Abstract
Background
Infection with the hepatitis B virus (HBV) is an independent risk factor for liver cirrhosis and hepatocellular carcinoma, polymorphisms in HLA-DQB1 play an important role in HBV infections.
Methods
This study examined the relationships between HLA-DQB1 alleles and HBV infection susceptibility among 256 HBV carriers and 433 healthy controls. Venous blood samples were subjected to DQB1 high-resolution typing and testing for interferon-gamma, interleukin-4 (IL-4), interleukin-10, and DQB1 mRNA expression. A meta-analysis was also performed using relevant case–control studies that evaluated the associations of HLA-DQB1 alleles with HBV infection and clearance.
Results
We found that HLA-DQB1*06:03 protected against HBV infection. Levels of IFN-γ and IL-4 were significantly elevated in HBV cases with HLA-DQB1*06:05 (vs. HLA-DQB1*05:03), and the HBV group had higher DQB1 mRNA expression than the healthy control group with HLA-DQB1*05:03 and HLA-DQB1*06:02. The meta-analysis revealed that HLA-DQB1*04:01, HLA-DQB1*05:02, HLA-DQB1*05:03, and HLA-DQB1*06:01 were risk factors for HBV infection susceptibility, while HLA-DQB1*05:01, HLA-DQB1*06:03, and HLA-DQB1*06:04 protected against HBV infection. Spontaneous HBV clearance was associated withHLA-DQB1*06:04, while chronic HBV infection was associated with HLA-DQB1*02:01 and HLA-DQB1*05:02.
Conclusion
DBQ1 typing can be used to identify patients who have elevated risks of HBV infection (i.e., patients with HLA-DQB1*04:01, HLA-DQB1*05:02, HLA-DQB1*05:03, and HLA-DQB1*06:01) or elevated risks of chronic HBV infection (i.e., patients with HLA-DQB1*02:01 and HLA-DQB1*05:02).
Similar content being viewed by others
Background
Infection with the hepatitis B virus (HBV) is an independent risk factor for liver cirrhosis and hepatocellular carcinoma [1]. There are > 68 million HBV carriers in China, and a national survey revealed that the seroprevalence rate was 7.18% in 2006 [2]. The HBV vaccine became widespread in China during 1992, and the seroprevalence of hepatitis B surface antigen (HBsAg) has decreased to approximately 5.49% in 2015 [3]. Nevertheless, given the size of the Chinese population, approximately 93 million people carry the HBsAg and approximately 30 million people have HBV-related diseases [4]. Therefore, HBV infection is associated with a significant public health burden in China [5].
Multiple factors influence the risk of chronic HBV infection or spontaneous HBV clearance, such as age, location, sex, body mass index, ethnicity, viral mutation, HBV virus genotype, host genetic variations, and host immune responses. In this context, human leukocyte antigens (HLA) play key roles in mediating the immune response. For example, HLA-I is present on the surface of all nucleated cells, where it presents exogenous antigens and is recognized by T cell receptors, which facilitates the adaptive immune response. In addition, HLA-II is expressed on the surface of antigen-presenting cells (APCs), which present endogenous antigens and are recognized by T-cell receptors as part of the adaptive immune response. Several polymorphisms in the HLA loci, especially HLA-II, are related to HBV infection susceptibility or spontaneous HBV clearance [6,7,8,9,10]. Furthermore, HLA-DQB1 gene polymorphisms are associated with the progression of human immunodeficiency virus infection [11], HCV infection susceptibility and spontaneous clearance [12], coronary artery disease [13], rheumatoid arthritis [14], and other diseases.
