Abstract
Xanthopappus subacaulis (Asteraceae) is a medicinal plant endemic to the alpine meadows from the middle east region of the Qinghai-Tibet Plateau in China. In this study, we sequenced and analyzed its whole chloroplast (cp) genome using Next Generation Sequencing techniques, representing the first complete cp genome for the genus Xanthopappus. Our results show that the cp genome of X. subacaulis contains 153 297 base pairs (bp) with a relatively high A + T content (62.28%) and has a standard quadripartite structure, where the large (LSC, 84 142 bp) and small (SSC, 18 769 bp) single copy regions are separated by two copies of inverted repeats (IRs, 25 193 bp each). The cp genome encoded a total of 131 genes of which 87 are protein-coding genes, 36 are tRNA genes and eight are rRNA genes. The majority (85) of these genes occur within the LSC region. When comparing these results with the previously published cp genome of a related taxon from Asteraceae (genus Arctium), both species share similar number and type of genes, and A + T content. In addition, a Maximum Likelihood and Maximum Parsimony phylogenetic analyses based on 33 chloroplast genomes of Asteraceae suggested that X. subacaulis forms an independent and well supported clade paraphyletic to all the other members within tribes Carduinae, Carlininae and Centaureinae of Asteraceae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
The Qinghai-Tibet Plateau (QTP) is the largest, highest and youngest plateau in the world, covering 2.5 million km2 and an average elevation of approximately 4000 m (Zheng, 1996). The QTP and its adjacent regions comprise a major hotspot for global biodiversity, supporting numerous endemic species (Wu, 1988; Mittermeier et al., 2005). This wide variety of species has evolved in part due to the combination of rapid and extensive uplifts of the QTP and the climatic oscillations experienced in the region during the Pleistocene, which have promoted allopatric speciation (Wang et al., 2009a, 2009b; Jia et al., 2012). Xanthopappus subacaulis C. Winkl. (Asterales: Asteraceae) is an alpine-endemic monotypic species mainly found in dry hillsides, meadows and grasslands of northern and western Sichuan, southeast Gansu, northwest Yunnan, and most parts of Qinghai at the QTP in China, within an altitudinal range from 2400 to 4000 m (Liu and Shi, 1987; Liu, 1996). This species is adapted to resist the harsh environmental conditions from high mountain elevations such as drought, cold, barren and disease, and as reported in other lineages of Asteraceae such as Senecio, this tolerance could have been a key factor to promote its reproductive isolation and subsequent speciation (Chapman et al., 2016).
In China, the entire plant of X. subacaulis is used in the traditional herbal Tibetan medicine to treat a variety of conditions such as duodenal ulcers, food poisoning, gastric ulcer, hematemesis, idiopathic thrombocytopenic purpura, and uterine bleeding (Du, 2006). Previous research on the phytochemistry of this species has been limited and include a phytochemical report on the isolation of several thiophene acetylenes (Tian et al., 2006), which are aromatic compounds often found in the Asteraceae family (Bohlmann et al., 1973; Harborne, 1984). Thiophene acetylenes have been shown to have a broad diversity of pharmacological activities that involves antifungal, antibacterial (Young et al., 2006), antiviral (Rashid et al., 2001), phototoxic and insecticidal agents (Tian et al., 2006). A more recent study also reported thiophene acetylenes and furanosesquiterpenes from X. subacaulis and confirmed its antibacterial activities (Zhang et al., 2014).
In spite of its medicinal value, we still lack comprehensive genomic resources for X. subacaulis. The only sequences available to date for the genus are a handful of barcoding regions used in previous phylogenetic studies (Wang et al., 2007; Barres et al., 2013). Therefore, we lack studies characterizing the structure of the complete chloroplast (cp) genome of X. subacaulis, and subsequent phylogenetic analyses based on it.
The typical complete chloroplast genome in plants has separate transcription and transport systems, and a covalently closed circular structure of about 120–160 kilobases in length (Gray, 1989; Howe et al., 2003). Because the structure and sequence composition of cp genomes are highly conserved and have low evolution rates, they can provide a rich source of genetic information.
