Abstract
Conyza blinii H.Lév. is a traditional medicine for the treatment of bronchitis cough. Triterpene saponins (conyzasaponins) are major pharmacological components in C. blinii. Here, two C. blinii genes coding for the important regulatory enzymes of the conyzasaponins biosynthetic pathway, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) and squalene synthase (SQS), were cloned and characterized. The open reading frames of CbHMGR and CbSQS were 1740 bp and 1257 bp in length, respectively. Gene structure display server analysis revealed that there were four introns in the gDNA of CbHMGR and no intron in the gDNA of CbSQS. The analyses of their deduced amino acid sequences showed that these two genes had the typical domains shown in homologous proteins. The phylogenetic relationship between the two genes and their homologous genes were consistent with their natural evolution. The GCMS results of enzymatic activity assays showed that CbHMGR catalysed the formation of mevalonate from HMG-CoA. CbSQS catalysed the synthesis of squalene with farnesyl diphosphate as a substrate. These findings will provide a sound base for further research on the conyzasaponins biosynthetic pathway and may have applications in the synthetic biology of conyzasaponins production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Isoprenoids in plants composed of primary metabolites and secondary metabolites. The primary metabolites are important in the basic life activities of plants. For example, sterol is involved in biofilm construction; ubiquinone is involved in respiration; carotenoids and chlorophyll are involved in photosynthesis; and gibberellins, abscisic acid, cytokines and brassinolide are involved in plant growth and development. Secondary metabolites play less essential roles but are important in regulating the relationship between plants and the ecological environment. Moreover, secondary metabolites usually have commercial value. They are used as pharmaceuticals, agrochemicals, solvents and food additives (Roberts 2007).
Conyza blinii H.Lév. is a folk herb that used in western Sichuan, for its treatment of asthmatic cough and other inflammatory conditions (Chinese Pharmacopoeia Commission 2015). Its main secondary metabolites are isoprenoids, including blinin, α-amyrin, β-amyrin, oleanolic acid, ursolic acid, conyzasaponins and so on (Xu et al. 1999; Su et al. 2001a, b, 2003). The entirety of the plant can be medicinally prepared and the most popular C. blinii extract product is “Conyza blinii extract tablets”, which consists of conyzasaponins (Li 1980). In addition, conyzasaponins have anticancer activity (Ma et al. 2016). Thus the conyzasaponins are responsible for C. blinii major pharmacologically bioactivity. However, the conyzasaponins content in C. blinii is low, which are insufficient to meet the demand for pharmaceutical preparations. Hence, methods that improve conyzasaponins content are the focal point in studies on C. blinii.
To better regulate the synthesis of target isoprenoids, it is essential to understand their biosynthetic pathways. Previous studies have suggested that isoprenoids are synthesized by the MVA (mevalonic acid) pathway or the MEP (methylerythritol phosphate) pathway (Lichtenthaler et al. 1997; Lichtenthaler 1999). Conyzasaponins are oleanane-type pentacyclic triterpene saponins, which are synthesized via the MVA pathway (Fig. 1). That is a complex and multi-branched pathway. Identification of the key enzyme genes is one of the important aspects of studying these complex, multi-branched metabolic processes.
The conyzasaponins biosynthetic pathway. AACT, acetoacetyl-coa thiolase; HMGS, 3-hydroxy-3-methylglutaryl coenzyme A synthase; CbHMGR, C. blinii 3-hydroxy-3-methylglutaryl coenzyme A reductase; MVK, mevalonate kinase; PMK, phosphor mevalonate kinase; MVD, mevalonate diphosphate decarboxylase; IPI, IPP isomerase; FPPS, farnesyl diphosphate synthase; CbSQS, C. blinii squalene synthase; SQE, squalene epoxidase; βAS, β-amyrin synthase; P450s: cytochrome P450 monooxygenases; GTs: Glycosyltransferases (Hsieh et al. 2011)
3-Hydroxy-3-methylglutaryl-CoA reductase (HMGR, EC: 1.1.1.34) is the first rate-limiting enzyme in the MVA pathway (Rodwell et al. 1976; Bach 1986; Stermer et al. 1994). It catalyses irreversible conversion of HMG-CoA into mevalonate, the precursor of the isoprenoids (Chappell et al. 1995). Due to its significance in isoprenoid metabolism, HMGR has been isolated and characterized from many high plants. Cao et al. (2010) isolated a new HMGR gene from young leaves of Euphorbia Pekinensis by RACE. And a functional colour complementation assay in Escherichia coli was operated to prove that EpHMGR could catalyse the biosynthesis of carotenoids. Kalita et al. (2015) reported the full length cDNA cloning of HMGR and its characterization from Centella asiatica. Most recently, genes encoding HMGRs have been cloned from Cymbopogon winterianus (Devi et al. 2017), Gossypium (Liu et al. 2018), Pogostemon cablin (Zhang et al. 2019), Ginkgo biloba (Rao et al. 2019) and Andrographis paniculata (Srinath et al. 2020).