Previous studies [15] have demonstrated that the beta 1 subunit of the HLA-DQ surface receptor (HLA-DQB1) is associated with the immunological response to the hepatitis B vaccine. For example, increased antibody responses were observed in cases with HLA-DQB1*05:01 and HLA-DQB1*0602, while decreased antibody responses were observed in cases with HLA-DQB1*02. Furthermore, cases with HLA-DRB1*1301 and HLA-DQB1*0602 had high response specificity but low response sensitivity. Moreover, a previous meta-analysis [16] has indicated that some HLA-DQB1 polymorphisms (HLA-DQB1*02:01, HLA-DQB1*03:01, and HLA-DQB1*05:02) are associated with susceptibility to chronic hepatitis B (CHB), while decreased susceptibility to CHB was observed for HLA-DQB1*03:03 and HLA-DQB1*06:04. However, that study did not evaluate the association of HLA-DQB1 polymorphisms with spontaneous HBV clearance, and also did not include a large recent case–control study. Thus, it is unclear why some alleles are risk factors or protective factors for HBV infection. The present study aimed to examine the different HLA-DQB1 alleles, their associations with various cytokines (interferon-gamma [IFN-γ], interleukin-4 [IL-4], and interleukin-10 [IL-10]), and the various HLA-DQB1 mRNA expressions among HBV carriers and healthy controls. Furthermore, we performed a meta-analysis of the various alleles’ associations with HBV infection susceptibility and spontaneous HBV clearance.
Methods
Samples
The present study included samples from 433 healthy individuals who provided blood to the Deyang central blood station. A total of 256 patients with HBV infection were also enrolled during physical examinations at the Deyang People’s Hospital, Sichuan, China. Patients with HBV infection were identified has on serological results for HBsAg, antibodies to HBsAg, antibodies to Hepatitis Be, and antibodies to the HBV core antigen. Healthy controls (HCs) were seronegative for any HBV biomarkers. HBsAg/HBeAg/HBcAb/HBeAb/HBcAb-seropositive volunteers or patients were considered to be HBV carriers and all the patients were enrolled according to the related diagnostic criteria of 2010 “Chronic hepatitis B prevention and treatment guidelines” [17], HBV surface markers were used to determine the infected patients and the healthy controls. Since our research only aimed to explore the relationship between the DQB1 gene and HBV susceptibility, the relationship between DQB1 and the HBV infection process was not a main concern for us and therefore HCC and LC were not considered in this research. All blood samples were negative for the hepatitis C virus and human immunodeficiency virus. Asymptomatic carriers were defined as patients who were seropositive for HBsAg, had normal serum alanine aminotransferase and aspartate transaminase levels (< 35 U/L), and had no clear clinical symptoms without using antiretroviral drugs.
HLA-DQB1 genotyping
The HLA-DQB1 genotyping was performed using peripheral blood samples that were collected into EDTA-coated tubes. High-quality genomic DNA were extracted from 400 µL of blood using a DNA extraction kit (Tiangen, Beijing, China), according to the manufacturer’s instructions. The DNA concentrations were 20–50 ng/µL, based on A260/280 values of 1.8–2.0. The genotyping was performed using PCR sequence-based typing and an ABI 3730 DNA Sequencer (Applied Biosystems, Foster City, CA). Amplification and sequencing primers were created using the sequences that were reported by van Dijk et al. [18]. The HLA-DQB1 alleles were identified based on the samples’ sequences using Utype software (Thermofisher, Waltham, USA).
HLA-DQB1 mRNA measurements
The RNA were prepared from suspensions of freshly isolated peripheral blood mononuclear cells using a TRIzol method (Invitrogen, USA). The samples were treated using RNase-free DNase I (Qiagen, Germany) to eliminate contaminant genomic DNA, and then quantified using A260/280 measurements. Reverse transcription was used to obtain cDNA, based on 1 µg of total RNA and High Capacity cDNA Reverse Transcription Kits (Invitrogen), according to the manufacturer’s instructions. The mRNA expressions were quantified using SYBR green quantitative PCR and a CFX96 Touch PCR machine (Bio-Rad), based on the threshold cycle method. The forward primer sequence was TGGAGCACCCCAGCCT and the reverse primer sequence was ATSAGCCCCAGCACGAA. Each PCR tube included 12.5 µL of FastStart Universal SYBR Green Master (Roche, Switzerland), 200 nM of the primers, and 2.5 µL of cDNA in a total volume of 25 µL. The specificity of the reaction was confirmed using melt curve analysis for the dissociation step after the recommended Roche qPCR protocol, and all reactions were standardized using the expression of GAPDH.
