Abstract
Background
Pathological response to neoadjuvant chemotherapy (NAC) is critical in prognosis and selection of systemic treatments for patients with triple-negative breast cancer (TNBC). The aim of this study is to identify gene expression-based markers to predict response to NAC.
Patients and Methods
A survey of 43 publicly available gene expression datasets was performed. We identified a cohort of TNBC patients treated with NAC (n = 708). Gene expression data from different studies were renormalized, and the differences between pretreatment (pre-NAC), on-treatment (post-C1), and surgical (Sx) specimens were evaluated. Euclidean statistical distances were calculated to estimate changes in gene expression patterns induced by NAC. Hierarchical clustering and pathway enrichment analyses were used to characterize relationships between differentially expressed genes and affected gene pathways. Machine learning was employed to refine a gene expression signature with the potential to predict response to NAC.
Results
Forty nine genes consistently affected by NAC were involved in enhanced regulation of wound response, chemokine release, cell division, and decreased programmed cell death in residual invasive disease. The statistical distances between pre-NAC and post-C1 significantly predicted pathological complete response [area under the curve (AUC) = 0.75; p = 0.003; 95% confidence interval (CI) 0.58–0.92]. Finally, the expression of CCND1, a cyclin that forms complexes with CDK4/6 to promote the cell cycle, was the most informative feature in pre-NAC biopsies to predict response to NAC.
Conclusions
The results of this study reveal significant transcriptomic changes induced by NAC and suggest that chemotherapy-induced gene expression changes observed early in therapy may be good predictors of response to NAC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Triple-negative breast cancer (TNBC) accounts for 10–20% of breast cancer and is characterized by the absence of expression of estrogen receptor (ER), progesterone receptor (PgR), and human epidermal growth factor receptor 2 (HER2).1 Due to the absence of therapeutic targets, anthracycline- and taxane-based neoadjuvant chemotherapy (NAC) has become part of the standard of care for patients with stage I–III TNBC. However, 60–70% of TNBC patients who receive NAC will not have pathological complete response (pCR), which is associated with higher risk of recurrence and shorter overall survival (OS).2 The remaining viable tumor cells may be inherently refractory or have acquired resistance to chemotherapy. Additionally, NAC may exert a selective pressure that promotes drug resistance, which enhances the ability of tumor cells to metastasize.3,4 However, clinical–demographic and molecular features that predict poor response are yet to be characterized.
Seminal attempts to characterize informative biomarkers have focused on the study of molecular features of pretreatment biopsies (pre-NAC).5,6 Yet, this strategy does not consider the impact of molecular alterations induced by therapeutic agents. Studies comparing paired pre-NAC and post-NAC specimens showed significant gene expression changes influenced by chemotherapeutic agents.7,8,9,10,–11 Therefore, the identification of molecular features after early exposure to NAC would provide critical information to aid clinical decision-making. This has been investigated in patients with hormone receptor-positive breast cancers treated with neoadjuvant endocrine therapy (NET), where changes in proliferation markers (i.e., Ki-67)12,13 or gene expression signatures14,15 between pre-NET and on-treatment biopsies were shown to be better predictors than evaluation of pre-NET specimens alone. However, informative gene expression-based markers predictive of response to NAC in TNBC patients remain poorly developed.
Here, we explored three approaches to assess gene expression patterns to more efficiently predict the response to NAC in TNBC patients. First, we compared changes to gene expression programs induced by NAC. Second, we investigated the utility of assessing the gene expression changes after a single cycle of NAC to predict pCR. Finally, using machine learning, we constructed and validated classifiers to identify patients who are likely to respond to treatment using pre-NAC specimens.