In fact, cp genomes are considered to be the most promising resource to obtain phylogenetically informative sites (e.g., barcoding regions) suitable for molecular systematics, especially to elucidate relationships among plant species (Odintsova and Yurina, 2006; Jansen et al., 2007). Up to May 31th, 2020, the number of cp genomes of green plants uploaded to NCBI was 4115 of which almost 3625 represent angiosperms. About half (57.5%) of these cp genomes comprise dicotyledons, and only 5.9% are from Asteraceae. As expected, the cp genome of a rare monotypic genus such as Xanthopappus has not been sequenced yet. Therefore, having available a complete and well-characterized cp genome for this taxon has theoretical and practical significances because will reveal its genomic structure, characteristics and functions, and can serve as base to explore functional molecular mechanisms, among others. In addition, the plastid sequences of Xanthopappus will be valuable to conduct phylogenetic analyses to help resolve its position within Asteraceae and to prepare future studies on the biogeography and evolution of the genus.
Thus, our objective here is to report the sequencing, assembly, and annotation of the complete cp genome of X. subacaulis using Next Generation Sequencing techniques. This resource will not only help to promote future studies on the genetic diversity on the species, but will also serve as an important reference to develop genetic markers to assess the genetic structure of populations and to design effective conservation plans to mitigate any overharvesting for its medicinal uses.
MATERIALS AND METHODS
Plant Material
We sampled fresh and healthy leaves from a single representative individual of X. subacaulis present at Xinghai County in Qinghai, China (35.88° N, 96.68° E; alt. 3856 m). The reference specimen collected (voucher no. X. Su 2019H7) was deposited at the Herbarium of the Northwest Plateau Institute of Biology (HNWP), Chinese Academy of Science, Xining, Qinghai Province, China.
DNA Extraction, Sequencing, Assembly and Annotation
First, we extracted total genomic DNA from silica gel-dried leaves using a modified 2× CTAB procedure (Doyle and Doyle, 1990). We sheared the total DNA to fragments between 400–500 bp using a Covaris M220 Focused-ultrasonicator (Covaris, Woburn, MA, USA). Then, we prepared the DNA library with a TruSeqTM DNA Sample Prep Kit using dual-indexing TruSeq HT, and performed a 150 bp paired-end sequencing of the library using an Illumina HiSeq 4000 platform (HiSeq 3000/4000 SBS Kits).
The Illumina sequencer produced 7 576 864 raw paired-end reads, which were quality-trimmed using Trimmomatic (version 0.33) (Bolger et al., 2014). From the clean reads, we assembled the complete cp genome using a de novo approach with Novoplasty software (Dierckxsens et al., 2017) using the complete cp genome sequence of Synurus deltoides (MN518847) as reference. The resultant circular plastome was annotated with GeSeq (Tillich et al., 2017) and AGORA (Jung et al., 2018) using again the cp genome of S. deltoides as reference. The results of both annotations were compared and visually verified in Geneious Prime (2019.1.3). Finally, the annotated circular cp genome of X. subacaulis was drawn with OGDRAW (Greiner et al., 2019) and submitted to GenBank under the accession number MT643189.
Phylogenetic Analysis
We estimated the phylogenetic position of X. subacaulis using a multiple sequence alignment of 33 complete cp genomes from species within tribe Cynareae (Asteraceae: subfamily Carduoideae) and using three genera from other tribes of Asteraceae as outgroups (Anaphalis sinica Hance [tribe Gnaphalieae], Aster tataricus L. f. [tribe Astereae], and Cichorium intybus L. [tribe Cichorieae]). All sequences were aligned with the MAFFT (Katoh and Standley, 2013) plugin implemented in Geneious (version 10.0.5). A Maximum-Parsimony (MP) phylogenetic tree was constructed with PAUP* (version 4.0b10) (Swofford, 2002), while a Maximum-Likelihood (ML) phylogenetic analysis was performed in RAxML (Stamatakis, 2014) under the GTR-GAMMA model with 1000 bootstrap replicates.
