Abstract
Background
Over 100 mutations in the SRD5A2 gene have been identified in subjects with 46,XY disorder of sex development (DSD). Exploration of SRD5A2 mutations and elucidation of the molecular mechanisms behind their effects should reveal the functions of the domains of the 5α-reductase 2 enzyme and identify the cause of 46,XY DSD. Previously, we reported a novel compound heterozygous p.Q6X/p.H232R mutation of the SRD5A2 gene in a case with 46,XY DSD. Whether the compound heterozygous p.Q6X/p.H232R mutation in this gene causes 46,XY DSD requires further exploration.
Methods
The two 46,XY DSD cases were identified and sequenced. In order to identify the source of the compound heterozygous p.Q6X/p.H232R mutation, the parents, maternal grandparents, and maternal uncle were sequenced. Since p.Q6X mutation is a nonsense mutation, p.H232R mutation was transfected into HEK293 cells and dihydrotestosterone (DHT) production were analyzed by liquid chromatography–mass spectrometry (LC–MS) for 5α-reductase 2 enzyme activities test. Apparent michaelis constant (Km) were measured of p.H232R mutation to analyze the binding ability change of 5α-reductase 2 enzyme with testosterone (T) or NADPH.
Results
The sequence results showed that the two 46,XY DSD cases were the compound heterozygous p.Q6X/p.H232R mutation, of which the heterozygous p.Q6X mutation originating from maternal family and heterozygous p.H232R mutation originating from the paternal family. The function analysis confirmed that p.H232R variant decreased the DHT production by LC–MS test. The Km analysis demonstrated that p.H232R mutation affected the binding of SRD5A2 with T or NADPH.
Conclusions
Our findings confirmed that the compound heterozygous p.Q6X/p.H232R mutation in the SRD5A2 gene is the cause of 46,XY DSD. p.H232R mutation reduced DHT production while attenuating the catalytic efficiency of the 5α-reductase 2 enzyme.
Similar content being viewed by others
Background
46,XY disorder of sex development (DSD) is characterized by incomplete gonadal development and a female or ambiguous phenotype in individuals with a normal 46,XY karyotype [1]. The main clinical phenotypes of 46,XY DSD include bilateral undescended testes and female external genitalia, but without uterus or ovaries [2,3,4]. The predicted morbidity among subjects with 46,XY DSD is up to 1:20,000. Among affected cases, the morbidity of hypospadias and cryptorchidism can reach 1:200 to 1:300 [5, 6]. Many etiological factors can give rise to 46,XY DSD, with the common causes including androgen insensitivity syndrome (AIS) [7,8,9] and steroid 5α-reductase type 2 deficiency [10, 11]. AIS is an X-linked recessive genetic disorder that is caused by androgen receptor dysfunction [3]. 5α-Reductase type 2 deficiency is an autosomal recessive disorder caused by decrease or loss of 5α-reductase type 2 function [12].
5α-Reductase type 2, which is coded by the SRD5A2 gene, is mainly expressed in epididymides, seminal vesicles, prostate, and genital skin [13]. The function of 5α-reductase type 2 is to convert Tto DHT, which is responsible for male sexual development [14, 15]. The loss of function of 5α-reductase type 2 leads to a failure of normal differentiation into the external genitalia, urethra, and prostate and causes 5α-reductase type 2 deficiency. To date, over 100 mutations of the SRD5A2 gene have been identified [16].
Recently, we identified a compound heterozygous mutation (p.H232R/p.Q6X) of the SRD5A2 gene in two cases from a single family diagnosed with 46,XY DSD [17]. DNA sequencing showed that the heterozygous p.H232R mutation in the SRD5A2 gene originated from the paternal side of the family and the heterozygous p.Q6X mutation in the SRD5A2 gene originated from the maternal side. To elucidate the effect of the p.H232R mutant on the activity of the 5α-reductase type 2 enzyme, HEK293 cells were transfected with p.H232R mutant. LC–MS showed that the mutation clearly decreased DHT production, indicating that this mutation reduced catalytic efficiency. Hence, our results confirmed compound heterozygous mutation (p.Q6X/p.H232R) as the cause of the 46,XY DSD and that p.H232R mutation decreased the catalytic efficiency of the 5α-reductase type 2 enzyme.