Another important regulatory enzyme, squalene synthase (SQS, EC: 2.5.1.21) is the first committed enzyme in the sterol and triterpenoid biosynthesis. It converts two molecules of FPP into squalene, a commom precursor of sterols and triterpenes (Brown and Goldstein 1980; Abe et al. 1993). Similarly, SQS has been cloned and characterized from many plants, such as Arabidopsis thaliana (Kribii et al. 1997), Centella asiatica (Kim et al. 2005), Taxus cuspidate (Huang et al. 2007), Siraitia grosvenorii (Su et al. 2017), Taraxacum koksaghyz (Unland et al. 2018), Medicago sativa (Kang et al. 2019) and Camellia sinensis (Fu et al. 2019).
However, the HMGR and SQS genes involved in conyzasaponins biosynthetic pathway have not been identified. In this study, we report the isolation and molecular characterization of CbHMGR and CbSQS genes from C. blinii transcript tags. And the biological function of the two genes were verified by in vitro enzymatic activity assays. The results will enable us to map and regulate the important steps involved in conyzasaponin biosynthetic pathway at the level of molecular genetics in the future.
Materials and methods
Plant material
C. blinii plants were collected from 101°46′~102°30′ E, 26° N at an altitude of 1680~2100 m in Panzhihua, Sichuan, China.
RNA and DNA isolation
Leaves collected from C. blinii were used to isolate RNA and DNA. A RNAprep pure Plant Kit (TIANGEN) was used to isolate the total RNA. Single-stranded cDNA was prepared using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara). A Plant Genomic DNA Kit (TIANGEN) was used to extract genomic DNA.
Gene cloning
The candidate HMGR and SQS genes were searched based on the in-text gene names and functional annotations of unique annotated genes from the C. blinii transcriptome annotation library (Sun et al. 2015). According to the selected tag sequences, specific primers (Table 1) were designed. PrimeSTAR Max DNA Polymerase premix (2×) (Takara) was used to amplify sequences. The TIANquick Mini Purification Kit (TIANGEN) was used to purify PCR products. The pMD19-T simple vector (TaKaRa) was used as a cloning vector and Escherichia coli strain DH5α (stored in the laboratory) was used as the cloning host strain. Finally, the PCR products sequences were sequenced by Invitrogen trading.
Bioinformatics analysis
Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) was used to analyse the genomic DNA sequence features (Hu et al. 2014). The alignment of multiple sequence were analysed by DNAMAN software. The SOPMA server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) was used to determine the secondary structure. Transmembrane domains were analysed with the TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Protein sequences were used to construct the phylogenetic tree by MEGA 7 software with Neighbor-Joining method and 1000 bootstrap replications (Tamura et al. 2011).
Construction of expression vectors and heterologous expression
The specific primers with restriction sites (Table 1) were used to amplify the coding sequences. The CbHMGR and CbSQS PCR products were digested with BamH I and Xho I restriction enzymes. Then inserted the digested products into the corresponding sites of the pYES2/NT B vector. Colony PCR, restriction digestion and sequencing were used to confirm positive clones. Subsequently, the positive plasmids were transformed into the Saccharomyces cerevisiae strain INVSc1. After cultivation for 3~4 days, single clones containing positive plasmids or empty vectors were inoculated in 15 mL of SC-U medium (synthetic complete medium without uracil). To induce gene expression, 2% galactose and 1% raffinose were used to replace glucose. Cultures were grown for 18 h at 30 °C with shaking at 200 rpm.