RESULTS AND DISCUSSION
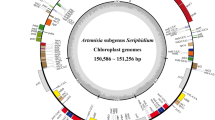
Our results of the assembled complete cp genome of X. subacaulis indicate a length of 153 297 bp (Fig. 1) with an overall A + T content of 62.28% (Table 1). Chloroplast genomes of other angiosperms have been reported with consistent higher A + T content and lower G + C content (Feng et al., 2020; Tian et al., 2020). This situation has been also observed in the cp genome of Arctium lappa L. (MH375874), a related genus also from tribe Cynareae (Xing et al., 2019). The overall structure of the cp genome is a standard quadripartite comprising a large (LSC, 84 142 bp) and a small (SSC, 18 769 bp) single copy regions separated by two inverted repeat regions (IRA and IRB, 25 193 bp each) (Fig. 1). The A + T content values in the LSC, SSC and IR regions were 64.16, 68.35 and 56.89%, respectively. The A + T content value in the IR region is lower than that both LSC and SSC region, which is mainly caused by rRNA genes with four high G + C content (Table 1).
Gene map of the chloroplast genome of Xanthopappus subacaulis (Asteraceae: tribe Cynareae). Gene colors indicate their functional groups, and the inner circle shows their locations within the LSC, SSC, IRA and IRB regions of the plastome. The dark-gray bars in the inner circle corresponds to G + C content, while the light-gray ones show the A + T content.
The whole cp genome encoded a total of 131 genes distributed into 87 protein-coding genes (PCGs), 36 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. Among them, the LSC region encoded 85 genes of which 64 are PCGs and 21 are tRNA genes. In SSC region, 12 PCGs and one tRNA genes were present. Similarly, there are 17 genes encoded in IR regions containing six PCGs, seven tRNA genes and four rRNA genes (Fig. 1).
Among the 131 encoded genes, 98 were found in the single copy region, whereas four protein-coding genes (ndhB, rpl2, rpl23, rps7), seven tRNA genes (trnA-UGC, trnE-UUC, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, trnV-GAC), four rRNA genes (rrn4.5S, rrn5S, rrn16S, rrn23S) and two unknown functional genes (ycf2, ycf15) were duplicated within the IRs (Fig. 1). In addition, the cp genome of X. subacaulis can be divided into four categories, depending on gene functions: (1) 66 genes related with expression; (2) 45 genes for photosynthesis; (3) four genes for open reading frame and other protein coding activities; and (4) five genes of unknown function (Table 2). Moreover, the genes related with expression can be further divided into six sub-categories such as tRNA genes, translation initiation factor, etc., while those for photosynthesis can be catalogued into seven sub-categories (Table 2).
The MP analysis identified 927 195 trees with 1162 steps, a consistency index (CI) of 0.96, and a retention index (RI) of 0.98. The strict MP consensus tree (Fig. 2) was generally congruent with the ML tree (–lnL = 4536.1006 for the best model, GTR + G + I) (Fig. 2). The resulting phylogenetic tree revealed that all species of Cynareae were clustered into a well-supported (BP = 100%) monophyletic clade comprising three subclades with a high bootstrap value (BP = 99–100%).
Consensus cladogram from the combined Maximum Likelihood and Maximum Parsimony trees indicating the inferred position of Xanthopappus subacaulis(Asteraceae).The ML and MP trees were constructed using 33 complete chloroplast genomes from tribe Cynareae, and rooted using three outgroups from other tribes within Asteraceae (Anaphalis, Aster, and Cichorium). Bootstrap support values from the ML and MP analyses are shown above branches (ML/MP). Stars (“★”) represents nodes with 100% bootstrap support, while dashes (“–”) represent nodes supported by bootstrap values of less than 80%.
Clade I consisted of 27 taxa from subtribes Carduinae (genera Arctium, Cynara, Cirsium, Saussurea and Silybum) and Centaureinae (genera Carthamus, Synurus and Centaurea); Clade II only contains the monotypic genus Xanthopappus; and Clade III comprise two species from genus Atractylodes. Interestingly, Clade II with its sole representative, X. subacaulis, is placed in a basal position to subtribes Carduninae and Centaureinae (i.e., Clade I), bringing a possible answer to the long-raised question on the identity of the sister lineage to these subtribes (Fig. 2) (Häffner and Hellwig, 1999).
The phylogenetic position of the genus Xanthopappus has important repercussions as seems to be basal to the lineage of thistles, which are ecologically and economically important herbs. Therefore, Xanthopappus can be used as a suitable outgroup in phylogenetic studies of thistles. This illustrates the potential of using cp genomes to infer phylogenies, and provide an important reference for future studies on the systematics and evolution on the hyper-diverse Asteraceae family. Future studies adding more taxa are essential to confirm the results we report here, as not all genera from tribe Cynareae and related groups were included in this phylogeny.