Results
Clinical phenotype of 46,XY DSD patients
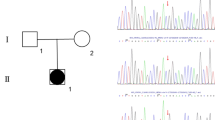
Case 1 (III-1): We previously reported a 2-year-old female diagnosed with 46,XY DSD. Clinical examination showed that this proband had normal female external genitalia, but with a blind-ending vagina. An ultrasound test revealed that she had testis in the left labium majus pudendi and right groin, but did not have a uterus or ovaries (Fig. 1a). Karyotype analysis indicated that the proband had a 46,XY karyotype, with no apparent anomalies in chromosome number or structure (Fig. 1b). The proband showed lower levels of follicle-stimulating hormone (FSH), luteinizing hormone (LH), and T (< 0.1 ng/ml) than normal males (Table 1). Family members of the proband were also examined and a pedigree was generated. Two members of this family were diagnosed with 46,XY DSD (Fig. 1c). The sequencing results showed that case 1(III-1) having 46,XY DSD was compound heterozygous for a previously reported nonsense mutation p.Q6X (c.16C > T) (Fig. 1d) [17] and a novel missense SRD5A2 mutation p.H232R (c.695A > G) (Fig. 1e).
Clinical features of the case 1 (III-1). a. B-ultrasound analysis of the case’s internal reproductive organs showed the absence of a uterus and ovaries, but testes on the left labia and right groin. b. Karyotype analysis revealed that the case’s karyotype was 46,XY. c. Pedigree of the case’s family; Males, females and the patient are indicated by squares, circles, filled circle, respectively. the case 1 (III-1) and her sister (case 2, III-2) were 46,XY DSD patients, while the other relatives did not suffer from 46,XY DSD. d. Sequencing analysis of the SRD5A2 gene. The heterozygous mutation 16C > T found in the patient lead to a stop codon. e. Sequencing analysis of the SRD5A2 gene. The heterozygous mutation 695A > G was found, causing amino acid 232 to change from histidine to arginine. Red arrows indicate mutated nucleotide
Case 2 (III-2): At 21 weeks’ and 25 weeks’ gestation, case 1’s mother came to our hospital for a B-ultrasound examination. The results showed that the fetus did not have male external genitalia (Fig. 2a). Cytogenetic analysis revealed that the child had a 46,XY karyotype, with no apparent anomalies in chromosome number or structure (Fig. 2b). The results of FISH assay to test chromosomes 13/16/18/21/22/X/Y revealed that the child had normal X and Y chromosomes (Fig. 2c). These results indicated that the fetus had 46,XY DSD. Considering the fetus may face difficulty of being unable to treat after birth, the mother decided to terminate the pregnancy. According to the appearance, we found that the external genitalia of the fetus is not a normal male genitalia(Fig. 2d). However, through histopathological examination of the external genitalia, the epididymis and testicular tissues were found (Fig. 2e). Sequencing analysis was performed and showed that Case 2 (III-2) also had the nonsense SRD5A2 mutation p.Q6X (Fig. 2f) and the novel missense SRD5A2 mutation p.H232R (Fig. 2g).
Clinical features of the case 2 (III-2). a. Prenatal B-ultrasound testing of the case 2 at 21 and 25 weeks revealed no abnormalities. b. Chromosome karyotype analysis showed a normal 46, XY karyotype. c. Fluorescence in situ hybridization (FISH) prenatally diagnosed 13/16/18/21/22/X/Y chromosomes in the case 2. d. The physical examination of the fetus aborted at 25 weeks’ gestation displayed female external genitalia, but with blind ending vagina. e. HE staining showed the fetus had epididymis tissue and testis tissue. f. Sequencing analysis of the SRD5A2 gene and the p.Q6X (c.16C > T) mutation were found. g. Sequencing analysis of the SRD5A2 gene and the p.H232R (c.695A > G) mutation were found. Red arrows indicate mutated nucleotide
Genetic analysis identified the sources of SRD5A2 mutations
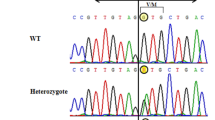
To find the familial sources of the SRD5A2 gene mutations, Sanger sequencing was performed in six members of the cases’ family. The sequencing results showed that cases’ father II-1 and younger sister III-3 had no the SRD5A2 mutation p.Q6X, while their mother II-2 had (Fig. 3a). The cases’ father II-1 and younger sister III-3 had the SRD5A2 mutation p.H232R, but their mother II-2 had no (Fig. 3b). These results demonstrated that the missense mutation p.Q6X of 46,XY DSD cases was derived from the maternal side of the family, while the novel SRD5A2 mutation p.H232R was from the paternal side.
