Abstract
Hepatitis B virus (HBV) affects approximately 68 million people in China, and 10–15% of adults infected with HBV develop chronic hepatitis B, liver cirrhosis, liver failure or hepatocellular carcinoma (HCC). HLA-DPB1 gene polymorphism and expression have been shown to be associated with HBV infection susceptibility and spontaneous clearance. The aim of this study is to evaluate the role of HLA-DPB1 gene polymorphism in HBV infection. HLA-DPB1 and rs9277535 polymorphisms were investigated in 259 patients with HBV infection and 442 healthy controls (HCs) using sequence-based typing. The mRNA of HLA-DPB1 was measured by real-time polymerase chain reaction. HLA-DPB1 genes and rs9277535 polymorphisms were all associated with HBV infection in the Sichuan Han population. rs9277535A and HLA-DPB1*04:02 played a protective role against HBV infection. rs9277535G and DPB1*05:01 were associated with susceptibility to HBV infection. rs9277535GG had significantly higher HLA-DPB1 mRNA expression in the HBV infection group compared with the HC group. HLA-DPB1*05:01 and HLA-DPB1*21:01 had significantly lower mRNA expression in the HBV infection group compared with the HC group. The meta-analysis revealed that HLA-DPB1*02:01, HLA-DPB1*02:02, HAL-DPB1*04:01 and HLA-DPB1*04:02 protected against HBV infection, while HLA-DPB1*05:01, HLA-DPB1*09:01, and HLA-DPB1*13:01 were risk factors for susceptibility to HBV infection. HLA-DPB1*02:01, HLA-DPB1*02:02, and HLA-DPB1*04:01 were associated with HBV spontaneous clearance, while HLA-DPB1*05:01 was associated with chronic HBV infection. HLA-DPB1 alleles and rs9277535 have a major effect on the risk of HBV infection, and HBV infection is associated with lower HLA-DPB1 expression. HLA-DPB1 alleles have an important role in HBV susceptibility and spontaneous clearance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There is a high prevalence of hepatitis B virus (HBV) in China, with an estimated 5.49% seropositivity for hepatitis B surface antigen (HBsAg) in 2015 (Schweitzer et al. 2015), meaning that ~ 93 million people were infected by HBV at that time. Around 10–20% of adults with HBV infection progress to chronic hepatitis B (CHB), alone or in combination with liver cirrhosis and/or hepatocellular carcinoma (Cui and Jia 2013). This leads to a heavy public health burden of HBV-related liver disease in China (Wang et al. 2014).
Multiple factors contribute to the risk of chronic HBV infection, such as age, sex, body mass index, ethnicity, viral mutation and genotypes, host genetic variations, and related host immune responses. A genome-wide association study (Kamatani et al. 2009) in Japanese and other Asian populations found a significant association between CHB and several polymorphisms of the HLA loci, including HLA-DPB1 and associated single nucleotide polymorphism (SNP) rs9277535A/G, and DPB1-rs9277535A/G haplotypes. Subsequently, they revealed risk alleles HLA-DPB1*05:01 and HLA-DPB1*03:01, and protective alleles HLA-DPB1*04:02 and HLA-DPB1*04:01. Many researchers (Donaldson et al. 2001; Nishida et al. 2015; Cho et al. 2008; Katrinli 2017; Nishida et al. 2016; Zhu et al. 2016; Nishida et al. 2014) have verified these results in different ethnic genetic populations. They have revealed that HLA-DPB1 and rs9277535A/G are related to susceptibility to HBV infection and whether the outcome is spontaneous clearance or CHB. Not only DPB1 gene, the DRB1 (Doganay et al. 2014; Yan et al. 2012) and DQB1 (Nishida et al. 2016; Ou et al. 2018) gene and the DRB1-DQB1-DPB1 haplotypes (Liao et al. 2014; Hu et al. 2012; Nishida et al. 2018) were also related HBV infection.
Recent research (O'Brien et al. 2011) has found that risk alleles of HLA-DP decrease liver mRNA expression of HLA-DP in HBV-infected patients, suggesting that expression of HLA-DP genes is important for control of HBV in non-Hispanic European populations. Thomas and colleagues found (Thomas et al. 2012) that the rs9277534A/G variant distinguishes the most protective HLA-DPB1 allele (HLA-DPB1*04:01) from the most susceptible (HLA-DPB1*01:01) after HBV infection. They also showed that healthy individuals with rs9277534GG had significantly higher levels of HLA-DPB1 surface protein and mRNA expression compared with individuals with rs9277534AA genotype, and that rs9277534A/G could be an HLA-DPB1 expression marker (Schone et al. 2018). Decreased expression of HLA-DPB1 mRNA is associated with HBV reactivation in patients treated with immunomodulatory agents (Matsuda et al. 2018), and increased HCV-related liver disease and correlated with HCV-related disease progression (Hiramatsu et al. 2017). These results suggest that differences in HLA-DPB1 alleles and HLA-DPB1 expression may influence the risk of persistent HBV infection.