Patients and Methods
Patients, Samples, and Clinical Data
We identified 4341 patients from 43 publicly available gene expression datasets from Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and European Genome-phenome Archive (EGA; https://www.ebi.ac.uk/ega/home) including specimens from breast cancer patients. Patients with non-TNBC (n = 1964), no NAC administered (n = 82), or absence of hormone receptor [estrogen receptor (ER), progesterone receptor (PgR)] and/or human receptor growth factor-2 (HER2) statuses (n = 883) were excluded. Patients without information about NAC regimens (n = 165), samples without pathologic response evaluation (n = 102), and duplicated samples (n = 74) were also excluded. NAC regimens in the datasets are summarized in Supplementary Tables 1 and 2. pCR was defined as absence of invasive disease in the breast and lymph nodes (ypT0/is, ypN0),2,16 while presence of invasive disease was considered residual disease (RD). Specimens were evaluated at different time points: pre-NAC, after the first and fourth NAC cycles (post-C1 and post-C4, respectively), and at surgery (Sx) in patients with RD. Specimens included in gene expression profiling were subjects of pathologist-guided microdissection or tumor enrichment processes. In addition, effective tumor purity of samples based on gene expression patterns was assessed using the ESTIMATE() v1.0.13 R package.
Data Access and Normalization
The raw intensity data (i.e., CEL files) were obtained for each GEO accession number using functions within the GEOquery() v2.50.5 R/Bioconductor package.17 Samples were normalized using the rma() function in the oligo v1.46.0R/Bioconductor package.18
Gene Expression Analyses
To identify gene expression changes influenced by NAC, paired TNBC specimens (GSE32603, GSE18728, and GSE21974; n = 102)7,8,10 were evaluated to identify differences between pre-NAC and during NAC. Paired TNBC specimens (GSE3260 and GSE18728; n = 88) were evaluated to determine differences between pre-NAC and Sx. Paired pre-NAC and post-C1 biopsies (GSE3260 and GSE18728; n = 25) were employed to generate the Euclidean metric distances using the expression of all genes (Supplementary Information), by applying the hierarchical clustering (HCL) function of the MeV v4.9.0 software. These distances were used to generate phylogenetic trees with the FigTree v1.4.3 software.
Classifier Construction to Predict Pathologic Response from Pretreatment Specimens
We employed gene expression profiles from pre-NAC TNBC biopsies from 16 datasets (Supplementary Table 2) to construct and validate machine learning-based transcriptomic classifiers to predict pCR. All samples were combined by microarray platform and normalized, resulting in three separate datasets. These three datasets were then merged, resulting in a final dataset with 708 pre-NAC samples with clinical annotation of response to NAC. To avoid potential biases related to different evaluation platforms, we performed a correction for batch effects from the various platforms and arrays (Supplementary Fig. 1). The batch-corrected dataset was used to construct a predictive model to distinguish patients who achieved pCR from those with RD after NAC, using the pre-NAC specimens. This cohort was split into training (80% of patients) and validation (20% of patients) datasets to train a random forest (RF) model.
Bioinformatics Analyses
Gene expression levels were compared between the groups using Student’s t test for unidimensional analyses. Genes with absolute fold change (fc) ≥ 2 and corrected false discovery rate (FDR) of p < 0.05 were considered significantly differentially expressed. Hierarchical clustering analyses using significant genes were employed to identify multidimensional associations between genes and specimens. Pathway enrichment analysis was performed to identify significant molecular functions affected by NAC using the Kyoto Encyclopedia of Genes and Genomes (KEGG), gene ontology (GO) molecular functions, and Reactome using GeneMania.19 The mean Euclidean distance between patients who reached pCR and patients who presented RD after completed NAC treatment were evaluated using Student’s t test.
Results
Neoadjuvant Chemotherapy Induces Gene Expression Changes in TNBC Tumors
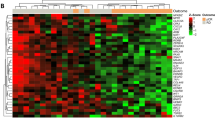
We analyzed paired tissue specimens before (pre-NAC), during (post-C1 or post-C4), and after (Sx) NAC from three different datasets to identify gene expression changes induced by chemotherapeutic agents in TNBC patients (n = 102; Fig. 1a). We identified 547 and 181 differentially expressed genes (p < 0.05) between pre-NAC and Sx tissues, and 972 differentially expressed genes (p < 0.05) between pre-NAC and post-C4 (Fig. 1b). Of these, 49 genes were consistently altered in at least two independent studies (Fig. 1c). Interestingly, the pathway enrichment analyses identified that NAC induced an enhanced regulation of wound response, chemokine release, cell division, and decreased programmed cell death (hypergeometric test, p < 0.05; Fig. 1d).
