to fill in terms of the mechanisms involved in normal L1 function as well as its dysfunction in pathophysiological processes underlying neurological and cancerous disorders. Investigating the context-dependent genetic and epigenetic mechanisms that regulate expression levels and function of L1 will not only increase our understanding of L1 function and dysfunction in the developing nervous system, but will also represent a key step toward in the development of therapeutic approaches to the treatment of human cancers.
References
Rathjen FG, Schachner M (1984) Immunocytological and biochemical characterization of a new neuronal cell surface component (L1 antigen) which is involved in cell adhesion. EMBO J 3:1–10
Maness PF, Schachner M (2007) Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci 10:19–26
Fransen E, Van Camp G, Vits L, Willems PJ (1997) L1-associated diseases: clinical geneticists divide, molecular geneticists unite. Hum Mol Genet 6:1625–1632
Kanemura Y, Okamoto N, Sakamoto H, Shofuda T, Kamiguchi H, Yamasaki M (2006) Molecular mechanisms and neuroimaging criteria for severe L1 syndrome with X-linked hydrocephalus. J Neurosurg Pediatr 105:403–412
Weller S, Gärtner J (2001) Genetic and clinical aspects of X-linked hydrocephalus (L1 disease): mutations in the L1CAM gene. Hum Mutat 18:1–12
Jouet M, Moncla A, Paterson J, McKeown C, Fryer A, Carpenter N, Holmberg E, Wadelius C, Kenwrick S (1995) New domains of neural cell-adhesion molecule L1 implicated in X-linked hydrocephalus and MASA syndrome. Am J Hum Genet 56:1304–1314
Cohen NR, Taylor JSH, Scott LB, Guillery RW, Soriano P, Furley AJW (1998) Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr Biol 8:26–33
Demyanenko GP, Tsai AY, Maness PF (1999) Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. J Neurosci 19:4907–4920
Dahme M, Bartsch U, Martini R, Anliker B, Schachner M, Mantei N (1997) Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat Genet 17:346–349
Schmid RS, Maness PF (2008) L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr Opin Neurobiol 18:245–250
Brümmendorf T, Kenwrick S, Rathjen FG (1998) Neural cell recognition molecule L1: from cell biology to human hereditary brain malformations. Curr Opin Neurobiol 8:87–97
Gil OD, Sakurai T, Bradley AE, Fink MY, Cassella MR, Kuo JA, Felsenfeld DP (2003) Ankyrin binding mediates L1CAM interactions with static components of the cytoskeleton and inhibits retrograde movement of L1CAM on the cell surface. J Cell Biol 162:719–730
Bechara A, Nawabi H, Moret F, Yaron A, Weaver E, Bozon M, Abouzid K, Guan J-L, Tessier-Lavigne M, Lemmon V, Castellani V (2008) FAK-MAPK-dependent adhesion disassembly downstream of L1 contributes to semaphorin3A-induced collapse. EMBO J 27:1549–1562
Kamiguchi H, Long KE, Pendergast M, Schaefer AW, Rapoport I, Kirchhausen T, Lemmon V (1998) The neural cell adhesion molecule L1 interacts with the AP-2 adaptor and is endocytosed via the clathrin-mediated pathway. J Neurosci 18:5311–5321
Schaefer AW, Kamei Y, Kamiguchi H, Wong EV, Rapoport I, Kirchhausen T, Beach CM, Landreth G, Lemmon SK, Lemmon V (2002) L1 endocytosis is controlled by a phosphorylation-dephosphorylation cycle stimulated by outside-in signaling by L1. J Cell Biol 157:1223–1232
Nakata A, Kamiguchi H (2007) Serine phosphorylation by casein kinase II controls endocytic L1 trafficking and axon growth. J Neurosci Res 85:723–734
Schaefer AW, Kamiguchi H, Wong EV, Beach CM, Landreth G, Lemmon V (1999) Activation of the MAPK signal cascade by the neural cell adhesion molecule L1 requires L1 internalization. J Biol Chem 274:37965–37973