Recent research has found (Niehrs et al. 2019) that a subset of HLA-DP molecules, such as HLA-DP401, which interact with NKp44, trigger functional natural killer (NK) cell responses. This interaction between DP alleles and NKp44 implicates HLA class II as a component of the innate immune response, much like HLA-C and KIR molecular. It may provide a potential mechanism (Niehrs and Altfeld 2020) for the relationship between HLA-DP alleles and disease outcomes, including for HBV infection. It is speculated that during acute HBV infection, NKp44 interacts with HLA-DP401 allele expressed on the surface of infected hepatocytes, and interferon-γ is secreted by T-helper 1 and NK cells that contribute to lysis of HBV-infected cells. In HBV-infected individuals carrying HLA-DP301, NKp44 is unable to bind to HLA-DP301 molecules, resulting in inefficient lysis of infected hepatocytes by NK cells, and this leads to a higher risk of chronic HBV infection.
The distribution of HLA-DPB1 molecules in Chinese populations is similar to that in other Asian populations, but HLA-DPB1 expression in Asian populations with HBV infection has not been reported yet. In this study, we typed HLA-DPB1 and associated rs9277535A/G in a population-based case–control study in the Chinese Han population in Sichuan Province, including 259 patients with HBV infection and 441 healthy controls (HCs). We also identified the different HLA-DPB1 expression in different alleles of these two groups. Meta-analysis was also used to comprehensively evaluate the correlation between HLA-DPB1 alleles and HBV infection susceptibility and HBV spontaneous clearance.
Materials and methods
Samples
The study samples were from the Physical Examination Center of Deyang People’s Hospital, Sichuan Province, China. There were 499 participants, including 259 patients with CHB and 441 HCs. HBV infection status was based on the serological results for hepatitis B surface antigen (HBsAg), hepatitis B surface antibody, hepatitis B e antigen (HBeAg), hepatitis B e antibody (HBeAb), and hepatitis B core antibody (HBcAb). Volunteers with seropositive for HBsAg combine with seropositive for HBeAg and HBcAb or HBeAb and HBcAb were considered to be HBV carriers. The HCs were seronegative for both HBsAg and HBeAg. All blood samples were negative for hepatitis C virus and human immunodeficiency virus. Subjects who were seropositive for HBsAg, had normal serum alanine aminotransferase levels (< 35 U/L) Supplementary Table 1, and had no obvious clinical symptoms were considered to be asymptomatic carriers and had not used any antiretroviral drugs. All subjects gave written informed consent before enrolment.
rs9277535 and HLA-DPB1 genotyping
Peripheral blood sampling and DNA extraction were performed as previously reported (Ou et al. 2018, 2019). Genotyping was performed using polymerase chain reaction (PCR) sequence-based typing. Amplification primers were as follows: rs9277535 forward, 5′-TAACTGTGTGTGTTGCTGC-3′, reverse, 5′-CTGCGTGGTGAGAAAACAGG-3′, and HLA-DPB1 exon 2 forward, 5′-CTGCGTGGTGAGAAAACAGG-3′, 5′-CCTGACAAGCTCCAGATGGG-3′, and reverse, 5′-TTCCTTTATGGTGTTGCTCCT-3′. Each PCR mix (total volume, 10 μL) contained 1 μL DNA and 5 μM primers and 5 μL GoTaq Green Master Mix (Promega, Madison, WI, USA). The DPB1 ambiguities were resolved by group-specific sequencing primers sequencing, and each primer was referenced by the EBI-IMGT database. The primers are shown in Table 1. Thermal cycling conditions were: 96 °C for 3 min, 30 cycles at 95 °C for 20 s, 62 °C for 15 s, 72 °C for 1 min, and 72 °C for 5 min. The PCR products were analyzed by an ABI 3730 DNA Sequencer (Applied Biosystem, Foster City, CA, USA).
Measurement of HLA-DPB1 mRNA level
RNA was prepared from suspensions of freshly isolated peripheral blood mononuclear cells using a TRIzol method (ThermoFisher, USA). RNA extraction and cDNA preparation were performed as previously reported (Ou et al. 2018, 2019).