This first whole cp genome from Xanthopappus not only enriches the cp genomic database of angiosperms and Asteraceae, but also provides a framework for the comprehensive evaluation of germplasm resources of Cynareae aimed to deepens our understanding on the genetic diversity, species delimitation, biogeographical history, conservation genetics, and medicinal evaluation of X. subacaulis and other alpine herbs endemic to the Qinghai-Tibet Plateau.
REFERENCES
Barres, L., Sanmartin, I., Anderson, C.L., et al., Reconstructing the evolution and biogeographic history of tribe Cardueae (Compositae), Am. J. Bot., 2013, vol. 100, no. 5, pp. 867–882.
Bohlmann, F., Burkhardt, T., and Zdero, C., Naturally Occurring Acetylenes, London: Academic, 1973.
Bolger, A.M., Lohse, M., and Usadel, B., Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, vol. 30, no. 15, pp. 2114–2120.
Chapman, M.A., Hiscock, S.J., and Filatov, D.A., The genomic bases of morphological divergence and reproductive isolation driven by ecological speciation in Senecio (Asteraceae), J. Evol. Biol., 2016, vol. 29, no. 1, pp. 98–113.
Dierckxsens, N., Mardulyn, P., and Smits, G., Novoplasty: de novo assembly of organelle genomes from whole genome data, Nucleic Acids Res., 2017, vol. 45, no. 4, art. e18.
Doyle, J.J. and Doyle, J.L., Isolation of plant DNA from fresh tissue, Focus, 1990, vol. 12, no. 1, pp. 13–15.
Du, P., Gannan Tibetan Flora, vol. 12 of Qinghai–Tibet Plateau, Lanzhou: Gansu Science and Technology Press, 2006, vol 12, pp. 248–249.
Feng, H., Xu, H., Chen, J.Y., et al., Characterization of the complete chloroplast genome of Chrysanthemum oreastrum (Asteraceae) as a traditional medicinal plant to China, Mitochondrial DNA, B, 2020, vol. 5, no. 2, pp. 1172–1173.
Gray, J.C., Genetic manipulation of the chloroplast genome, Biotechnology, 1989, vol. 12, pp. 317–335.
Greiner, S., Lehwark, P., and Bock, R., OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes, Nucleic Acids Res., 2019, vol. 47, vol. W1, pp. W59–W64.
Häffner, E. and Hellwig, F.H., Phylogeny of the tribe Cardueae (Compositae) with emphasis on the subtribe Carduinae: an analysis based on ITS sequence data, Willdenowia, 1999, vol. 29, nos. 1/2, pp. 27–39.
Harborne, J.B., Phytochemical Methods, 2nd ed., New York: Chapman and Hall, 1984.
Howe, C.J., Barbrook, A.C., Koumandou, V.L., et al., Evolution of the chloroplast genome, Philos. Trans. R. Soc. London, Ser. B, 2003, vol. 358, no. 1429, pp. 99–107.
Jansen, R.K., Cai, Z.Q., Raubeson, L.A., et al., Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns, Proc. Natl. Acad. Sci. U. S. A., 2007, vol. 104, no. 49, pp. 19369–19374.
Jia, D.R., Abbot, R.J., Liu, T.L., et al., Out of the Qinghai–Tibet Plateau: evidence for the origin and dispersal of Eurasian temperate plants from a phylogeographic study of Hippophae rhamnoides (Elaeagnaceae), New Phytol., 2012, vol. 194, no. 4, pp. 1123–1133.
Jung, J., Kim, J.I., Jeong, Y.S., et al., AGORA: organellar genome annotation from the amino acid and nucleotide references, Bioinformatics, 2018, vol. 34, no. 15, pp. 2661–2663.
Katoh, K. and Standley, D.M., MAFFT multiple sequence alignment software version 7: improvements in performance and usability, Mol. Biol. Evol., 2013, vol. 30, no. 4, pp. 772–780.
Lin, R. and Shi, Z., Flora of China, Beijing: Science Press, 1987, vol 78, no 1, p. 61.
Liu, S.W., Flora of Qinghai, Xining: Qinghai People’s Publishing House, 1996, vol 3, no 1, p 437.
Mittermeier, R.A., Gil, P.R., Hoffman, M., et al., Hotspots Revisited: Earth’s Biologically Richest And Most Endangered Terrestrial Ecoregions, Chicago, ILL University of Chicago Press, 2005.