Thelen K, Kedar V, Panicker AK, Schmid RS, Midkiff BR, Maness PF (2002) The neural cell adhesion molecule L1 potentiates integrin-dependent cell migration to extracellular matrix proteins. J Neurosci 22:4918–4931
Fogel M, Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Smirnov A, Edler L, Ben-Arie A, Huszar M, Altevogt P (2003) L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet 362:869–875
Gavert N, Conacci-Sorrell M, Gast D, Schneider A, Altevogt P, Brabletz T, Ben-Ze’ev A (2005) L1, a novel target of {beta}-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J Cell Biol 168:633–642
Muerkoster S, Werbing V, Sipos B, Debus MA, Witt M, Grobmann M, Leisner D, Kotteritzsch J, Kappes H, Kloppel G, Altevogt P, Folsch UR, Schafer H (2006) Drug-induced expression of the cellular adhesion molecule L1CAM confers anti-apoptotic protection and chemoresistance in pancreatic ductal adenocarcinoma cells. Oncogene 26:2759–2768
Arlt MJE, Novak-Hofer I, Gast D, Gschwend V, Moldenhauer G, Grunberg J, Honer M, Schubiger PA, Altevogt P, Kruger A (2006) Efficient inhibition of intra-peritoneal tumor growth and dissemination of human ovarian carcinoma cells in nude mice by anti-L1-cell adhesion molecule monoclonal antibody treatment. Cancer Res 66:936–943
Gast D, Riedle S, Issa Y, Pfeifer M, Beckhove P, Sanderson MP, Arlt M, Moldenhauer G, Fogel M, Kruger A, Altevogt P (2007) The cytoplasmic part of L1-CAM controls growth and gene expression in human tumors that is reversed by therapeutic antibodies. Oncogene 27:1281–1289
Knogler K, Grünberg J, Zimmermann K, Cohrs S, Honer M, Ametamey S, Altevogt P, Fogel M, Schubiger PA, Novak-Hofer I (2007) Copper-67 radioimmunotherapy and growth inhibition by anti-L1-cell adhesion molecule monoclonal antibodies in a therapy model of ovarian cancer metastasis. Clin Cancer Res 13:603–611
Haspel J, Grumet M (2003) The L1CAM extracellular region: a multi-domain protein with modular and cooperative binding modes. Front Biosci 8:1210–1225
Herron L, Hill M, Davey F, Gunn-Moore F (2009) The intracellular interactions of the L1 family of cell adhesion molecules. Biochem J 1:519–531
Raveh S, Gavert N, Ben-Ze’ev A (2009) L1 cell adhesion molecule (L1CAM) in invasive tumors. Cancer Lett 282:137–145
Gu S-M, Orth U, Zankl M, Schröder J, Gal A (1997) Molecular analysis of the L1CAM gene in patients with X-linked hydrocephalus demonstrates eight novel mutations and suggests non-allelic heterogeneity of the trait. Am J Med Genet 71:336–340
Kenwrick S, Watkins A, Angelis ED (2000) Neural cell recognition molecule L1: relating biological complexity to human disease mutations. Hum Mol Genet 9:879–886
Kaepernick L, Legius E, Higgins J, Kapur S (1994) Clinical aspects of the MASA syndrome in a large family, including expressing females. Clin Genet 45:181–185
Dobson CB, Villagra F, Clowry GJ, Smith M, Kenwrick S, Donnai D, Miller S, Eyre JA (2001) Abnormal corticospinal function but normal axonal guidance in human L1CAM mutations. Brain 124:2393–2406
Vos YJ, Hofstra RMW (2010) An updated and upgraded L1CAM mutation database. Hum Mutat 31:E1102–E1109
Yamasaki M, Thompson P, Lemmon V (1997) CRASH syndrome: mutations in L1CAM correlate with severity of the disease. Neuropediatrics 28:175–178
Wong EV, Cheng G, Ross Payne H, Lemmon V (1995) The cytoplasmic domain of the cell adhesion molecule L1 is not required for homophilic adhesion. Neurosci Lett 200:155–158
Nagaraj K, Kristiansen LV, Skrzynski A, Castiella C, Garcia-Alonso L, Hortsch M (2009) Pathogenic human L1-CAM mutations reduce the adhesion-dependent activation of EGFR. Hum Mol Genet 18:3822–3831
Godenschwege TA, Kristiansen LV, Uthaman SB, Hortsch M, Murphey RK (2006) A conserved role for Drosophila neuroglian and human L1-CAM in central-synapse formation. Curr Biol 16:12–23