Sequence analysis of SRD5A2 in the cases’ father II-1, mother II-2 and younger sister III-3. a. The heterozygous mutation p.Q6X (c.16C > T) in SRD5A2 gene was only found in their mother, not found in their father and younger sister. b. The heterozygous mutation p.H232R (c.695A > G) in SRD5A2 gene was found in their patient's father and younger sister, not found in their mother. Red arrows indicate mutated nucleotide. Green arrows indicate unmutated nucleotide
To further search for the sources of the mutation p.Q6X/p.H232R, Sanger sequencing was performed on cases’ maternal uncle II-3, grandfather I-1, and grandmother I-2. As expected, the maternal uncle II-3 and grandfather I-1 had the nonsense mutation p.Q6X (Fig. 4a), but not the novel mutation p.H232R (Fig. 4b). Cases’ grandmother had neither the mutation p.Q6X (Fig. 4a) nor the mutation p.H232R (Fig. 4b). These results confirmed that the nonsense SRD5A2 mutation p.Q6X originated from cases’ maternal grandfather pedigree.
Sequencing analysis of SRD5A2 gene in the cases’ grandfather I-1, grandmother I-2 and maternal uncle II-3. a. The heterozygous mutation p.Q6X (c.16C > T) in SRD5A2 gene was found in the cases’ grandfather and maternal uncle, but not found in their grandmother. b. The heterozygous mutation p.H232R (c.695A > G) in SRD5A2 gene was not found in the cases’ grandfather, grandmother and maternal uncle. Red arrows: mutated nucleotides. Green arrows: unmutated nucleotides
Pedigree analysis clearly demonstrated that the SRD5A2 mutations exhibited autosomal recessive inheritance. Neither the novel SRD5A2 mutation p.H232R nor the nonsense mutation p.Q6X alone caused 46,XY DSD to develop, but their co-occurrence gave rise to this condition.
p.H232R mutation reduced the catalytic efficiency of the 5α-reductase type 2 enzyme
To investigate the effect of p.H232R mutation on the activity of the enzyme encoded by the SRD5A2 gene, we transfected wild-type SRD5A2, p.H232R mutant SRD5A2, or control pcDNA3.1-GFP plasmids into HEK293 cells. DNA sequencing confirmed that p.H232R mutation was present in the cells transfected with p.H232R mutant SRD5A2 (Fig. 5a). qPCR and western blot confirmed that wild-type or p.H232R mutant SRD5A2 transfection upregulated the SRD5A2 expression in HEK293 cells (Fig. 5b, c). LC–MS analysis showed that p.H232R mutation in the SRD5A2 gene obviously decreased DHT production when cultured with various concentration of testosterone. (Fig. 5d). Using GraphPad Prism 8.0, we found a Km of 12.5 μM, maximum velocity of enzyme-catalyzed reaction (Vm) of 1843 nmol DHT/mg protein/h, and catalytic efficiency (Vmax/Km) of 147.4 nmol DHT/mg protein/h/(μmol/L). In contrast, the cells transfected with plasmid vectors expressing wild-type SRD5A2 had a Km of 9.3 μM, Vmax of 3352 nmol DHT/mg protein/h, and catalytic efficiency (Vmax/Km) of 360.4 nmol DHT/mg protein/h/(μmol/L). These results indicated that p.H232R mutation affected the binding of SRD5A2 with T. In addition, with various concentration of NADPH, LC–MS analysis confirmed that p.H232R mutation decreases the catalytic activity of 5α-reductase 2 (Fig. 5e). Using GraphPad Prism 8.0, we found that the p.H232R mutant enzyme had a Km 11.3 μM, a Vm of 18,038 nmol DHT/mg protein/h, and a catalytic efficiency of 1596.3 nmol DHT/mg protein/h/(μmol/L). In contrast, in the wild-type SRD5A2, the enzyme activity possessed a Km of 1.5 μM, a Vm of 11,077 nmol DHT/mg protein/h, and the catalytic efficiency was 7384.67 nmol DHT/mg protein/h/(μmol/L). These resluts showed that p.H232R mutation affected the binding of SRD5A2 with NADPH. Our mutagenesis studies demonstrated that p.H232R mutation reduced the 5α-reductase 2 enzymatic activity via disrupting the binding of SRD5A2 with T or NADPH.