Expression of HLA-DPB1 was quantified by SYBR Green quantitative PCR using the threshold cycle method in a CFX96 Touch PCR machine (Bio-Rad, USA) with the following primers: forward, 5′-GTGCATTGCAGAAGGTCAGA-3′, and reverse, 5′-CTGGTGATAGGCCATCAGGT-3′. Each PCR was run in triplicate, including 12.5 μL FastStart Universal SYBR Green Master (Roche, USA), 200 nM primers, and 2.5 μL cDNA in a total volume of 25 μL. The quantitative PCR protocol was that recommended by the Roche specification. The specificity of the DPB1 primers was confirmed by melt curve analysis using a single-peak dissociation step. All reactions were standardized to expression of the reference gene GAPDH.
Linkage of HLA-DPB1 and SNPs
Genotype frequency was tested for Hardy–Weinberg equilibrium using Arlequin version 3.5.1.2. D’ was calculated as described by (Paximadis et al. 2012). The maximum-likelihood method was used to estimate the linkage disequilibrium (LD). An expectation–maximization algorithm for multilocus genotypic data was used for the observed data with an unknown gametic phase. An ELB algorithm was used to estimate the gametic phase of the genotype data generated at all the polymorphic positions within the gene region when the recessive alleles were present. The statistical significance of the LD between each of the allele pairs was evaluated by the approximate χ2 test as described previously (Ou et al. 2019). All statistical significance was defined as P < 0.05. (Supplementary Material)
Meta-analysis study
In order to demonstrate the influence of HLA-DPB1 on the susceptibility to HBV infection and its spontaneous clearance, we performed a meta-analysis. Relevant studies were identified by a computerized literature search of electronic databases, including PubMed, EBSCO, Elsevier, and Web of Science, with English as the language restriction. The following index terms were used: hepatitis B and HLA-DP. The inclusion criteria were as follows: (1) genotype frequencies of HLA-DPB1 were obtained in HCs, HBV carriers, and individuals with chronic HBV infection and HBV spontaneous clearance; (2) the study included specific criteria for enrolling the samples; (3) the study had a case–control design; and (4) the numbers of cases and controls, and the alleles frequencies were clearly stated. Studies that did not meet these criteria were excluded, details of the mata-analysis were in the Supplementary Materials.
Statistical analysis
The Hardy–Weinberg equilibrium of the genotype distributions and the LD of the SNPs were examined using Arliquin version 3.5 software. The differences in categorical variables and continuous variables were compared using the χ2 test and Student’s t test, respectively. We used a logistic regression model to calculate the age- and sex-adjusted odds ratios (ORs) and 95% confidence intervals (CIs) between the HLA-DPB1 variants and the risk of HBV infection. The meta-analysis was performed by Review Manager version 5.2 software, and ORs with 95% CIs were used to assess the strength of HLA-DPB1 polymorphisms in HBV susceptibility. The I2 test was used for heterogeneity examine between the studies, and the fixed-effect model (F) was used if there was < 50% heterogeneity; otherwise, the random-effect model (R) was used. We added it in the manuscript. P < 0.05 was considered statistically significant.
Results
Relationship between HLA-DPB1 and rs9277535A/G in HBV infection
The carriage of HLA-DPB1 rs9277535A indicated stronger protection against HBV infection (OR = 0.52, 95% Cl 0.41–0.65), while rs9277535G indicated susceptibility to HBV infection (OR = 1.94, 95% Cl 1.54–2.46) (Table 2).
HLA-DPB1 was highly polymorphic, and the most prevalent allele in our population was HLA-DPB1*05:01, which was significantly associated with HBV susceptibility in the Sichuan Han population (OR = 1.41, 95% Cl 1.14–1.76). Both HLA-DPB1*04:02 were significantly associated with protection against HBV infection in our study population (OR = 0.31, 95% Cl 0.16–0.59) (Table 3).
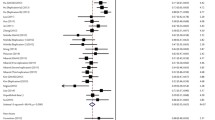
After meta-analysis, we found that HLA-DPB1*02:01, HLA-DPB1*02:02, HLA-DPB1*04:01, and HLA-DPB1*04:02 were paly protect role in HBV infection; however, HLA-DPB1*05:01, HLA-DPB1*09:01, and HLA-DPB1*13:01 were associated with HBV susceptibility. HLA-DPB1*02:01, HLA-DPB1*02:02, and HLA-DPB1*04:01 were associated with HBV spontaneous clearance, while HLA-DPB1*05:01 was associated with chronic HBV infection as shown in Tables 5-8 and Supplementary Tables 2-3 and Figs. 2-11.