De Angelis E, Watkins A, Schafer M, Brummendorf T, Kenwrick S (2002) Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum Mol Genet 11:1–12
Itoh K, Cheng L, Kamei Y, Fushiki S, Kamiguchi H, Gutwein P, Stoeck A, Arnold B, Altevogt P, Lemmon V (2004) Brain development in mice lacking L1-L1 homophilic adhesion. J Cell Biol 165:145–154
Tapanes-Castillo A, Weaver E, Smith R, Kamei Y, Caspary T, Hamilton-Nelson K, Slifer S, Martin E, Bixby J, Lemmon V (2010) A modifier locus on chromosome 5 contributes to L1 cell adhesion molecule X-linked hydrocephalus in mice. Neurogenetics 11:53–71
Wang S, Fenglei H, Wei X, Shuping G, Hongbin L, Tao Z, Xueyan Y, Yiping C (2007) Polycomblike-2-deficient mice exhibit normal left-right asymmetry. Dev Dyn 236:853–861
Charness ME, Safran RM, Perides G (1994) Ethanol inhibits neural cell-cell adhesion. J Biol Chem 269:9304–9309
Ramanathan R, Wilkemeyer MF, Mittal B, Perides G, Charness ME (1996) Alcohol inhibits cell-cell adhesion mediated by human L1. J Cell Biol 133:381–390
Yeaney NK, He M, Tang N, Malouf AT, O’Riordan MA, Lemmon V, Bearer FC (2009) Ethanol inhibits L1 cell adhesion molecule tyrosine phosphorylation and dephosphorylation and activation of pp60(src). J Neurochem 110:779–790
Tang N, He M, O’Riordan MA, Farkas C, Buck K, Lemmon V, Bearer CF (2006) Ethanol inhibits L1 cell adhesion molecule activation of mitogen-activated protein kinases. J Neurochem 96:1480–1490
Bearer CF, Swick AR, O′Riordan MA, Cheng G (1999) Ethanol inhibits L1-mediated neurite outgrowth in postnatal rat cerebellar granule cells. J Biol Chem 274:13264–13270
Chen SY, Wilkemeyer MF, Sulik KK, Charness ME (2001) Octanol antagonism of ethanol teratogenesis. FASEB J 15:1649–1651
Wilkemeyer MF, Chen SY, Menkari CE, Brenneman DE, Sulik KK, Charness ME (2003) Differential effects of ethanol antagonism and neuroprotection in peptide fragment NAPVSIPQ prevention of ethanol-induced developmental toxicity. Proc Natl Acad Sci U S A 100:8543–8548
Vallejo Y, Hortsch M, Dubreuil RR (1997) Ethanol does not inhibit the adhesive activity of Drosophila neuroglian or human L1 in Drosophila S2 tissue culture cells. J Biol Chem 272:12244–12247
Hoffman EJ, Mintz CD, Wang S, McNickle DG, Salton SRJ, Benson DL (2008) Effects of ethanol on axon outgrowth and branching in developing rat cortical neurons. Neuroscience 157:556–565
Arevalo E, Shanmugasundararaj S, Wilkemeyer MF, Dou X, Chen S, Charness ME, Miller KW (2008) An alcohol binding site on the neural cell adhesion molecule L1. Proc Natl Acad Sci U S A 105:371–375
Schurmann G, Haspel J, Grumet M, Erickson HP (2001) Cell adhesion molecule L1 in folded (horseshoe) and extended conformations. Mol Biol Cell 12:1765–1773
Freigang J, Proba K, Leder L, Diederichs K, Sonderegger P, Welte W (2000) The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion. Cell 101:425–433
Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, Schmucker D (2007) Structural basis of Dscam isoform specificity. Nature 449:487–491
Gouveia RM, Gomes CM, Sousa M, Alves PM, Costa J (2008) Kinetic analysis of L1 homophilic interaction: role of the first four immunoglobulin domains and implications on binding mechanism. J Biol Chem 283:28038–28047
Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X, Griseri P, Brooks AS, Antinolo G, de Pontual L, Clement-Ziza M, Munnich A, Kashuk C, West K, Wong KKY, Lyonnet S, Chakravarti A, Tam PKH, Ceccherini I, Hofstra RMW, Fernandez R; Hirschsprung Disease Consortium (2008) Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet 45:1–14
Heanue TA, Pachnis V (2007) Enteric nervous system development and Hirschsprung’s disease: advances in genetic and stem cell studies. Nat Rev Neurosci 8:466–479