Validation of SRD5A2 wild-type (WT) and p.H232R mutant HEK293 cell models. a. The HEK293 cells were transiently transfected with WT or p.H232R (c.695A > G) mutant SRD5A2 plasmids. Sequencing analysis showed the mutation c.695A > G in SRD5A2 in HEK-293 cells transfected with p.H232R mutant plasmids, causing amino acid 232 to change from histidine to arginine. b. The transcription of the SRD5A2 gene in HEK-293 cells transfected with WT and p.H232R mutant plasmids using qRT-PCR. The β-actin gene was used as an internal control. The amounts of SRD5A2 transcripts were calculated by the standard 2 − ΔΔCt method and were made into a histogram. c. Western blot results of SRD5A2 protein in HEK-293 cells transfected with WT-Flag and p.H232R-Flag mutant plasmids. β-Actin was used as an internal control. d. Enzyme activity analysis of SRD5A2 WT and mutants H232R binding with T by LC–MS. e. Enzyme activity analysis of SRD5A2 WT and mutants H232R binding with NADPH by LC–MS
Conservation analysis of SRD5A2 in humans and other species
We used MEGA software to analyze the conservation of residues 6 (Fig. 6a) and 232 (Fig. 6b) (marked by red boxes) of the SRD5A2 gene. The H232 amino acid was found to be highly conserved among different species, including human, mouse, rat, cow, nematode, zebrafish, chicken, macaque, chimpanzee, and clawed frog. This suggested that the H232 amino acid is important in organisms and that mutation at this site has a major effect on gene expression.
In silico prediction of pathogenicity of human SRD5A2 p.H232R mutation
Because the identified mutation p.H232R is novel and its pathogenicity is unknown, we first predicted its pathogenicity using software. Three widely used software tools for in silico prediction were used to predict the effect of the human SRD5A2 p.H232R variant: Polyphen2 [18] score was 1, the SIFT [19] score was 0.007, and the PROVEAN [20] score was − 7.330. These scores indicated that the human SRD5A2 p.H232R mutation is highly likely to be pathogenic (for details, see Table 2).
Discussion
In this study, we first found that two 46,XY DSD patients in a single family had a compound SRD5A2 mutation (p.H232R/p.Q6X) [17]. Further sequencing within this family demonstrated that the novel SRD5A2 p.H232R mutation or the nonsense p.Q6X mutation alone would not result in the development of 46,XY DSD. LC–MS showed that the novel mutation (p.H232R) clearly decreased DHT production and SRD5A2 catalytic efficiency.
Mutations in the SRD5A2 gene are common genetic defects causative of 46,XY DSD, which include point mutations, deletions, and insertions [21]. SRD5A2 functions in converting T into DHT [22]. During early development, DHT is responsible for the formation of the male external genitalia, urethra, and prostate [23, 24]. Some mutations in the SRD5A2 gene cause 5α-reductase-2 deficiency syndrome [12]. In one Thai patient with male pseudohermaphroditism, sequencing showed the compound mutation p.Q6X/p.G203S in the SRD5A2 gene [25]. Moreover, in one Chinese patient with hypospadias, sequencing revealed the compound mutation p.G203S/p.Q6X in the SRD5A2 gene [26]. In our research, the two 46,XY DSD cases had the compound heterozygous SRD5A2 mutation p.H232R/p.Q6X. The findings showed that the heterozygous SRD5A2 mutation (p.H232R) or the heterozygous SRD5A2 mutation (p.Q6X) alone would not give rise to 46,XY DSD. Our and others’ researches support that the compound heterozygous SRD5A2 mutation (p.H232R/p.Q6X) results in 46,XY DSD.
The SRD5A2 gene, located on chromosome 2p23.1, encodes a 254-amino-acid protein and contains five exons and four introns [24]. The p.Q6X mutation in the SRD5A2 gene, reported only in Asians, creates a drastically truncated protein with complete loss of enzymatic activity due to the lack of both the T- and the NADPH-binding domains [26, 27]. Histidine 232 is located in a stretch of three histidines (residues 230–232) in the SRD5A2 enzyme. H230P mutation in the SRD5A2 protein inactivates its enzyme activity [28, 29]. Meanwhile, H231R mutation in the SRD5A2 protein impairs its isozyme activity since the mutation primarily affects the ability of the enzyme to bind T [29, 30]. Therefore, we speculate that H232R mutation in the SRD5A2 protein could also inactivate or impair SRD5A2 enzyme activity. Taking this previous work together with our findings, the compound heterozygous SRD5A2 mutation p.H232R/p.Q6X could inhibit DHT formation and cause 46,XY DSD.
Most identified SRD5A2 mutations can reduce DHT levels and cause deficiency of 5α-reductase, which is an autosomal recessive disorder [31, 32]. The p.H232R mutation, located in exon 4 of SRD5A2, is in the T-binding domain and most likely affects the binding of 5α-reductase 2 to T, rendering the former less active. Indeed, it was reported that p.H231R mutation in exon 4 of the SRD5A2 gene causes 46,XY DSD via a mechanism associated with impaired binding of T [30]. Because amino acids 230, 231 and 232 are located in exon 4 and involve the same mutation of histidine to proline or arginine, the mutational impacts are likely to be similar, both preventing binding of the enzyme to its substrate or coenzyme [28, 29]. Our analysis of the enzymatic activity of the SRD5A2 p.H232R mutation showed that the Vmax/Km value was lower than that of WT SRD5A2 and reduced the SRD5A2 catalytic efficiency, suggesting that the p.H232R mutation contributes to the development of genital ambiguity.