Neoadjuvant chemotherapy induces transcriptomic changes. a Diagram describing patient selection criteria and treatment regimens of each dataset (n = 102). b Hierarchical clustering analysis indicating differential gene expression between the paired specimens within each dataset. c Venn diagrams representing genes with consistent differential expression (n = 49 genes). d Molecular functions determined to be affected by NAC using pathway enrichment analysis
Transcriptomic Variations During Neoadjuvant Chemotherapy are Associated with Pathological Response
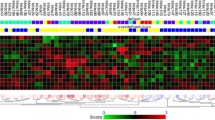
Based on the premise that TNBC tumors responding to NAC will experience early changes in gene expression patterns, we utilized the Euclidean statistical distance between paired pre-NAC, post-C1, and Sx as a method for assessing the impact of NAC (Fig. 2a). We observed that post-C1 biopsies were generally located between the pre-NAC and the Sx specimens, but the distances between pre-NAC and post-C1 were variable among the patients (Supplementary Fig. 2). Notably, we identified that a larger Euclidean distance between the pre-NAC and post-C1 biopsies was significantly associated with pCR to NAC (n = 25; Student’s t test; p = 0.02; Fig. 2b), and showed significantly good accuracy in predicting pCR (AUC = 0.75; p = 0.003; 95% CI 0.58–0.92; Fig. 2c). We additionally identified 21 genes that had a unique expression pattern in each time point of the treatment [one-way analysis of variance (ANOVA); p < 0.01; Fig. 2d, Supplementary Table 3]. Phylogenetic trees using the 21-gene signature showed a separation of the specimens according to tissue types that resemble the time points of tissue collection during NAC treatment (Fig. 2e).
Euclidean distance for prediction of pathological response to NAC. a Representative Euclidean distances between pre-NAC, post-C1, and Sx specimens for GSM808117. b Boxplot representing the Euclidean distances between pre-NAC and post-C1 in TNBC patients with RD versus pCR (n = 25). c Receiver-operating curve (ROC) showing accuracy in predicting pCR using Euclidean distance between pre-NAC and post-C1 specimens. d Heatmap representing 21-gene signature specific to each specimen type. e Phylogenetic tree of representative patients (n = 5) based on the 21-gene signature
Gene Expression-Based Classifiers Predict pCR to NAC Using Pretreatment Specimens
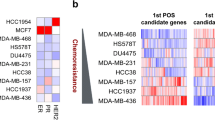
To expand the prediction performance of the transcriptomic profiling, we evaluated a large collection of pre-NAC biopsies from TNBC patients with and without pCR at NAC completion (n = 708; Fig. 3a, Supplementary Table 2). Using random forest, a machine learning algorithm, we constructed a gene expression classifier using a training cohort of patients and evaluated the ability to predict response to NAC using an independent validation cohort (baseline characteristics of both cohorts are summarized in Table 1). Of note, the overall accuracy of predicting pCR was 81% (95% CI 74–88%), with sensitivity of 63% and specificity of 89%. The most informative genes of this classifier were CCND1, SCD5, and ILF2 (Fig. 3b) and differed significantly in expression between pre-NAC tumor biopsies from patients with pCR and patients with RD (Fig. 3c). Using the top 200 most informative genes, we generated gene network analyses and identified significant enrichments of the G1/S transition in mitosis, DNA strand elongation in DNA replication, and response to transforming growth factor (TGF)-β processes (Fig. 3d, Supplementary Table 4). These gene pathways, frequently involved in oncogenic processes, may offer additional insights regarding the mechanisms behind the poor response to NAC in TNBC patients.
Transcriptome-based classifier model of NAC response using pre-treatment specimens. a Diagram showing the sample filtering strategy used to generate the 708 TNBC samples used to build and test the random forest-based classifier model. b The top 20 most informative features in the final trained classifier model. Array probe IDs have been replaced with gene symbols. c Boxplots of gene expression for the top three most informative genes. ***Wilcoxon rank sum test p value < 0.01. d Gene network showing the relationships among informative features and shared gene ontologies. Shaded circles indicate genes that are included in the top 200 most informative features of the classifier. Nonshaded circles indicate genes identified in the network analysis that are not features of the classifier. Colors indicate a genes association with a particular gene ontology
Discussion
To date, there are no efficient predictors of overall outcome in TNBC other than the evaluation of pathological response after NAC. Our study shows that NAC induces significant changes in gene expression patterns, some of which specifically affect tumor-related pathways that may influence response to this treatment. Data presented here indicate that transcriptomic profiling can accurately predict pCR, both before initiation of treatment and after a single cycle of NAC, offering additional information to guide treatment decision-making.