Heterologous protein extraction
A One Step Yeast Active Protein Extraction Kit (Sangon Biotech) was used to extract the heterologous proteins from S. cerevisiae. An ultrafiltration tube (Millipore) was used to exchange buffer and concentrate protein.
Enzyme assays
A CbHMGR enzyme activity assay was carried out as described by Gu et al. (2015). The 1 mL reaction mixture contained 50 mmol/L KCl, 25 mmol/L K2HPO4 (pH=7.2), 1 mmol/L EDTA, 5 mmol/L DTT, 100 μL CbHMGR crude protein (100 μg/mL), 0.3 mmol/L NADPH (Roche), 0.3 mmol/L HMG-CoA (Sigma-Aldrich) and ddH2O. After incubation at 30 °C for 30 min, terminate the reaction by adding 100 μL of 6 mol/L HCl. Then, the reaction was stored at 25 °C for 1~2 h. Finally, the reaction product was extracted with two volumes of ethyl acetate. The extracts were analysed by gas chromatography coupled to mass spectrometry (GC-MS) under the same conditions as those described by Gu et al. (2015). The product was identified with NIST software.
A CbSQS enzyme activity assay was carried out as described by Ye et al. (2014) with some modifications. The 500 μL reaction mixture contained 40 mmol/L MgCl2, 100 mmol/L Tris-HCl (pH 7.5), 0.1 mmol/L FPP (Sigma-Aldrich), 4 mmol/L DTT, 30 mmol/L BSA, 0.2 mmol/L NADPH and 220 μL of CbSQS crude protein (100 μg/mL). The mixture was incubated at 32 °C for 10 h. Then two volumes of hexane were used to extract the reaction product. Finally the concentrated organic phase was analysed by GC-MS under the same conditions as those described by Ye et al. (2014). The squalene was identified with NIST software.
Results
Sixteen sequences predicted as candidate HMGRs were obtained from the C.blinii transcript tags (Table S1). However twelve among them is too short, which only encode peptide less than 100 aa. c29868 and c45602 encode the same peptide. c29868 (c45602) and c38514 encode peptide about 330 aa. c29574 encode the full length HMGR protein. According to the FPKM value of these tags indicated that c29574 tag is the highest expressed one (Table S1). Therefore, c29574 was selected as the CbHMGR gene for further research.
The CbHMGR gene has a 1740 bp long coding sequence and encodes a peptide of 580 aa. Its GenBank accession number is KX907777. A BLASTp search revealed that CbHMGR has the highest similarity to HMGR from Chamaemelum nobile. The protein conserved domain prediction analysis predicted that the CbHMGR belongs to HMG-CoA_reductase_classI. This HMGR class catalyses the synthesis of coenzyme A and mevalonate in isoprenoid synthesis (Choi et al. 1992). The calculated molecular mass of CbHMGR is 62.17 kDa, and its isoelectric point is 6.61. GSDS analysis revealed four introns (1383 bp, 1165 bp, 470 bp and 189 bp) in the genomic DNA of CbHMGR (Fig. 2).
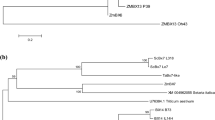
Sequence alignment showed that mature CbHMGR contains two HMG-CoA binding motifs (EMPVGYVQIP and TTEGCLVA) and two NADP(H) binding motifs (DAMGMNM and GTVGGGT), which were the four highly conserved motifs in all plant HMGRs and function as the catalytic active sites of the HMGR protein (Fig. 3). The secondary structure of CbHMGR was composed of 40.76% alpha helices, 33.51% random coils, 17.10% extended strands and 8.64% beta turns. The results of the phylogenetic analysis (Fig. 4) showed that CbHMGR is homologous to HMGR from C. nobile, which is in accordance with the BLAST results.
To examine the function of CbHMGR, the pYES-CbHMGR recombinant plasmid was constructed, then expressed in INVSc1 yeast. GC analysis of reaction products of 18-h-old pYES-CbHMGR strain revealed a single peak at 8.0 min, which was absent in the empty pYES2/NT B vector strain and the blank control. The MS data indicated that the particular peak detected in pYES-CbHMGR strain (8.0 min) was mevalonic acid lactone (Fig. 5). Consequently, we conclude that CbHMGR is indeed a 3-hydroxy-3-methylglutaryl-CoA reductase.