Meta-analysis
The effects of the HLA-DQB1 alleles on HBV infection susceptibility was evaluated using a meta-analysis. Relevant reports were identified using searches of the PubMed, EBSCO, Elsevier, and Web of Science databases up to September 2017. Reports in all languages were considered eligible for inclusion, and the search terms were set to ‘Hepatitis B’ and ‘HLA-DQB1’ without any other limits. The inclusion criteria were: (a) studies that analysed the genotype frequencies of HLA-DQB1 in HBV carriers and healthy HBV-uninfected controls, (b) studies that used a case–control design, (c) reports that provided the specific enrolment criteria, and (d) reports that included the number of cases, number of controls, and allele frequencies. All other studies were excluded.
Statistical analysis
The Hardy–Weinberg equilibrium of the genotype distributions and the linkage disequilibrium of the single-nucleotide polymorphisms SNPs were examined using Arliquin software (version 3.5). Categorical and continuous variables were compared using the χ2 test and Student’s t test, respectively,multiple comparisons were adjusted by Bonferroni correction. The meta-analysis was performed using Review Manager Software (version 5.2), and the results were reported as ORs and 95% CIs. Heterogeneity between the studies was examined using the I2 test. The fixed-effect model was used if there was < 50% heterogeneity, otherwise the random-effect model was used. All tests were two-sided, and P-values of < 0.05 were considered statistically significant.
Results
Participant characteristics
The HBV carriers and healthy controls had similar values for age, sex, alanine transaminase, aspartate transaminase, albumin, and total bilirubin (Additional file 1: Table S1).
Relationship between HLA-DQB1 and HBV infection susceptibility
The distributions of the HLA-DQB1 alleles in the HBV and healthy control groups were fit using the Hardy–Weinberg equilibrium. The results indicated that carriage of HLA-DQB1*06:03 protected against chronic HBV infection (odds ratio [OR] 0.009, 95% confidence interval [CI] 0.02–0.86) (Table 1).
Levels of IFN-γ, IL-4, and IL-10 in patients with HBV infection
Patients with HBV infection had significantly lower levels of IFN-γ, IL-4, and IL-10. In the HBV group, significantly higher levels of IFN-γ and IL-4 were associated with HLA-DQB1*05:01 and *06:05, compared to HLA-DQB1*05:03 (Figs. 1, 2). However, no significant differences in IL-10 levels were observed between the various HLA-DQB1 alleles (Additional file 1: Fig S1).
HLA-DQB1 mRNA expression
Compared to the healthy control group, HLA-DQB1*05:03 and DQB1*06:02 in the HBV group were associated with significantly elevated HLA-DQB1 mRNA expression (Fig. 3).
Meta-analysis of HLA-DQB1 alleles and HBV susceptibility
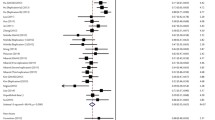
The meta-analysis evaluated 6 case–control studies regarding HLA-DQB1 and HBV infection susceptibility, which included 3459 cases of HBV infection and 7424 healthy controls (Table 2 and Additional file 1: Table S2). The risk factors for HBV infection susceptibility were HLA-DQB1*04:01 (OR 1.33, 95% CI 1.02–1.73, P = 0.03), HLA-DQB1*05:02 (OR 1.22, 95% CI 1.04–1.42, P = 0.01), HLA-DQB1*05:03 (OR 1.16, 95% CI 1.02–1.31, P = 0.02), and HLA-DQB1*06:01 (OR 1.32, 95% CI 1.02–1.70, P = 0.03). The factors that protected against HBV infection were HLA-DQB1*05:01 (OR 0.59, 95% CI 0.51–0.69, P < 0.00001), HLA-DQB1*06:03 (OR 0.28, 95% CI 0.08–1.00, P = 0.05), and HLA-DQB1*06:04 (OR 0.31, 95% CI 0.20–0.49, P < 0.00001) (Table 3). The forest plots are shown in Additional file 1: Figures S2–S8.