Evaluation of gene expression changes in paired samples collected before and on treatment has been shown to be a useful predictor of response to NAC in patients with breast cancer. Korde et al. identified 45 gene pathways in which the change in expression after one cycle of docetaxel/capecitabine significantly differed between responders and non-responders.7 Stickeler et al. found that upregulation of HER4 after four cycles of epirubicin/cyclophosphamide was associated with pCR,8 and Magbanua et al. observed a significant downregulation of proliferation and immune-related genes after one cycle of anthracycline-based chemotherapy.10 Our study explored gene expression changes induced by NAC specifically in TNBC patients, and identified alterations of cellular functions, including wound healing response, chemokine release, cell division, and decreased programmed cell death, mechanisms that are involved in TNBC drug resistance. For example, enhanced regulation of wound response, which involves matrix remodeling and angiogenesis, has been shown to be associated with worse prognosis, especially in patients with basal-like breast tumors.20
While the vast majority of transcriptomic studies have considered all breast cancer subtypes in pre-NAC specimens,5,6,21,22,–23 here we explored global gene expression variations in TNBC tumors during NAC treatment. This approach, based on a simple and unbiased statistical comparison (Euclidean distances), allowed us to establish a new approach to predict response to NAC early in the treatment regimen. Upon validation of this novel method in an expanded prospective cohort, we believe that this could be used to monitor, in real time, the response to specific therapeutic interventions, since a lack of early response may suggest that changes should be made to the chemotherapy regimen, or anticipate surgical procedures in patients with operable TNBC.
Implementation of machine learning approaches allowed us to further refine gene expression signatures in TNBC tissues before NAC that can efficiently predict the pathological response to NAC. In addition to reducing the number of genes that need to be assessed, while enhancing the predictive potential, this method allows for the design of cost-effective laboratory assays. Indeed, therein lies the clinical applicability of this classifier. As we have previously shown for breast cancer patients with brain metastases, the most informative genes can be evaluated through polymerase chain reaction (PCR).24 Thus, this approach, combined with the assessment of the Euclidean distances, can significantly contribute to comprehensive monitoring of treatment efficacy. In addition to the predictive potential of the machine learning-based classifiers, the most informative genes (CCND1, SCD5, and ILF2) have previously been associated with chemotherapy resistance.25,26,–27 Interestingly, genes involved in cell-cycle control, such as CCND1, are associated with response to NAC,28 which has prompted investigations into markers of sensitivity for response to CDK4/6 inhibitors in preclinical studies.29
While the large number of specimens evaluated allowed us to identify significant transcriptomic changes, the differences in individual study design involving time points of sampling, methods for examining gene expression, and the different chemotherapy regimens utilized in each study limit definitive conclusions. This issue was specifically reflected in the data batch effects that required a renormalization of the raw data to unify different gene expression platforms. While, in general, the cohorts evaluated allowed for the identification of statistically significant changes, sample size was limited in the analysis of paired pre-NAC and post-C1 biopsies (n = 25). However, these data generated significant and promising results that would justify additional and specific validation studies, mainly focused on specific chemotherapy regimens. Despite these limitations, the results presented in this study provide additional approaches to assess the impact of NAC in TNBC tumors and novel insights about the variable, and still unpredictable response to NAC.
Conclusions
Our study shows that neoadjuvant chemotherapy induces changes in gene expression patterns in TNBC that can be employed to predict the pathological response employing pretreatment and early on-treatment tumor biopsies. Validation of both the Euclidean metric distances and the machine learning-based classifiers will provide the basis for clinical assays that can aid clinical decision-making.
References
Foulkes WD, Smith IE, and Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948.
Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172.
Obenauf AC, Zou Y, Ji AL, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520(7547):368–372.
Schmitt MW, Loeb LA, and Salk JJ. The influence of subclonal resistance mutations on targeted cancer therapy. Nat Rev Clin Oncol. 2016;13(6):335–347.
Ayers M, Symmans WF, Stec J, et al. Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J Clin Oncol. 2004;22(12):2284–2293.
Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244.