GC-MS results of CbHMGR in vitro enzymatic activity assay. a GC chromatogram of the CbHMGR group reaction products (a and b), the empty pYES2/NT B vector group reaction product (c) and the blank control group reaction product (d). b The MS spectrum of line a in A. c The MS spectrum and the structure of mevalonic acid lactone
According to the transcriptome analysis, only one tag corresponded to SQS gene (Table S1). Therefore, we selected it as the CbSQS gene for further research. CbSQS has a 1257-bp coding sequence which encodes 418 amino acid residues. Its GenBank accession number is KX907779. A BLAST search indicated that CbSQS shares 90% identity with the SQS from Artemisia annua. The calculated molecular mass of CbSQS is 47.96 kDa, and its isoelectric point is 8.61. GSDS analysis indicated that there are no introns in the gDNA of CbSQS. The result of sequence alignment showed that CbSQS contains four highly conserved motifs and two poor conserved motifs (Fig. 6). Domain A is an extended hydrophobic domain bounded on domain B. Domain B and domain D are two aspartate-rich domains, which constitute the two sets of substrate binding sites for allylic. Domain C is a partially conserved phytoene synthetases motif that is essential for catalysis. Domain E is present only in squalene synthetases. Domain F is the transmembrane domain of CbSQS.
Alignment analysis of C. blinii SQS and the SQS sequences from Eleutherococcus senticosus (AEA41712.1), Gossypium raimondii (XP_012449773.1), Vitis vinifera (XP_002266150.1) and Artemisia annua (AAR20329.1). Black: 100% homologous residues; Gray: ≥ 75% homologous residues. a: Hydrophobic motif; b and d: Two aspartate-rich motifs; c: Phytoene synthetases motif; e: Squalene synthetases peculiar motif; f: Transmembrane domain
The CbSQS secondary structure primarily include alpha helices (61.24%), with some random coils (21.05%), extended strands (9.57%) and beta turns (8.13%). The results of the phylogenetic analysis (Fig. 7) showed that CbSQS has the closest genetic relationship with the SQS from A. annua, which concurs with the BLAST results.
To examine the catalytic activity of CbSQS in squalene production, active proteins from pYES-CbSQS transgenic yeast were incubated with FPP for 10 h at 32 °C. Analysis of GC retention times revealed that there was a peak for the pYES-CbSQS strain at 11.5 min, while there were no peaks for the empty pYES2/NT B vector strain and the blank control group. After searching the NIST database the peak detected in the transgenic strain was confirmed as squalene (Fig. 8). This result suggests that CbSQS catalyses conversion of FPP to squalene.
GC-MS results of CbSQS in vitro enzymatic activity assay. a GC chromatogram of the CbSQS group reaction product (a), the empty pYES2/NT B vector group reaction product (b) and the blank control group reaction product (c). b The MS spectrum of line a in A. c The MS spectrum and the structure of squalene
Discussion
Conyza blinii is a rare Chinese herb endemic to southwest China that is commonly called Jin Long Dan Cao. According to the records of the Chinese Pharmacopoeia, it has anti-inflammatory, antitussive, anti-asthmatic and expectorant effects (Chinese Pharmacopoeia Commission 2015). The pharmacological effects of medicinal plants are mediated by secondary metabolites, which are the main sources of natural medicines. However the contents of metabolites in the natural plants are usually low, which hampered the applications of the pharmacologically active compounds (Misawa 2011). Overexpressing the biosynthesis pathway genes is an effective way to enhance the yield of metabolites (Lu et al. 2016). For example, Deng et al. (2017) reported co-overexpression of PnHMGR and PnSS could remarkably enhance the accumulation of total saponins in Panax notoginseng cells, which was 3-fold higher than those in control. Overexpression of Panax ginseng HMGR resulted in 1.1- to 1.6-fold increase of phytosterol and triterpene in hairy root cultures of Platycodon grandiflorum (Kim et al. 2013). Conyzasaponins are oleanane-type triterpene saponins from C. blinii, which are responsible for C. blinii major pharmacologically bioactivity. Nevertheless, the use of conyzasaponins is hampered by their low levels in C. blinii and by the lack of information about their biosynthetic pathway. To date there are only two researches on conyzasaponins pathway, which are identifying the CbSQE and CbβAS genes involved in conyzasaponins biosynthetic pathway (Sun et al. 2016; Sun et al. 2017). However the upstream genes have not been identified, which are also important in conyzasaponins biosynthetic pathway.