Conclusions
Our research strongly suggested that the compound heterozygous SRD5A2 mutation p.H232R/p.Q6X leads to 46,XY DSD and revealed the effect of the SRD5A2 p.H232R mutation on SRD5A2 catalytic efficiency. Further research is needed to construct a mouse model of 46,XY DSD with the SRD5A2 p.H232R mutation, replicate the 46,XY DSD disease phenotype, and investigate the regulatory mechanism of p.H232R on steroid 5α-reductase 2 activity in vivo.
Methods
Patients
Case 1, a 2-year-old girl with 46,XY DSD, attended our hospital. Physical examination of the female external genitalia was performed. B-ultrasound was carried out to detect male or female internal genital organs, including testis, uterus, and ovaries. In addition, hormone assay and karyotype analysis were performed, along with the sequencing of 219 DSD-related genes.
Case 2, case 1’s younger sibling, was suspected of having 46,XY DSD at 25 weeks’ gestation. Type B ultrasonic test, karyotype analysis, and SRD5A2 gene sequencing for 46,XY DSD were performed. Upon abortion of the fetus, physical examinations were performed for female external genitalia and HE staining for testis tissue and epididymis tissue.
Written informed consent for the genetic studies was obtained from the family, and all analyses were approved by the Medical Ethics Committee of the People’s Hospital of Xingtai City.
Hormone assays
Serum levels of FSH, LH, estradiol (E2), progestin (PROG), T, and prolactin (PRL) of the case 1 (III-1) were measured by radioimmunoassay.
Karyotype analysis
Peripheral blood cells from the proband were cultured in RPMI-1640 medium supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, and 10% fetal bovine serum (FBS) for 72 h. Colchicine (20 μg/ml) was added for 2 h to arrest cells in metaphase and inhibit spindle body formation. Samples were incubated for 20 min with 75 mM potassium chloride to spread out the spindle body and fixed in Carnoy’s solution. The fixed cells were dropped onto glass slides and placed in an incubator at 75 °C for 3 h to air-dry. Giemsa solution was used to stain the G-bands of the chromosomes.
FISH analysis
The uncultured amniotic fluid cells from the aborted sibling of the case 1 (III-1) were obtained for FISH analysis. The probes were used to test chromosomes 13, 16, 18, 21, 22, X, and Y.
Genetic analysis
Peripheral blood samples of case 1, her aborted sibling, surviving sibling, father, mother, maternal uncle, grandfather, and grandmother were obtained for DNA sequencing. Genomic DNA was extracted using the TIANamp Blood DNA Kit (Tiangen), in accordance with the manufacturer’s instructions. Sequencing of 219 of the case 1 (III-1)’s DSD-related genes was performed by BGI using Illumina Genome Analyzer IIx (Supplementary Table 1). The mutated genes in case 1(III-1) and her relatives as listed above were subjected to Sanger sequencing. Mutations were identified by comparing the sequencing results of the case to the UCSC reference genome using the BWA tool.
Human SRD5A2 site-directed mutagenesis
Human SRD5A2 cDNA was kindly provided by Professor Jiahuai Han (Xiamen University) and was subcloned into vector pcDNA3.1 (with 3Flag tag, purchased from Life Technologies). To generate 695A > G mutation in the SRD5A2 gene, the pcDNA3.1-SRD5A2 plasmid was used as a template and the QuikChange II Site-Directed Mutagenesis Kit (Catalog #200,523; Agilent) was used to produce the mutation site, in accordance with the manufacturer’s instructions. The primers for cloning the 695A > G mutation in the SRD5A2 gene were as follows: SRD5A2 mF-695: gcgagcttttcaccaccGtaggttctacctcaagatgtttg, and SRD5A2 mR-695: catcttgaggtagaacctaCggtggtgaaaagctcgcag.
Transfection assay
HEK293 cells (purchased from the Chinese Academy of Sciences Cell Bank in Shanghai) were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (Gibco) and 1% streptomycin/penicillin (Gibco). The cells were transiently transfected with 2.0 μg of wild-type or p.H232R mutant SRD5A2 plasmids in each well of a six-well plate using TurboFect™ Transfection Reagent (Thermo Scientific), in accordance with the manufacturer’s protocol. The transfected HEK293 cells were cultured in 5% CO2 at 37 °C for 48 h before the assays.