Korde LA, Lusa L, McShane L, et al. Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer. Breast Cancer Res Treat. 2010;119(3):685–699.
Stickeler E, Pils D, Klar M, et al. Basal-like molecular subtype and HER4 up-regulation and response to neoadjuvant chemotherapy in breast cancer. Oncol Rep. 2011;26(4):1037–1045.
Gonzalez-Angulo AM, Iwamoto T, Liu S, et al. Gene expression, molecular class changes, and pathway analysis after neoadjuvant systemic therapy for breast cancer. Clin Cancer Res. 2012;18(4):1109–1119.
Magbanua MJ, Wolf DM, Yau C, et al. Serial expression analysis of breast tumors during neoadjuvant chemotherapy reveals changes in cell cycle and immune pathways associated with recurrence and response. Breast Cancer Res. 2015;17:73.
Gruosso T, Mieulet V, Cardon M, et al. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol Med. 2016;8(5):527–549.
Dowsett M, Smith IE, Ebbs SR, et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007;99(2):167–170.
Ellis MJ, Suman VJ, Hoog J, et al. Ki67 proliferation index as a tool for chemotherapy decisions during and after neoadjuvant aromatase inhibitor treatment of breast cancer: results from the American College of Surgeons Oncology Group Z1031 trial (Alliance). J Clin Oncol. 2017;35(10):1061–1069.
Turnbull AK, Arthur LM, Renshaw L, et al. Accurate prediction and validation of response to endocrine therapy in breast cancer. J Clin Oncol. 2015;33(20):2270–2278.
Patani N, Dunbier AK, Anderson H, et al. Differences in the transcriptional response to fulvestrant and estrogen deprivation in ER-positive breast cancer. Clin Cancer Res. 2014;20(15):3962–3973.
Bossuyt V, Provenzano E, Symmans WF, et al. Recommendations for standardized pathological characterization of residual disease for neoadjuvant clinical trials of breast cancer by the BIG-NABCG collaboration. Ann Oncol. 2015;26(7):1280–1291.
Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847.
Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367.
Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46(W1):W60–W64.
Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102(10):3738–3743.
Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881.
Iwamoto T, Bianchini G, Booser D, et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst. 2011;103(3):264–272.
Prat A, Galvan P, Jimenez B, et al. Prediction of response to neoadjuvant chemotherapy using core needle biopsy samples with the prosigna assay. Clin Cancer Res. 2016;22(3):560–566.
Orozco JIJ, Knijnenburg TA, Manughian-Peter AO, et al. Epigenetic profiling for the molecular classification of metastatic brain tumors. Nat Commun. 2018;9(1):4627.
Feng Z, Guo W, Zhang C, et al. CCND1 as a predictive biomarker of neoadjuvant chemotherapy in patients with locally advanced head and neck squamous cell carcinoma. PLoS One. 2011;6(10):e26399.
Marchesini M, Ogoti Y, Fiorini E, et al. ILF2 is a regulator of RNA splicing and DNA damage response in 1q21-amplified multiple myeloma. Cancer Cell. 2017;32(1):88–100.e106.
Chen YZ, Xue JY, Chen CM, et al. PPAR signaling pathway may be an important predictor of breast cancer response to neoadjuvant chemotherapy. Cancer Chemother Pharmacol. 2012;70(5):637-644.
Witkiewicz AK, Balaji U, Knudsen ES. Systematically defining single-gene determinants of response to neoadjuvant chemotherapy reveals specific biomarkers. Clin Cancer Res. 2014;20(18):4837-4848.
Asghar US, Barr AR, Cutts R, et al. Single-cell dynamics determines response to CDK4/6 inhibition in triple-negative breast cancer. Clin Cancer Res. 2017;23(18):5561-5572.
Acknowledgement
This study was supported by the Associates for Breast and Prostate Cancer Studies (ABCs) Foundation, the Fashion Footwear Association of New York (FFANY) Foundation, and the John Wayne Cancer Institute Translational Research Fund.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosures
The authors have no conflict of interest disclosures to report.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Orozco, J.I.J., Grumley, J.G., Matsuba, C. et al. Clinical Implications of Transcriptomic Changes After Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Ann Surg Oncol 26, 3185–3193 (2019). https://doi.org/10.1245/s10434-019-07567-6
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-019-07567-6