HMGR is the first key enzyme in the MVA pathway (Chappell 1995). In this study, a HMGR gene of C. blinii, namely, CbHMGR, was cloned and identified. The open reading frame length (1740 bp) of cloned CbHMGR is similar to that of HMGRs from other plants. As previously reported that there are two distinct classes of HMGRs: HMGRs class I and HMGRs class II (Bochar et al. 1999). Class I HMGRs contain N-terminal membrane domains involved in the membrane localization and the sterol-regulated degradation of HMGR molecules (Caelles et al. 1989; Denbow et al. 1996). The results of TMHMM analysis indicated that CbHMGR, like that of all known plant HMGRs, contains two transmembrane domains (40–62 and 83–105), which is consistent with conserved domain prediction results.
Additionally, SQSs were studied as a key enzyme for the biosynthesis of squalene as an intermediate for the production triterpenoids. The cloned CbSQS sequence show the same characteristics as known SQS sequences. CbSQS sequence codes for 418 aa with a 47.96 kDa molecular mass. These results are in accordance with those of previous reports, which have indicated that the SQS protein is approximately 410 to 461 aa long with a molecular mass in the 42.9~52.5 kDa range (Hanley and Chappell 1992; Robinson et al. 1993; Okada et al. 2000). And as other SQSs, six conserved regions are present in the CbSQS. These consensus regions are predicted or even have been proven to be important for the SQS activity (Gu et al. 1998; Pandit et al. 2000). In summary, these results provide new information about previously unannotated genes of conyzasaponins biosynthesis pathway.
Furthermore, in this study, we investigated the in vitro enzymatic activity of these genes. Through yeast expression analysis, CbHMGR was characterized as a reductase that produces mevalonic acid from HMG-CoA. CbSQS was characterized as a synthase, catalyses the reductive dimerization of farnesyl pyrophosphate. These results can not only help to increase understanding of the conyzasaponins biosynthesis pathway, but also provide a foundation for biotechnological improvement of the conyzasaponins content. However, further research on their function in conyzasaponins biosynthesis is still requiring. For example, overexpressing these genes in homologous and ectopic plants, knock out these genes in C. blinii or co-expressing these genes with other genes involved in conyzasaponins pathway to produce conyzasaponins using synthetic biology.
Abbreviations
- GC-MS:
-
Gas chromatography coupled to mass spectrometry
- HMG-CoA:
-
3-Hydroxy-3-methylglutaryl Coenzyme A
- RACE:
-
Rapid amplification of cDNA end
- FPP:
-
Farnesyl diphosphate
- DTT:
-
Dithiothreitol
- FPKM:
-
Expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced
References
Abe I, Rohmer M, Prestwich GD (1993) Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem Rev 93:2189–2206. https://doi.org/10.1021/cr00022a009
Bach TJ (1986) Hydroxymethylglutaryl-CoA reductase, a key enzyme in phytosterol synthesis? Lipids 21:82–88. https://doi.org/10.1007/BF02534307
Bochar DA, Freisen JA, Stauffacher CV, Rodwell VW (1999) Biosynthesis of mevalonic acid from acetyl-CoA. In: Barton D, Nakanishi K (eds) Comprehensive natural caritinoids and steroids. Elsevier Science Ltd., London, pp 15–44
Brown MS, Goldstein JL (1980) Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res 21:505–517
Caelles C, Ferrer A, Balcells L, Hegardt FG, Boronat A (1989) Isolation and structural characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Mol Biol 13:627–638. https://doi.org/10.1007/BF00016018
Cao XY, Zong ZM, Ju XY, Sun Y, Dai CC, Liu Q, Jiang JH (2010) Molecular cloning, characterization and function analysis of the gene encoding HMG-CoA reductase from Euphorbia Pekinensis Rupr. Mol Biol Rep 37:1559–1567. https://doi.org/10.1007/s11033-009-9558-7