DNA sequencing of transfection plasmids
HEK293 cells were transfected with wild-type or p.H232R mutant SRD5A2 plasmids for 48 h and Sanger sequencing was used to assess the DNA sequence. Briefly, the DNA samples were isolated by TIANamp Genomic DNA kit (Cat# DP304-02; TIANGEN, China). The concentration and purity of the DNA were determined by NanoDrop (at 260/280 nm, ND2000C; Thermo). The DNA samples were amplified by polymerase chain reaction (PCR) and sequenced by Genewiz Corporation (Suzhou, China). Primer sequences for the PCR were as follows: SRD5A2-Fseq: AGCCCGTTAAGCAGTTGAGG, and SRD5A2-Rseq: CGGCTTCTTCCGCTTCTTGA.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed to assess the mRNA expression after transfection of the wild-type or p.H232R mutant SRD5A2 plasmid for 48 h. Total RNA was isolated by RNAiso Plus (Cat# 9109; TaKaRa) and reverse transcription was performed using the PrimeScript™ RT Reagent Kit (RR047A; Takara), in accordance with the manufacturer’s protocol. qPCR was performed using PrimeScript™ RT Reagent Kit (RR037A; Takara) with the ViiA7 Real-time PCR System (ABI). The PCR schedule was as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. For the relative quantification of SRD5A2 mRNA, the 2−ΔΔCt method was performed. The primer sequences for qPCR were as follows: SRD5A2F: GCCACTTTGGTCGCCCTT, SRD5A2R: CTCCGTGTGCTTCCCGTAG, β-actinF: AGAGCTACGAGCTGCCTGAC, and β-actinR: AGCACTGTGTTGGCGTACAG.
Western blot
Western blot was performed as previously described [33]. Briefly, after HEK293 cells has been transfected with wild-type or p.H232R mutant SRD5A2 plasmids for 48 h, they were collected from six-well plates and lysed with RIPA buffer (P0013; Beyotime Biotechnology). The whole-protein lysates were separated by 12% SDS-PAGE and then transferred to nitrocellulose membranes. After blocking with 5% milk, the membranes were incubated with the primary antibodies monoclonal anti-flag (F9291; Sigma) and β-actin (sc-69879; Santa) at 4°C overnight. After rinsing with Tris-buffered saline containing 1% Tween-20 (TBST), the membrane was then incubated with appropriate HRP-conjugated secondary antibodies (sc516102; Santa Cruz) and detected with an ECL Plus kit (P1050; Applygen).
Kinetic assays
Kinetic assays were performed to assess the influence of p.H232R variant on the binding of 5α-reductase 2 enzyme to testosterone or NADPH. The HEK293 cells transfected with control vector GFP, SRD5A2 or SRD5A2 pH232R was cultured with 500 μl of fresh medium containing 500 μM NADPH (CAS: 100929–71-3, N302057; Aladdin) and various concentrations of T (0.25–8.0 μmol/L, CAS: 58–18-4, M163044; Aladdin). Or the HEK293 cells transfected with control vector GFP, SRD5A2 or SRD5A2 pH232R was cultured with 500 μl of fresh medium containing 10 μM T and various concentrations of NADPH (0.0625–2 mM/L). After incubation for 4 h, the medium was collected. The steroids were extracted with chloroform, condensed by a freeze-drying apparatus, and re-dissolved in chromatographic methanol forLC-MS analysis, using buffer A containing 1 mM formate (F112034; Aladdin) and buffer B containing 100% methanol. DHT was quantified according to the concentration and peak area of the standard DHT [CDCT-C10255010; ANPEL Laboratory Technologies (Shanghai) Inc.]. The protein concentration was determined using BCA assays (P0012S; Beyotime). The rate of enzyme production (nmol/mg protein/h) was calculated as an indicator of enzyme activity. The data were processed by Prism8 (GraphPad). The experiments on each group were repeated three times. The data are presented as mean ± SEM.
Sequence alignment of SRD5A2 in humans and other species
FASTA format SRD5A2 sequences of homologous species were downloaded from the NCBI database. Protein sequences of SRD5A2 were aligned between humans and the other homologous species using MEGA software.
Analysis of the pathogenicity of human SRD5A2 p.H232R mutation
The pathogenicity of the human SRD5A2 p.H232R mutation was analyzed using the following bioinformatic programs: Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.jcvi.org), and PROVEAN (http://provean.jcvi.org/seq_submit.php).
Availability of data and materials
The data that support the findings of this study are available on request from the corresponding author.