Meta-analysis of HLA-DQB1 alleles and spontaneous HBV clearance
The meta-analysis evaluated 7 case–control studies regarding HLA-DQB1 and spontaneous HBV clearance, which included 602 cases with spontaneous HBV clearance and 954 cases with chronic HBV infection (Table 4 and Additional file 1: Table S3). Spontaneous HBV clearance was significantly associated with HLA- DQB1*06:04 (OR 2.00, 95% CI 1.07–3.74, P = 0.03). Chronic HBV infection was significantly associated with HLA-DQB1*02:01 (OR 0.77, 95% CI 0.60–0.98, P = 0.04) and HLA-DQB1*05:02 (OR 0.44, 95% CI 0.27–0.70, P = 0.0006) (Table 5). The forest plots are shown in Additional file 1: Figures S9–S11.
Discussion
The associations between HLA expression and HBV infection or clearance have been intensively assessed across various populations[19−21]. For example, Nishda et al. [22] evaluated 1033 Japanese patients with HBV infection and 942 healthy controls, and found that HLA-DQB1*06:01 had the strongest association with CHB susceptibility. Mbarek et al. [20] also examined three independent Japanese cohorts (2209 CHB cases and 4440 controls), and found that HLA-DQA1∗01:02-DQB1∗06:04 and HLA-DQA1∗01:01-DQB1∗05:01 protected against CHB, while HLA-DQA1∗01:02-DQB1∗03:03 and HLA-DQA1∗03:01-DQB1∗06:01 were risk factors for CHB. Furthermore, Zhang et al. [19]. found that HLA-DQB1*03:01 was closely related to CHB susceptibility, while Park et al. [23] reported that HLA-DQB1*04:02 and HLA-DQB1*06:04 protected against CHB. Chen et al. [24] confirmed that HLA-DQ1*06:04 was associated with susceptibility to HBV infection among Caucasian individuals. However, in our Sichuan population, we found that HLA-DQB1*06:03 independently protected against HBV infection. Given the varying results in the different studies and patient populations, we performed a meta-analysis of 6 reports that included Chinese, Japanese, Korean, and Caucasian patients. Among the 3459 cases of HBV infection and 7424 healthy controls, susceptibility to HBV infection was associated with HLA-DQB1*04:01, HLA-DQB1*05:02, HLA-DQB1*05:03, and HLA-DQB1*06:01. However, HLA-DQB1*05:01, HLA-DQB1*06:03, and HLA-DQB1*06:04 protected against HBV infection.
The associations of HLA expression with spontaneous HBV clearance has also been investigated in several studies. For example, Zhou et al. [19] found that HLA-DQB1*02:01 protected against HBV infection, while HLA-DQB1*03:01 was associated with chronic HBV infection in Xinjiang. Cho et al. [25] have also found that HLA-DQB1*06:09 was unexpectedly and strongly associated with HBV clearance. Furthermore, Liu et al. [26] demonstrated that HLA-DQB1*02:01 and HLA-DQB1*06:01 conferred susceptibility to chronic HBV infection. Zhu et al. [21] have also reported that HLA-DQB1*05:02 was independently associated with the outcomes of HBV infection. Thus, we performed a meta-analysis of the 7 reports that included Chinese, Korean, African American, and Caucasian patients. This analysis revealed that spontaneous HBV clearance was significantly associated with HLA- DQB1*06:04, while chronic HBV infection was significantly associated with HLA-DQB1*02:01 and HLA-DQB1*05:02. An easy conclusion is that DQB1*05:02 was not only associated with susceptible of HBV infection, but also with persistent chronic HBV infection. The DQB1*06:04, on the other hand, was related to HBV protection and HBV spontaneous clearance. We also found that some risk alleles or protect alleles varies between different researches. This heterogeneity was also shown in the meta-analysis: the heterogeneity (I2 > 50%) between DQB1 alleles and HBV susceptibility were obviously for DQB1*04:01, 06:01, 06:03 and 06:04. As host-virus immune interaction might be influenced by viral genotype. Doganay et al. reported [27] that DQB1 05:01 was associated with chronic active disease where all patients were exclusively genotype D. As we know, genotype B and C were dominantly prevalent in Chinese HBV infections while the HBV genotype in European and American infections were mainly A and D. Therefore, the difference in the HBV virus genotype in different regions may also one important factor for HBV susceptibility and could account for the heterogeneity of the DQB1 gene polymorphism and HBV infection outcomes.