Odintsova, M.S. and Yurina, N.P., Chloroplast genomics of land plants and algae, in Biotechnological Applications of Photosynthetic Protein: Biochips, Biosensors and Biodevices, Springer, US, 2006, pp. 57–72.
Rashid, M.A., Gustafson, K.R., Cardellina, J.H., et al., Absolute stereochemistry and anti-HIV activity of minquartynoic acid, a polyacetylene from Ochanostachys amentacea, Nat. Prod. Lett., 2001, vol. 15, no. 1, pp. 21–26.
Stamatakis, A., RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies, Bioinformatics, 2014, vol. 30, no. 9, pp. 1312–1313.
Swofford, D.L., PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods), 4th ed., Sunderland, MA: Sinauer Associates, 2002.
Tian, Y., Liu, J., and Zhe, X., The complete chloroplast genome of Cerasus humilis (Bge.) Sok., Mitochondrial DNA, B, 2020, vol. 5, no. 2, pp. 1210–1212.
Tian, Y.Q., Wei, X.Y., and Xu, H.H., Photoactivated insecticidal thiophene derivatives from Xanthopappus subacaulis, J. Nat. Prod., 2006, vol. 69, no. 8, pp. 1241–1244.
Tillich, M., Lehwark, P., Pellizzer, T., et al., GeSeq-versatile and accurate annotation of organelle genomes, Nucleic Acids Res., 2017, vol. 45, no. W1, pp. W6–W11.
Wang, Y.J., Liu, J.Q., and Miehe, G., Phylogenetic origins of the Himalayan endemic Dolomiaea, Diplazoptilon and Xanthopappus (Asteraceae: Cardueae) based on three DNA regions, Ann. Bot., 2007, vol. 99, no. 2, pp. 311–322.
Wang, L.Y., Abbott, R.J., Zheng, W., et al., History and evolution of alpine plants endemic to the Qinghai–Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae), Mol. Ecol., 2009a, vol. 18, no. 4, pp. 709–721.
Wang, Y.J., Susanna, A., von Raab-Straube, E., et al., Island like radiation of Saussurea (Asteraceae: Cardueae) triggered by uplifts of the Qinghai–Tibetan Plateau, Bot. J. Linn. Soc., 2009b, vol. 97, no. 4, pp. 893–903.
Wu, C.Y., Hengduan Mountains flora and their significance, Jpn. J. Bot., 1988, vol. 63, no. 9, pp. 297–311.
Xing, Y.P., Xu, L., Chen, S.Y., et al., Comparative analysis of complete chloroplast genomes sequences of Arctium lappa and A. tomentosum, Biol. Plant., 2019, vol. 63, pp. 565–574.
Young, K., Jayasuriya, H., Ondeyka, J.G., et al., Discovery of FabH/FabF inhibitors from natural products, Antimicrob. Agents Chemother., 2006, vol. 50, no. 2, pp. 519–526.
Zhang, L., Chen, C.J., Chen, J., et al., Thiophene acetylenes and furanosesquiterpenes from Xanthopappus subacaulis and their antibacterial activities, Phytochemistry, 2014, vol. 106, pp. 134–140.
Zheng, D., The system of physico-geographical regions of the QinghaiTibet (Xizang) Plateau, Sci. China Ser. D, 1996, vol. 39, no. 4, pp. 410–417.
ACKNOWLEDGMENTS
The authors thank anonymous reviewers for their comments on the manuscript.
Funding
This study was financially supported by the National Natural Science Foundation of China (31800310), the Key Laboratory of Medicinal Animal and Plant Resources of the Qinghai-Tibet Plateau in Qinghai Province (2020-ZJ-Y40), the “High-end Innovative Talents Thousands of People Plan” in Qinghai Province and the “135 High-level Personnel Training Project” in Qinghai Province.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
About this article
Cite this article
Ma, ZL., Xu, SS., Zhang, Y. et al. Characterization and Phylogenetic Analysis of the Complete Chloroplast Genome of Xanthopappus subacaulis (Asteraceae), an Endemic Species from the Qinghai-Tibet Plateau in China. Cytol. Genet. 56, 77–83 (2022). https://doi.org/10.3103/S0095452722010078
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452722010078