Chappell J (1995) Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu Rev Plant Physiol Plant Mol Biol 46:521–547. https://doi.org/10.1146/annurev.pp.46.060195.002513
Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C (1995) Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme a reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol 109:1337–1343. https://doi.org/10.1104/pp.109.4.1337
Chinese Pharmacopoeia Commission (2015) Conyza herba. In: pharmacopoeia of the People's Republic of China, vol 1. People's Medical Publishing House, Beijing, p 217
Choi D, Ward BL, Bostock RM (1992) Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme a reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell 4:1333–1344. https://doi.org/10.1105/tpc.4.10.1333
Denbow CJ, Lång S, Crame CL (1996) The N-terminal domain of tomato 3-hydroxy-3-methylglutaryl-CoA reductases. Sequence, microsomal targeting, and glycosylation. J Biol Chem 271:9710–9715. https://doi.org/10.1074/jbc.271.16.9710
Deng B, Zhang P, Ge F, Liu DQ, Chen CY (2017) Enhancement of triterpenoid saponins biosynthesis in Panax notoginseng cells by co-overexpressions of 3-hydroxy-3-methylglutaryl CoA reductase and squalene synthase genes. Biochem Eng J 122:38–46. https://doi.org/10.1016/j.bej.2017.03.001
Devi K, Patar L, Modi MK, Sen P (2017) An insight into structure, function, and expression analysis of 3-hydroxy-3-methylglutaryl-CoA reductase of Cymbopogon winterianus. Bioinform Biol Insights 11:1–11. https://doi.org/10.1177/1177932217701735
Fu JY, Liu GH, Yang M, Wang XC, Chen XL, Chen F, Yang YJ (2019) Isolation and functional analysis of squalene synthase gene in tea plant Camellia sinensis. Plant Physiol Biochem 142:53–58. https://doi.org/10.1016/j.plaphy.2019.06.030
Gu P, Ishii Y, Spencer TA, Shechter I (1998) Function-structure studies and identification of three enzyme domains involved in the catalytic activity in rat hepatic squalene synthase. J Biol Chem 273:12515–12525. https://doi.org/10.1074/jbc.273.20.12515
Gu W, Geng C, Xue WD, Wu QN, Chao JG, Xu F, Sun HM, Jiang L, Han Y, Zhang SQ (2015) Characterization and function of the 3-hydroxy-3-methylglutaryl-CoA reductase gene in Alisma orientale (Sam.) Juz. And its relationship with protostane triterpene production. Plant Physiol Biochem 97:378–389. https://doi.org/10.1016/j.plaphy.2015.10.031
Hanley K, Chappell J (1992) Solubilization, partial purification, and immunodetection of squalene synthetase from tobacco cell suspension cultures. Plant Physiol 98:215–220. https://doi.org/10.1104/pp.98.1.215
Hsieh FL, Chang TH, Ko TP, Wang AHJ (2011) Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol 155:1079–1090. https://doi.org/10.1104/pp.110.168799
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Huang ZS, Jiang KJ, Pi Y, Hou R, Liao ZH, Cao Y, Han X, Wang Q, Sun XF, Tang KX (2007) Molecular cloning and characterization of the yew gene encoding squalene synthase from Taxus cuspidata. J Biochem Mol Biol 40:625–635. https://doi.org/10.5483/BMBRep.2007.40.5.625
Kalita R, Patar L, Shasany AK, Modi MK, Sen P (2015) Molecular cloning, characterization and expression analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Centella asiatica L. Mol Biol Rep 42: 1431–1439. https://doi.org/10.1007/s11033-015-3922-6
Kang JM, Zhang QY, Jiang X, Zhang TJ, Long RC, Yang QC, Wang Z (2019) Molecular cloning and functional identification of a squalene synthase encoding gene from alfalfa (Medicago sativa L.). Int J Mol Sci 20:4499. https://doi.org/10.3390/ijms20184499