APCs express HLA-II molecules (HLA-DR, HLA-DQ, and HLA-DP) to activate CD4+ helper T-cells, which play a crucial role in the immune response to HBV infection. The present study revealed that HBV infection was associated with significantly lower levels of INF-γ, IL-4, and IL-10, while previous studies have not examined this issue. Furthermore, we observed that HLA-DQB1*05:03 in the HBV group was associated with significantly lower IFN-γ and IL-4 levels, and the meta-analysis revealed that HLA-DQB1*05:03 was associated with HBV infection. Thus, the lower levels of IFN-γ and IL-4 may reflect a reduced immune response in cases with this allele, which could increase susceptibility to HBV.
O’Brien et al. [28] have reported that the rs3077TT genotype was associated with higher HLA-DPB1 mRNA expression in the liver, which was also associated with a lower risk of chronic HBV infection. In addition, Thomas et al. [9] demonstrated that the rs9277534GG genotype confers susceptibility to chronic HBV infection, with significantly decreased levels of HLA-DP transcripts and surface protein in HBV donors. In contrast, the present study revealed that plasma expression of HLA-DQB1 mRNA was elevated in cases with HLA-DQB1*05:03, which was associated with HBV infection susceptibility. A previous study [29] also detected elevated HLA-DQB1 mRNA expression in cases with HLA-DQB1*06:02, which was associated with narcolepsy, We also found that the risk allele DQB1*05:03 have significantly lower IFN-γ and IL-4 levels compared than protect allele DQB1*05:01, at the same time, the immune negative regulatory factor IL-10 have no difference between risk alleles and protect alleles in HBV group. From these results, we could postulate that some risk DQB1 alleles may increase DQB1 mRNA expression, resulting in decreased immunity related cytokines. Thus, additional studies and large population studies are needed to understand the relevance of HLA-DQB1 expression, especially in the context of APCs and different HBV genotype-specific antigens.
Conclusion
HBV infection susceptibility was associated with HLA-DQB1*04:01, HLA-DQB1*05:02, HLA-DQB1*05:03, and HLA-DQB1*06:01, while protection against HBV infection was associated with HLA-DQB1*05:01, HLA-DQB1*06:03, and HLA-DQB1*06:04. Spontaneous HBV clearance was associated with HLA-DQB1*02:01 and HLA-DQB1*05:02, while chronic HBV infection was associated with HLA-DQB1*06:04. In cases of HBV infection, HLA-DQB1*05:03 was associated with low levels of IFN-γ andIL-4 secretion, and with increased HLA-DQB1 mRNA expression.
Abbreviations
- HBV:
-
hepatitis B virus
- IFN-γ:
-
interferon-gamma
- IL-4:
-
interleukin-4
- IL-10:
-
interleukin-10
- HBsAg:
-
hepatitis B surface antigen
- HLA:
-
human leukocyte antigens
- APCs:
-
antigen-presenting cells
References
Budny A, Kozlowski P, Kaminska M, Jankiewicz M, Kolak A, Budny B, Budny W, Niemunis-Sawicka J, Szczypior G, Kurniawka B, Burdan F. Epidemiology and risk factors of hepatocellular carcinoma. Pol Merkur Lekarski. 2017;43:133–9.
Liang X, Bi S, Yang W, Wang L, Cui G, Cui F, Zhang Y, Liu J, Gong X, Chen Y, et al. Epidemiological serosurvey of hepatitis B in China—declining HBV prevalence due to hepatitis B vaccination. Vaccine. 2009;27:6550–7.
Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–55.
Cui Y, Jia J. Update on epidemiology of hepatitis B and C in China. J Gastroenterol Hepatol. 2013;28(Suppl 1):7–10.
Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099–108.
Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591–5.
An P, Winkler C, Guan L, O’Brien SJ, Zeng Z, Consortium HBVS. A common HLA-DPA1 variant is a major determinant of hepatitis B virus clearance in Han Chinese. J Infect Dis. 2011;203:943–7.
Li J, Yang D, He Y, Wang M, Wen Z, Liu L, Yao J, Matsuda K, Nakamura Y, Yu J, et al. Associations of HLA-DP variants with hepatitis B virus infection in southern and northern Han Chinese populations: a multicenter case–control study. PLoS ONE. 2011;6:e24221.