Garcia-Barcelo MM, Tang CS, Ngan ES, Lui VC, Chen Y, So MT, Leon TY, Miao XP, Shum CKY, Liu FQ, Yeung MY, Yuan ZW, Guo WH, Liu L, Sun XB, Huang LM, Tou JF, Song YQ, Chan D, Cheung KM, Wong KKY, Cherny SS, Sham PC, Tam PKH (2009) Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung’s disease. Proc Natl Acad Sci U S A 106:2694–2699
Garcia-Barcelo MM, Fong PY, Tang CS, Miao XP, So MT, Yuan ZW, Li L, Guo WH, Liu L, Wang B, Sun XB, Huang LM, Tou JF, Wong KK, Ngan ES, Lui VC, Cherny SS, Sham PC, Tam PK (2008) Mapping of a Hirschsprung’s disease locus in 3p21. Eur J Hum Genet 16:833–840
Ikawa H, Kawano H, Takeda Y, Masuyama H, Watanabe K, Endo M, Yokoyama J, Kitajima M, Uyemura K, Kawamura K (1997) Impaired expression of neural cell adhesion molecule L1 in the extrinsic nerve fibers in Hirschsprung’s disease. J Pediatr Surg 32:542–545
Parisi MA, Kapur RP, Neilson I, Hofstra RMW, Holloway LW, Michaelis RC, Leppig KA (2002) Hydrocephalus and intestinal aganglionosis: is L1CAM a modifier gene in Hirschsprung disease? Am J Med Genet 108:51–56
Basel-Vanagaite L, Straussberg R, Friez MJ, Inbar D, Korenreich L, Shohat M, Schwartz CE (2006) Expanding the phenotypic spectrum of L1CAM-associated disease. Clin Genet 69:414–419
Griseri P, Vos Y, Giorda R, Gimelli S, Beri S, Santamaria G, Mognato G, Hofstra RMW, Gimelli G, Ceccherini I (2008) Complex pathogenesis of Hirschsprung’s disease in a patient with hydrocephalus, vesico-ureteral reflux and a balanced translocation t(3;17)(p12;q11). Eur J Hum Genet 17:483–490
Okamoto N, Wada Y, Goto M (1997) Hydrocephalus and Hirschsprung’s disease in a patient with a mutation of L1CAM. J Med Genet 34:670–671
Nakakimura S, Sasaki F, Okada T, Arisue A, Cho K, Yoshino M, Kanemura Y, Yamasaki M, Todo S (2008) Hirschsprung’s disease, acrocallosal syndrome, and congenital hydrocephalus: report of 2 patients and literature review. J Pediatr Surg 43:e13–e17
Turner KN, Schachner M, Anderson RB (2009) Cell adhesion molecule L1 affects the rate of differentiation of enteric neurons in the developing gut. Dev Dyn 238:708–715
Anderson RB, Turner KN, Nikonenko AG, Hemperly J, Schachner M, Young HM (2006) The cell adhesion molecule L1 Is required for chain migration of neural crest cells in the developing mouse gut. Gastroenterology 130:1221–1232
Liebau M, Gal A, Superti-Furga A, Omran H, Pohl M (2007) L1CAM mutation in a boy with hydrocephalus and duplex kidneys. Pediatr Nephrol 22:1058–1061
Debiec H, Kutsche M, Schachner M, Ronco P (2002) Abnormal renal phenotype in L1 knockout mice: a novel cause of CAKUT. Nephrol Dial Transplant 17:42–44
Allory Y, Matsuoka Y, Bazille C, Christensen EI, Ronco P, Debiec H (2005) The L1 cell adhesion molecule is induced in renal cancer cells and correlates with metastasis in clear cell carcinomas. Clin Cancer Res 11:1190–1197
Debiec H, Christensen EI, Ronco PM (1998) The cell adhesion molecule L1 Is developmentally regulated in the renal epithelium and is involved in kidney branching morphogenesis. J Cell Biol 143:2067–2079
Knops NBB, Bos KK, Kerstjens M, van Dael K, Vos YJ (2008) Nephrogenic diabetes insipidus in a patient with L1 syndrome: a new report of a contiguous gene deletion syndrome including L1CAM and AVPR2. Am J Med Genet A 146A:1853–1858
Hubbe M, Kowitz A, Schirrmacher V, Schachner M, Altevogt P (1993) L1 adhesion molecule on mouse leukocytes: regulation and involvement in endothelial cell binding. Eur J Immunol 23:2927–2931
Zecchini S, Bianchi M, Colombo N, Fasani R, Goisis G, Casadio C, Viale G, Liu J, Herlyn M, Godwin AK, Nuciforo PG, Cavallaro U (2008) The differential role of L1 in ovarian carcinoma and normal ovarian surface epithelium. Cancer Res 68:1110–1118