Kim OT, Seong NS, Kim MY, Hwang B (2005) Isolation and characterization of squalene synthase cDNA from Centella asiatica (L) urban. J Plant Biol 48:263–269. https://doi.org/10.1007/BF03030521
Kim YK, Kim JK, Kim YB, Lee S, Kim SU, Park SU (2013) Enhanced accumulation of phytosterol and triterpene in hairy root cultures of Platycodon grandiflorum by overexpression of Panax ginseng 3-hydroxy-3-methylglutaryl-coenzyme a reductase. J Agric Food Chem 61:1928–1934. https://doi.org/10.1021/jf304911t
Kribii R, Arró M, Del AA, González V, Balcells L, Delourme D, Ferrer A, Karst F, Boronat A (1997) Cloning and characterization of the Arabidopsis thaliana SQS1 gene encoding squalene synthase--involvement of the C-terminal region of the enzyme in the channeling of squalene through the sterol pathway. Eur J Biochem 249:61–69. https://doi.org/10.1111/j.1432-1033.1997.00061.x
Li LX (1980) Comparison of Conyza blinii saponin tablets with doxycycline tablets in treating 310 cases of chronic tuberculosis. Journal of Chengdu University of Traditional Chinese Medicine 6:29–34
Lichtenthaler HK (1999) The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 50:47–65. https://doi.org/10.1146/annurev.arplant.50.1.47
Lichtenthaler HK, Rohmer M, Schwender J (1997) Two independent biochemical pathways for isopentenyl diphosphate and isoprenoid biosynthesis in higher plants. Physiol Plant 101:643–652. https://doi.org/10.1111/j.1399-3054.1997.tb01049.x
Liu W, Zhang ZQ, Li W, Zhu W, Ren ZY, Wang ZY, Li LL, Jia L, Zhu SJ, Ma ZB (2018) Genome-wide identification and comparative analysis of the 3-hydroxy-3-methylglutaryl coenzyme a reductase (HMGR) gene family in Gossypium. Molecules 23:193. https://doi.org/10.3390/molecules23020193
Lu X, Tang KX, Li P (2016) Plant metabolic engineering strategies for the production of pharmaceutical terpenoids. Front Plant Sci 7:1647. https://doi.org/10.3389/fpls.2016.01647
Ma L, Liu HY, Qin P, Hu CX, Man SL, Li YY, Liu Z, Liu ZX, Diao A (2016) Saponin fraction isolated from Conyza blinii H.Lév. Demonstrates strong anti-cancer activity that is due to its NF-κB inhibition. Biochem Biophys Res Commun 483:779–785. https://doi.org/10.1016/j.bbrc.2016.12.066
Misawa N (2011) Pathway engineering for functional isoprenoids. Curr Opin Biotechnol 22:627–633. https://doi.org/10.1016/j.copbio.2011.01.002
Okada S, Devarennne TP, Chappell J (2000) Molecular characterization of squalene synthase from the green microalga Botryococcus braunii, race B. Arch Biochem Biophys 373:307–317. https://doi.org/10.1006/abbi.1999.1568
Pandit J, Danley DE, Schulte GK, Mazzalupo S, Pauly TA, Hayward CM, Hamanaka ES, Thompson JF, Harwood HJ Jr (2000) Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis J Biol Chem 275:30610–30617. https://doi.org/10.1074/jbc.M004132200
Rao S, Meng XX, Liao YL, Yu T, Cao J, Tan JP, Xu F, Cheng SY (2019) Characterization and functional analysis of two novel 3-hydroxy-3-methylglutaryl-coenzyme a reductase genes (GbHMGR2 and GbHMGR3) from Ginkgo biloba. Sci Rep 9:14109. https://doi.org/10.1038/s41598-019-50629-8
Roberts SC (2007) Production and engineering of terpenoids in plant cell culture. Nat Chem Biol 3:387–395. https://doi.org/10.1038/nchembio.2007.8
Robinson GW, Tsay YH, Kienzle BK, Smith-Monroy CA, Bishop RW (1993) Conservation between human and fungal squalene synthetases: similarities in structure, function, and regulation. Mol Cell Biol 13:2706–2717. https://doi.org/10.1128/MCB.13.5.2706
Rodwell VW, Nordstrom JL, Mitschelen JJ (1976) Regulation of HMG-CoA reductase. Adv Lipid Res 14:1–74. https://doi.org/10.1016/b978-0-12-024914-5.50008-5
Srinath M, Bindu BBV, Shailaja A, Giri CC (2020) Isolation, characterization and in silico analysis of 3-Hydroxy-3-methylglutaryl-coenzyme a reductase (HMGR) gene from Andrographis paniculata (Burm.F) Nees. Mol Biol Rep 47:639–654. https://doi.org/10.1007/s11033-019-05172-0