Thomas R, Thio CL, Apps R, Qi Y, Gao X, Marti D, Stein JL, Soderberg KA, Moody MA, Goedert JJ, et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. 2012;86:6979–85.
Wong DK, Watanabe T, Tanaka Y, Seto WK, Lee CK, Fung J, Lin CK, Huang FY, Lai CL, Yuen MF. Role of HLA-DP polymorphisms on chronicity and disease activity of hepatitis B infection in Southern Chinese. PLoS ONE. 2013;8:e66920.
Li W, Li C, Xia W, Li X. HLA-DQB1*06 and breadth of Nef core region-specific T-cell response are associated with slow disease progression in antiretroviral therapy-naive Chinese HIV-1 subtype B patients. Hum Vaccin Immunother. 2017;13(10):2341–7.
Waldron PR, Belitskaya-Levy I, Chary A, Won J, Winters M, Monto A, Ryan J, Lazzeroni LC, Holodniy M. Genetic variation in the IL-6 and HLA-DQB1 genes is associated with spontaneous clearance of hepatitis C virus infection. J Immunol Res. 2016;2016:6530436.
Xiong Y, Wang L, Mo P, Huang G, Li A, Chai R, Lin X, Zhong Y, Liu B, Ou W, et al. Association between HLA-DQB1 alleles and susceptibility to coronary artery disease in Southern Han Chinese. Hum Immunol. 2017;78:540–6.
Wu J, Li J, Li S, Zhang TP, Li LJ, Lv TT, Pan HF, Ye DQ. Association of HLA-DQB1 polymorphisms with rheumatoid arthritis: a meta-analysis. Postgrad Med J. 2017;93:618–25.
Li ZK, Nie JJ, Li J, Zhuang H. The effect of HLA on immunological response to hepatitis B vaccine in healthy people: a meta-analysis. Vaccine. 2013;31:4355–61.
Huang J, Xiong L, Wang J, Liu Y, Zhu Q, Lei J, Zhou Z. Association between the HLA-DQB1 polymorphisms and the susceptibility of chronic hepatitis B: a comprehensive meta-analysis. Biomed Rep. 2016;4:557–66.
Chinese Society of Hepatology CMA. Chinese Society of Infectious Diseases CMA: [The guidelines of prevention and treatment for chronic hepatitis B]. Zhonghua Gan Zang Bing Za Zhi. 2005;13:881–91.
van Dijk A, Melchers R, Tilanus M, Rozemuller E. HLA-DQB1 sequencing-based typing updated. Tissue Antigens. 2007;69(Suppl 1):64–5.
Zhang Y, Zhao F, Lan L, Qin Z, Jun L. Correlation of HLA-DQB1 gene polymorphism of Xinjiang Uygur with outcome of HBV infection. Int J Clin Exp Med. 2015;8:6067–72.
Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, Takahashi A, Kamatani Y, Miki D, Abe H, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011;20:3884–92.
Zhu XL, Du T, Li JH, Lu LP, Guo XH, Gao JR, Gou CY, Li Z, Liu Y, Li H. Association of HLA-DQB1 gene polymorphisms with outcomes of HBV infection in Chinese Han population. Swiss Med Wkly. 2007;137:114–20.
Nishida N, Ohashi J, Khor SS, Sugiyama M, Tsuchiura T, Sawai H, Hino K, Honda M, Kaneko S, Yatsuhashi H, et al. Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis. Sci Rep. 2016;6:24767.
Park MH, Song EY, Ahn C, Oh KH, Yang J, Kang SJ, Lee HS. Two subtypes of hepatitis B virus-associated glomerulonephritis are associated with different HLA-DR2 alleles in Koreans. Tissue Antigens. 2003;62:505–11.
Chen DF, Kliem V, Endres W, Brunkhorst R, Tillmann HL, Koch KM, Manns MP, Stangel W. Relationship between human leukocyte antigen determinants and courses of hepatitis B virus infection in Caucasian patients with end-stage renal disease. Scand J Gastroenterol. 1996;31:1211–5.
Cho SW, Cheong JY, Ju YS, Oh DH, Suh YJ, Lee KW. Human leukocyte antigen class II association with spontaneous recovery from hepatitis B virus infection in Koreans: analysis at the haplotype level. J Korean Med Sci. 2008;23:838–44.