Kaifi JT, Reichelt U, Quaas A, Schurr PG, Wachowiak R, Yekebas EF, Strate T, Schneider C, Pantel K, Schachner M, Sauter G, Izbicki JR (2007) L1 is associated with micrometastatic spread and poor outcome in colorectal cancer. Mod Pathol 20:1183–1190
Boo YJ, Park JM, Kim J, Chae YS, Min BW, Um JW, Moon HY (2007) L1 expression as a marker for poor prognosis, tumor progression, and short survival in patients with colorectal cancer. Ann Surg Oncol 14:1703–1711
Gast D, Riedle S, Kiefel H, Müerköster SS, Schäfer H, Schäfer MKE, Altevogt P (2008) The RGD integrin binding site in human L1-CAM is important for nuclear signaling. Exp Cell Res 314:2411–2418
Müerköster SS, Kötteritzsch J, Geismann C, Gast D, Kruse M-L, Altevogt P, Fölsch UR, Schäfer H (2009) α5-integrin is crucial for L1CAM-mediated chemoresistance in pancreatic adenocarcinoma. Int J Oncol 34:243–253
He Z, Tessier-Lavigne M (1997) Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell 90:739–751
Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD (1997) Neuropilin is a semaphorin III receptor. Cell 90:753–762
Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M (1998) Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 92:735–745
Castellani V, De Angelis E, Kenwrick S, Rougon G (2002) Cis and trans interactions of L1 with neuropilin-1 control axonal responses to semaphorin 3A. EMBO J 21:6348–6357
Stoeck A, Schlich S, Issa Y, Gschwend V, Wenger T, Herr I, Marmé A, Bourbie S, Altevogt P, Gutwein P (2006) L1 on ovarian carcinoma cells is a binding partner for Neuropilin-1 on mesothelial cells. Cancer Lett 239:212–226
Castellani V, Chedotal A, Schachner M, Faivre-Sarrailh C, Rougon G (2000) Analysis of the L1-deficient mouse phenotype reveals cross-talk between Sema3A and L1 signaling pathways in axonal guidance. Neuron 27:237–249
Issa Y, Nummer D, Seibel T, Müerköster S, Koch M, Schmitz-Winnenthal F-H, Galindo L, Weitz J, Beckhove P, Altevogt P (2009) Enhanced L1CAM expression on pancreatic tumor endothelium mediates selective tumor cell transmigration. J Mol Med 87:99–112
Voura EB, Ramjeesingh RA, Montgomery AMP, Siu CH (2001) Involvement of integrin alpha vbeta 3 and cell adhesion molecule L1 in transendothelial migration of melanoma cells. Mol Biol Cell 12:2699–2710
Hall H, Hubbell JA (2004) Matrix-bound sixth Ig-like domain of cell adhesion molecule L1 acts as an angiogenic factor by ligating [alpha]v[beta]3-integrin and activating VEGF-R2. Microvasc Res 68:169–178
Robinson SD, Reynolds LE, Kostourou V, Reynolds AR, Graca da Silva R, Tavora B, Baker M, Marshall JF, Hodivala-Dilke KM (2009) Alphavbeta3-integrin limits the contribution of neuropilin-1 to VEGF-induced angiogenesis. J Biol Chem 284:33966–33981
Friedli A, Fischer E, Novak-Hofer I, Cohrs S, Ballmer-Hofer K, Schubiger PA, Schibli R, Grünberg J (2009) The soluble form of the cancer-associated L1 cell adhesion molecule is a pro-angiogenic factor. Int J Biochem Cell Biol 41:1572–1580
Kallunki P, Edelman GM, Jones FS (1997) Tissue-specific expression of the L1 cell adhesion molecule is modulated by the neural restrictive silencer element. J Cell Biol 138:1343–1354
De Angelis E, Brümmendorf T, Cheng L, Lemmon V, Kenwrick S (2001) Alternative use of a mini exon of the L1 gene affects L1 binding to neural ligands. J Biol Chem 276:32738–32742
Jacob J, Haspel J, Kane-Goldsmith N, Grumet M (2002) L1 mediated homophilic binding and neurite outgrowth are modulated by alternative splicing of exon 2. J Neurobiol 51:177–189
Takeda Y, Hiroaki A, Yoshinori M, Masayuki M, Masaaki K, Keiichi U (1996) A nonneuronal isoform of cell adhesion molecule L1: tissue-specific expression and functional analysis. J Neurochem 66:2338–2349