Abbreviations
- DSD:
-
Disorder of sex development
- DHT:
-
Dihydrotestosterone
- T:
-
Testosterone
- LC–MS:
-
Liquid chromatography–mass spectrometry
- Km:
-
Apparent michaelis constant
- AIS:
-
Androgen insensitivity syndrome
- FSH:
-
Follicle-stimulating hormone
- LH:
-
Luteinizing hormone
- Vm:
-
Maximum velocity of enzyme-catalyzed reaction
- E2:
-
Estradiol
- PROG:
-
Progestin
- PRL:
-
Prolactin
References
Berglund A, Johannsen TH, Stochholm K, Viuff MH, Fedder J, Main KM, et al. Morbidity, Mortality, and Socioeconomics in Females With 46, XY Disorders of Sex Development: A Nationwide Study. J Clin Endocrinol Metab. 2018;103(4):1418–28.
Gomez-Lobo V, AmiesOelschlager AM, P for North American Society, G Adolescent. Disorders of Sexual Development in Adult Women. Obstet Gynecol. 2016;128(5):1162–73.
Mongan NP, Tadokoro-Cuccaro R, Bunch T, Hughes IA. Androgen insensitivity syndrome. Best Pract Res Clin Endocrinol Metab. 2015;29(4):569–80.
Byers HM, Mohnach LH, Fechner PY, Chen M, Thomas IH, Ramsdell LA, et al. Unexpected ethical dilemmas in sex assignment in 46, XY DSD due to 5-alpha reductase type 2 deficiency. Am J Med Genet C Semin Med Genet. 2017;175(2):260–7.
Fabbri-Scallet H, de Mello MP, Guerra-Junior G, Maciel-Guerra AT, de Andrade JGR, de Queiroz CMC, et al. Functional characterization of five NR5A1 gene mutations found in patients with 46, XY disorders of sex development. Hum Mutat. 2018;39(1):114–23.
Lee PA, Nordenstrom A, Houk CP, Ahmed SF, Auchus R, Baratz A, et al. Global Disorders of Sex Development Update since 2006: Perceptions. Approach and Care Horm Res Paediatr. 2016;85(3):158–80.
Bashamboo A, Donohoue PA, Vilain E, Rojo S, Calvel P, Seneviratne SN, et al. A recurrent p.Arg92Trp variant in steroidogenic factor-1 (NR5A1) can act as a molecular switch in human sex development. Hum Mol Genet. 2016;25(16):3446–53.
Petroli RJ, Hiort O, Struve D, Gesing JK, Soardi FC, Spinola-Castro AM, et al. Functional Impact of Novel Androgen Receptor Mutations on the Clinical Manifestation of Androgen Insensitivity Syndrome. Sex Dev. 2017;11(5–6):238–47.
Jung EJ, Im DH, Park YH, Byun JM, Kim YN, Jeong DH, et al. Female with 46. XY karyotype Obstet Gynecol Sci. 2017;60(4):378–82.
Andonova S, Robeva R, Vazharova R, Ledig S, Grozdanova L, Stefanova E, et al. New Territory for an Old Disease: 5-Alpha-Reductase Type 2 Deficiency in Bulgaria. Sex Dev. 2017;11(1):21–8.
Bertelloni S, Baldinotti F, Russo G, Ghirri P, Dati E, Michelucci A, et al. 5alpha-Reductase-2 Deficiency: Clinical Findings, Endocrine Pitfalls, and Genetic Features in a Large Italian Cohort. Sex Dev. 2016;10(1):28–36.
Kang HJ, Imperato-McGinley J, Zhu YS, Rosenwaks Z. The effect of 5alpha-reductase-2 deficiency on human fertility. Fertil Steril. 2014;101(2):310–6.
Zhang K, Li Y, Mao Y, Ma M. Steroid 5-alpha-reductase type 2 (SRD5A2) gene V89L polymorphism and hypospadias risk: A meta-analysis. J Pediatr Urol. 2017;13(6):630.
Sata F, Kurahashi N, Ban S, Moriya K, Tanaka KD, Ishizuka M, et al. Genetic polymorphisms of 17 beta-hydroxysteroid dehydrogenase 3 and the risk of hypospadias. J Sex Med. 2010;7(8):2729–38.
Chan AO, But WM, Lee CY, Lam YY, Ng KL, Loung PY, et al. Aetiological bases of 46, XY disorders of sex development in the Hong Kong Chinese population. Hong Kong Med J. 2015;21(6):499–510.
Kim JH, Kang E, Heo SH, Kim GH, Jang JH, Cho EH, et al. Diagnostic yield of targeted gene panel sequencing to identify the genetic etiology of disorders of sex development. Mol Cell Endocrinol. 2017;444:19–25.