Stermer BA, Bianchini GM, Korth KL (1994) Regulation of HMG-CoA reductase activity in plants. J Lipid Res 35:1133–1140
Su YF, Koike K, Guo D, Satou T, Liu JS, Zheng JH, Nikaido T (2001a) New apiose-containing triterpenoid saponins from Conyza blinii. Tetrahedron 57:6721–6726. https://doi.org/10.1016/S0040-4020(01)00632-9
Su YF, Guo D, Guo HZ, Liu JS, Zheng JH, Koike K, Nikaido T (2001b) Four new triterpenoid saponins from Conyza blinii. J Nat Prod 64:32–36. https://doi.org/10.1021/np000310v
Su YF, Koike K, Nikaido T, Liu JH, Zheng JH, Guo D (2003) Conyzasaponins I-Q, nine new triterpenoid daponins from Conyza blinii. J Nat Prod 66:1593–1599. https://doi.org/10.1021/np030327o
Su HL, Liu YM, Xiao YL, Tan YL, Gu YY, Liang B, Huang HL, Wu YS (2017) Molecular and biochemical characterization of squalene synthase from Siraitia grosvenorii. Biotechnol Lett 39:1009–1018. https://doi.org/10.1007/s10529-017-2328-z
Sun R, Liu S, Tang ZZ, Jin HJ, Li CL, Chen H (2015) Study on transcriptome characteristic of genuine traditional Chinese medicine Conyza blinii H.Lév leaves of Sichuan. Molecular Plant Breeding 13:2754–2760
Sun R, Luo J, Liu S, Tang ZZ, Fan ML, Li CL, Chen H (2016) Cloning and prokaryotic expression of squalene epoxidase gene from Conyza blinii H.Lév. Genomics and Applied Biology 35:3141–3146
Sun R, Liu S, Tang ZZ, Zheng TR, Wang T, Chen H, Li CL, Wu Q (2017) β-Amyrin synthase from Conyza blinii expressed in Saccharomyces cerevisiae. FEBS Open bio 7:1575–1585. https://doi.org/10.1002/2211-5463.12299
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 10:2731–2739. https://doi.org/10.1093/molbev/msr121
Unland K, Pütter KM, Vorwerk K, van Deenen N, Twyman RM, Prüfer D, Gronover CS (2018) Functional characterization of squalene synthase and squalene epoxidase in Taraxacum koksaghyz. Plant Direct 2:e00063. https://doi.org/10.1002/pld3.63
Xu LP, Guo D, Liu JS, Zheng JH, Koike K, Jia ZH, Nikaido T (1999) A new trans-clerodane diterpene lactone from Conyza blinii. Heterocycles 51:605–609. https://doi.org/10.3987/COM-98-8408
Ye Y, Wang RF, Jin L, Shen JH, Li XT, Yang T, Zhou MZ, Yang ZF, Chen YQ (2014) Molecular cloning and differential expression analysis of a squalene synthase gene from Dioscorea zingiberensis, an important pharmaceutical plant. Mol Biol Rep 41:6097–6104. https://doi.org/10.1007/s11033-014-3487-9
Zhang G, Wu Y, Haq Muhammad ZU, Yang YZ, Yu J, Zhang JF, Yang DM (2019) cDNA cloning, prokaryotic expression and functional analysis of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) in Pogostemon cablin. Protein Expr Purif 163:105454. https://doi.org/10.1016/j.pep.2019.105454
Acknowledgments
We thank the Sichuan Province Science and Technology Support Program (Grant No. 2018HH0074) and Panzhihua University (Grant No. 035200167) for financial support. And we thank Prof. Meng-gen Ma (Sichuan Agricultural University) for the pYES2/NT B vector and INVSc1 strain.
Author contribution statement
HC participated in the design of the study. RS and SL performed gene cloning and bioinformatics analysis. RS drafted the manuscript. JLG performed enzyme assays. HC and SL contributed to revisions of the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The manuscript does not involve any animal study.
Conflict of interest
The authors declare that they have no conflict of interest in publishing this manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Sun, R., Gao, J., Liu, S. et al. Cloning and characterization of CbHMGR and CbSQS genes in Conyza blinii. Biologia 76, 2337–2347 (2021). https://doi.org/10.2478/s11756-020-00671-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11756-020-00671-z