Liu C, Cheng B. Association of polymorphisms of human leucocyte antigen-DQA1 and DQB1 alleles with chronic hepatitis B virus infection, liver cirrhosis and hepatocellular carcinoma in Chinese. Int J Immunogenet. 2007;34:373–8.
Doganay L, Fejzullahu A, Katrinli S, Yilmaz Enc F, Ozturk O, Colak Y, Ulasoglu C, Tuncer I, Dinler Doganay G. Association of human leukocyte antigen DQB1 and DRB1 alleles with chronic hepatitis B. World J Gastroenterol. 2014;20:8179–86.
O’Brien TR, Kohaar I, Pfeiffer RM, Maeder D, Yeager M, Schadt EE, Prokunina-Olsson L. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun. 2011;12:428–33.
Weiner Lachmi K, Lin L, Kornum BR, Rico T, Lo B, Aran A, Mignot E. DQB1*06:02 allele-specific expression varies by allelic dosage, not narcolepsy status. Hum Immunol. 2012;73:405–10.
Authors’ contributions
GO, HX, HY and XJ performed the experiments. JW and ZL designed the experiments and performed quality control. XL and LY collected and evaluated the samples. OG and ZL wrote the original draft of the manuscript. ZL designed the experiments, analysed the data, and contributed to the discussion. JW and ZL performed the data analysis, discussed the results, and substantially revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank the participants for generously providing the venous blood samples. We thank Yuan Xue for critical review and language proof of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Data availability
All the data and materials supporting the conclusions were included in the main paper.
Ethics approval and consent to participate
This study was approved by the ethic committees of the Institution of Blood Transfusion,CAMS & PUMC, and was conducted according to the principles of the Declaration of Helsinki. All participants provided written informed consent before enrolment, and the study’s protocol was approved by the ethic committees of the Institution of Blood Transfusion, CAMS & PUMC.
Funding
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) under contract 2016-I2 M-3-024, Funding of Sichuan Science and Technology Department under Contract 2017RZ0047 and Ministry of Science and Technology of China, Grant/Award Number: 2014EG150133. The CIFMS program provided us blood samples and relevant data, funding of Sichuan Science and Technology Department project provide us the cytokine detect methods and methods of HLA-DQB1 typing, program of Ministry of Science and Technology of China provide us HBV relevant detect methods.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Table S1
The characteristic of the participates. Table S2. Distribution of HLA-DQB1 alleles in different studies. Table S3. Distribution of the HLA-DQB1 polymorphism in HBV spontaneous clearance. Figure S1. IL-10 with different DQB1 alleles in HBV group. Figure S2. Meta-analysis of correlation of the HLA-DQB1*04:01 allele polymorphism in HBV susceptibility. Figure S3. Meta-analysis of correlation of the HLA-DQB1*05:01 allele polymorphism in HBV susceptibility. Figure S4. Meta-analysis of correlation of the HLA-DQB1*05:02 allele polymorphism in HBV susceptibility. Figure S5. Meta-analysis of correlation of the HLA-DQB1*05:03 allele polymorphism in HBV susceptibility. Figure S6. Meta-analysis of correlation of the HLA-DQB1*06:01 allele polymorphism in HBV susceptibility. Figure S7. Meta-analysis of correlation of the HLA-DQB1*06:03 allele polymorphism in HBV susceptibility. Figure S8. Meta-analysis of correlation of the HLA-DQB1*06:04 allele polymorphism in HBV susceptibility. Figure S9. Meta-analysis of correlation of the HLA-DQB1*02:01 allele polymorphism in HBV spontaneous clearance. Figure S10. Meta-analysis of correlation of the HLA-DQB1*05:02 allele polymorphism in HBV spontaneous clearance. Figure S11. Meta-analysis of correlation of the HLA-DQB1*06:04 allele polymorphism in HBV spontaneous clearance.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ou, G., Xu, H., Yu, H. et al. The roles of HLA-DQB1 gene polymorphisms in hepatitis B virus infection. J Transl Med 16, 362 (2018). https://doi.org/10.1186/s12967-018-1716-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-018-1716-z