Li SP, Li LW, Sun MX, Chen XX, Wang XF, Li ZK, et al. Identification of a novel mutation in the SRD5A2 gene of one patient with 46, XY disorder of sex development. Asian J Androl. 2018;20(5):518–9.
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;7(7):20.
Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31(16):2745–7.
Fu XH, Zhang WQ, Qu XS. Correlation of androgen receptor and SRD5A2 gene mutations with pediatric hypospadias in 46, XY DSD children. Genet Mol Res. 2016;15(1):15018232.
Audet-Walsh É, Yee T, Tam I, Giguère V. Inverse Regulation of DHT Synthesis Enzymes 5α-Reductase Types 1 and 2 by the Androgen Receptor in Prostate Cancer. Endocrinology. 2017;158(4):1015–21.
Kang H-J, Imperato-McGinley J, Zhu Y-S, Rosenwaks Z. The effect of 5α-reductase-2 deficiency on human fertility. Fertil Steril. 2014;101(2):310–6.
Okeigwe I, Kuohung W. 5-Alpha reductase deficiency: a 40-year retrospective review. Current opinion in endocrinology, diabetes, and obesity. 2014;21(6):483–7.
Sahakitrungruang T, Wacharasindhu S, Yeetong P, Snabboon T, Suphapeetiporn K, Shotelersuk V. Identification of mutations in the SRD5A2 gene in Thai patients with male pseudohermaphroditism. Fertil Steril. 2008;90(5):2015.
Zhang M, Yang J, Zhang H, Ning G, Li X, Sun S. A novel SRD5A2 mutation with loss of function identified in Chinese patients with hypospadias. Horm Res Paediatr. 2011;76(1):44–9.
Wang Y, Li Q, Xu J, Liu Q, Wang W, Lin Y, et al. Mutation analysis of five candidate genes in Chinese patients with hypospadias. Eur J Hum Genet. 2004;12(9):706–12.
Wilson JD, Griffin JE, Russell DW. Steroid 5 alpha-reductase 2 deficiency. Endocr Rev. 1993;14(5):577–93.
Wigley WC, Prihoda JS, Mowszowicz I, Mendonca BB, New MI, Wilson JD, et al. Natural mutagenesis study of the human steroid 5 alpha-reductase 2 isozyme. Biochemistry. 1994;33(5):1265–70.
Nordenskjold A, Ivarsson SA. Molecular characterization of 5 alpha-reductase type 2 deficiency and fertility in a Swedish family. J Clin Endocrinol Metab. 1998;83(9):3236–8.
Thigpen AE, Davis DL, Milatovich A, Mendonca BB, Imperato-McGinley J, Griffin JE, et al. Molecular genetics of steroid 5 alpha-reductase 2 deficiency. J Clin Invest. 1992;90(3):799–809.
Mendonca BB, Batista RL, Domenice S, Costa EM, Arnhold IJ, Russell DW, et al. Steroid 5alpha-reductase 2 deficiency. J Steroid Biochem Mol Biol. 2016;163:206–11.
Dou XW, Liang YK, Lin HY, Wei XL, Zhang YQ, Bai JW, et al. Notch3 Maintains Luminal Phenotype and Suppresses Tumorigenesis and Metastasis of Breast Cancer via Trans-Activating Estrogen Receptor-alpha. Theranostics. 2017;7(16):4041–56.
Acknowledgements
We thank the family and medical staff involved in this study.
Funding
This work was funded by the National Natural Science Foundation of China (No. 81860516), Science and Technology Foundation of Guizhou Health Committee (gzwjkj2019-1–036) the National Natural Science Fund Cultivation Project of Guizhou Medical University (20NSP025) and Xingtai Science and Technology Bureau (No. 2020ZC343).
Author information
Authors and Affiliations
Contributions
LW Li, JH Zhang, Q Li, L Qiao, PC Li, Y Cui, SJ Li, SR Hao, TQ Wu, LL Liu, JM Yin, and PS Hu carried out the molecular genetic studies, participated in the sequence alignment, and drafted the manuscript. SP Li and XW Dou carried out the immunoassay. H Yang participated in the sequence alignment. XW Dou and H Yang designed the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The experiment was approved by the Medical Ethics Committee of the People’s Hospital of Xingtai City. Parental/guardian consent was obtained.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, L., Zhang, J., Li, Q. et al. Mutational analysis of compound heterozygous mutation p.Q6X/p.H232R in SRD5A2 causing 46,XY disorder of sex development. Ital J Pediatr 48, 47 (2022). https://doi.org/10.1186/s13052-022-01243-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13052-022-01243-4