Miura M, Asou H, Kobayashi M, Uyemura K (1992) Functional expression of a full-length cDNA coding for rat neural cell adhesion molecule L1 mediates homophilic intercellular adhesion and migration of cerebellar neurons. J Biol Chem 267:10752–10758
Itoh K, Sakurai Y, Asou H, Umeda M (2000) Differential expression of alternatively spliced neural cell adhesion molecule L1 isoforms during oligodendrocyte maturation. J Neurosci Res 60:579–586
Palm K, Belluardo N, Metsis M, Timmusk To (1998) Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci 18:1280–1296
Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, Grooms SY, Regis R, Bennett MVL, Zukin RS (2003) Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci 23:2112–2121
Miyake K, Yamamoto W, Tadokoro M, Takagi N, Sasakawa K, Nitta A, Furukawa S, Takeo S (2002) Alterations in hippocampal GAP-43, BDNF, and L1 following sustained cerebral ischemia. Brain Res 935:24–31
Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Göttgens B, Buckley NJ (2004) Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A 101:10458–10463
Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, MacDonald M, Fossale E, Zeitlin S, Buckley N, Cattaneo E (2007) Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J Neurosci 27:6972–6983
Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, Rigamonti D, Cattaneo E (2003) Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet 35:76–83
Lepagnol-Bestel AM, Zvara A, Maussion G, Quignon F, Ngimbous B, Ramoz N, Imbeaud S, Loe-Mie Y, Benihoud K, Agier N, Salin PA, Cardona A, Khung-Savatovsky S, Kallunki P, Delabar JM, Puskas LG, Delacroix H, Aggerbeck L, Delezoide AL, Delattre O, Gorwood P, Moalic JM, Simonneau M (2009) DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum Mol Genet 18:1405–1414
Canzonetta C, Mulligan C, Deutsch S, Ruf S, O’Doherty A, Lyle R, Borel C, Lin-Marq N, Delom F, Groet J, Schnappauf F, De Vita S, Averill S, Priestley JV, Martin JE, Shipley J, Denyer G, Epstein CJ, Fillat C, Estivill X, Tybulewicz VLJ, Fisher EMC, Antonarakis SE, Nizetic D (2008) DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am J Hum Genet 83:388–400
Park J, Song WJ, Chung K (2009) Function and regulation of Dyrk1A: towards understanding Down syndrome. Cell Mol Life Sci 66:3235–3240
Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, DePinho RA, Chin L, Elledge SJ (2005) A genetic screen for candidate tumor suppressors identifies REST. Cell 121:837–848
Reddy BY, Greco SJ, Patel PS, Trzaska KA, Rameshwar P (2009) RE-1-silencing transcription factor shows tumor-suppressor functions and negatively regulates the oncogenic TAC1 in breast cancer cells. Proc Natl Acad Sci U S A 106:4408–4413
Moss AC, Jacobson GM, Walker LE, Blake NW, Marshall E, Coulson JM (2009) SCG3 transcript in peripheral blood is a prognostic biomarker for REST-deficient small cell lung cancer. Clin Cancer Res 15:274–283
Shtutman M, Levina E, Ohouo P, Baig M, Roninson IB (2006) Cell adhesion molecule L1 disrupts E-cadherin-containing adherens junctions and increases scattering and motility of MCF7 breast carcinoma cells. Cancer Res 66:11370–11380
Geismann C, Morscheck M, Koch D, Bergmann F, Ungefroren H, Arlt A, Tsao M-S, Bachem MG, Altevogt P, Sipos B, Folsch UR, Schafer H, Muerkoster SS (2009) Up-regulation of L1CAM in pancreatic duct cells is transforming growth factor {beta}1- and slug-dependent: role in malignant transformation of pancreatic cancer. Cancer Res 69:4517–4526
Meli ML, François C, Robert W, Hanspeter A, Nigel C, Rolf J, Holger M, Schubiger PA, Ilse NH (1999) Anti-neuroblastoma antibody chCE7 binds to an isoform of L1-CAM present in renal carcinoma cells. Int J Cancer 83:401–408
Izumoto S, Ohnishi T, Arita N, Hiraga S, Taki T, Hayakawa T (1996) Gene expression of neural cell adhesion molecule L1 in malignant gliomas and biological significance of L1 in glioma invasion. Cancer Res 56:1440–1444
Gast D, Svenja R, Svenja R, Heidi S, Sabine S, Annette S, Yasmin I, Alexander S, Mina F, Safwan J, Till W, Ingrid H, Paul G, Peter A (2005) L1 augments cell migration and tumor growth but not beta3 integrin expression in ovarian carcinomas. Int J Cancer 115:658–665
Kato K, Maesawa C, Itabashi T, Fujisawa K, Otsuka K, Kanno S, Tada H, Tatemichi Y, Kotani K, Oikawa H, Sugai T, Wakabayashi G, Masuda T (2009) DNA hypomethylation at the CpG island is involved in aberrant expression of the L1 cell adhesion molecule gene in colorectal cancer. Int J Oncol 35:467–476
Faissner A, Teplow DB, Kubler D, Keilhauer G, Kinzel V, Schachner M (1985) Biosynthesis and membrane topography of the neural cell-adhesion molecule L1. EMBO J 4:3105–3113
Wolff JM, Rathjen FG, Frank R, Roth S (1987) Biochemical characterization of polypeptide components involved in neurite fasciculation and elongation. Eur J Biochem 168:551–561
Burgoon MP, Hazan RB, Phillips GR, Crossin KL, Edelman GM, Cunningham BA (1995) Functional analysis of posttranslational cleavage products of the neuron-glia cell adhesion molecule, Ng-CAM. J Cell Biol 130:733–744
Kayyem JF, Roman JM, de la Rosa EJ, Schwarz U, Dreyer WJ (1992) Bravo/Nr-CAM is closely related to the cell adhesion molecules L1 and Ng-CAM and has a similar heterodimer structure. J Cell Biol 118:1259–1270
Nybroe O, Dalseg A-M, Bock E (1990) A developmental study of soluble L1. Int J Dev Neurosci 8:273–281
Sadoul K, Sadoul R, Faissner A, Schachner M (1988) Biochemical characterization of different molecular forms of the neural cell adhesion molecule L1. J Neurochem 50:510–521
Kalus I, Schnegelsberg B, Seidah NG, Kleene R, Schachner M (2003) The proprotein convertase PC5A and a metalloprotease are involved in the proteolytic processing of the neural adhesion molecule L1. J Biol Chem 278:10381–10388
Nayeem N, Silletti S, Yang X, Lemmon VP, Reisfeld RA, Stallcup WB, Montgomery AM (1999) A potential role for the plasmin(ogen) system in the posttranslational cleavage of the neural cell adhesion molecule L1. J Cell Sci 112:4739–4749
Matsumoto-Miyai K, Ninomiya A, Yamasaki H, Tamura H, Nakamura Y, Shiosaka S (2003) NMDA-dependent proteolysis of presynaptic adhesion molecule L1 in the hippocampus by neuropsin. J Neurosci 23:7727–7736
Nakamura Y, Tamura H, Horinouchi K, Shiosaka S (2006) Role of neuropsin in formation and maturation of Schaffer-collateral L1cam-immunoreactive synaptic boutons. J Cell Sci 119:1341–1349
Richter-Landsberg C, Lee VM, Salton SR, Shelanski ML, Greene LA (1984) Release of the NILE and other glycoproteins from cultured PC12 rat pheochromocytoma cells and sympathetic neurons. J Neurochem 43:841–848
Beer S, Oleszewski M, Gutwein P, Geiger C, Altevogt P (1999) Metalloproteinase-mediated release of the ectodomain of L1 adhesion molecule. J Cell Sci 112:2667–2675
Gutwein P, Oleszewski M, Mechtersheimer S, Agmon-Levin N, Krauss K, Altevogt P (2000) Role of Src kinases in the ADAM-mediated release of L1 adhesion molecule from human tumor cells. J Biol Chem 275:15490–15497
Montgomery AM, Becker JC, Siu CH, Lemmon VP, Cheresh DA, Pancook JD, Zhao X, Reisfeld RA (1996) Human neural cell adhesion molecule L1 and rat homologue NILE are ligands for integrin alpha v beta 3. J Cell Biol 132:475–485
Mechtersheimer S, Gutwein P, Agmon-Levin N, Stoeck A, Oleszewski M, Riedle S, Postina R, Fahrenholz F, Fogel M, Lemmon V, Altevogt P (2001) Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J Cell Biol 155:661–674
Maretzky T, Schulte M, Ludwig A, Rose-John S, Blobel C, Hartmann D, Altevogt P, Saftig P, Reiss K (2005) L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol Biol Cell 25:9040–9053
Duczmal A, Schöllhammer S, Katich S, Ebeling O, Schwartz-Albiez R, Altevogt P (1997) The L1 adhesion molecule supports [alpha]v[beta]3-mediated migration of human tumor cells and activated t lymphocytes. Biochem Biophys Res Commun 232:236–239
Lemmon V, Burden SM, Payne HR, Elmslie GJ, Hlavin ML (1992) Neurite growth on different substrates: permissive versus instructive influences and the role of adhesive strength. J Neurosci 12:818–826
Chang S, Rathjen FG, Raper JA (1990) Neurite outgrowth promoting activity of G4 and its inhibition by monoclonal antibodies. J Neurosci Res 25:180–186
Silletti S, Yebra M, Perez B, Cirulli V, McMahon M, Montgomery AMP (2004) Extracellular signal-regulated kinase (ERK)-dependent gene expression contributes to L1 cell adhesion molecule-dependent motility and invasion. J Biol Chem 279:28880–28888
Panicker AK, Buhusi M, Erickson A, Maness PF (2006) Endocytosis of [beta]1 integrins is an early event in migration promoted by the cell adhesion molecule L1. Exp Cell Res 312:299–307
Strekalova H, Buhmann C, Kleene R, Eggers C, Saffell J, Hemperly J, Weiller C, Muller-Thomsen T, Schachner M (2006) Elevated levels of neural recognition molecule L1 in the cerebrospinal fluid of patients with Alzheimer disease and other dementia syndromes. Neurobiol Aging 27:1–9
Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M, Altevogt P (2003) ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J 17:292–294
Riedle S, Kiefel H, Gast D, Bondong S, Wolterink S, Gutwein P, Altevogt P (2009) Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/gamma-secretase activity. Biochem J 420:391–402
Selkoe DJ, Wolfe MS (2007) Presenilin: running with scissors in the membrane. Cell 131:215–221
Landman N, Kim TW (2004) Got RIP?: Presenilin-dependent intramembrane proteolysis in growth factor receptor signaling. Cytokine Growth Factor Rev 15:337–351
Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O (2009) Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol 11:162–171
De Angelis E, MacFarlane J, Du JS, Yeo G, Hicks R, Rathjen FG, Kenwrick S, Brummendorf T (1999) Pathological missense mutations of neural cell adhesion molecule L1 affect hemophilic and heterophilic binding activities. EMBO J 18:4744–4753
Buhusi M, Schlatter MC, Demyanenko GP, Thresher R, Maness PF (2008) L1 Interaction with ankyrin regulates mediolateral topography in the retinocollicular projection. J Neurosci 28:177–188
Needham LK, Thelen K, Maness PF (2001) Cytoplasmic domain mutations of the L1 cell adhesion molecule reduce L1-ankyrin interactions. J Neurosci 21:1490–1500
Gavert N, Sheffer M, Raveh S, Spaderna S, Shtutman M, Brabletz T, Barany F, Paty P, Notterman D, Domany E, Ben-Ze’ev A (2007) Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res 67:7703–7712
Acknowledgment
The authors thank Dr. Sandra Dieni (Institute of Anatomy and Cell Biology, Freiburg, Germany) for helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
We dedicate this review to Melitta Schachner and co-workers for their outstanding contribution to the field of L1CAM research.
Rights and permissions
About this article
Cite this article
Schäfer, M.K.E., Altevogt, P. L1CAM malfunction in the nervous system and human carcinomas. Cell. Mol. Life Sci. 67, 2425–2437 (2010). https://doi.org/10.1007/s00018-010-0339-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0339-1