Expression of HLA-DPB1 mRNA levels in HBV infection
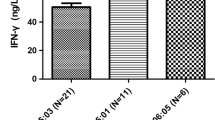
HLA-DPB1 mRNA expression was significantly lower in subjects with the rs9277535AA genotype compared with the rs9277535GG genotype in HC group. rs9277535GG genotype, HLA-DPB1*05:01, and HLA-DPB1*21:01 in the HBV infection group showed significantly lower HLA-DPB1 mRNA expression compared with the HC group, as shown in Figs. 1, 2 and 3, Supplementary Fig 1.
LD between rs9277535A/G and HLA-DPB1 alleles
The LD test indicated the association between HLA-DPB1 alleles and different rs9277535A/G that make up the haplotypes. HLA-DPB1*04:01 and HLA-DPB1*04:02 had strong linkage with rs9277535A, which had a protective role in HBV infection. HLA-DPB1*05:01 had strong linkage with rs9277535G, which was associated with susceptibility to HBV infection (Tables 4, 5, 6, 7 and 8).
Discussion
China is a high-endemic area for HBV infection. There are many HBV carriers, and patients with chronic hepatitis, cirrhosis, or hepatocellular carcinoma caused by HBV infection; all of which cause a heavy social burden (Shin et al. 2010). After the introduction of the HBV vaccine into the immunization program in China in 1992, the government administered free HBV vaccine to newborns to reduce the spread of the virus. At the same time, China adopted a variety of methods to prevent and control HBV, strengthening the standardized management of HBV screening and medical treatment for blood donors, and effectively controlling iatrogenic HBV infection. All the above measures have significantly reduced the infection rate of new HBV infections, and the total HBV prevalence rate in China had dropped to 5.49% by 2015 (Schweitzer et al. 2015). Therefore, it is important to study the infective mechanism of HBV for the prevention and treatment of hepatitis B.
HLA-II genes play a key role in viral antigen presentation to mediate cellular and humoral immune responses. A genome-wide association study of a large cohort with HBV infection showed that HLA-DPB1 and the related SNP rs9277535A/G are strong risk factors for persistent HBV infection (Kamatani et al. 2009). Thereafter, many studies focused on and successfully repeated the association between HLA-DPB1 and HBV infection and its outcomes (Yu et al. 2015). HLA-DPB1*09:01 increased HBV susceptibility, HLA-DPB1*02:01 was not related to susceptibility, and HLA-DPB1*02:01 was associated with reduced risk of HBV progression to CHB (Nishida et al. 2014). Studies in American populations found that HLA-DPB1*01:01 allele was associated with the greatest risk of HBV persistent infection, and HLA-DPB1*04:01 allele with the greatest protection against HBV persistent infection (Thomas et al. 2012). Subsequently, the genotypes of HLA-DPB1*05:01 and HLA-DPB1*09:01, and HLA-DPB1*02:01, HLA-DPB1*04:01, and HLA-DPB1*04:02 were found to be associated with susceptibility to HBV in Japanese (Nishida et al. 2016) and Chinese (Zhu et al. 2016; Huang et al. 2020) populations. Some HLA-DPB1 alleles and rs9277535G were related to weak HBV vaccine response (Wang et al. 2019), low sensitivity to anti-HBV drug therapy (Matsuda et al. 2018), incidence of occult HBV infection (Mardian et al. 2017), and susceptibility to hepatocellular carcinoma development (Zhang et al. 2013). In our study, we found a strong association between rs9277535 and HLA-DPB1 alleles and HBV infection susceptibility. We found that rs9277535A and HLA-DPB1*04:02 were protective factors against chronic HBV infection, and the results were according to the previous study. rs9277535G and HLA-DPB1*05:01 were significantly associated with HBV susceptibility. Our results suggest that HLA-DP has a key role in HBV infection progression.
The influence of the HLA-DPB1 region on HBV recovery results from levels of HLA-DPB1 expression and less likely from differences in the peptides presented by different HLA-DPB1 alleles. HLA-DPB1 expression differed significantly from that of rs9277535AA and rs9277535GG; the protective genotype 9277535AA had significantly lower HLA-DPB1 mRNA expression level compared with the high-risk genotype rs9277535GG in HCs. Previous research found (Petersdorf and Malkki 2015) rs9277534A with lower HLA-DPB1 expression than rs9277535G, the risk of acute GVHD was lower for recipients with rs9277534A-linked HLA-DPB1 mismatches compared than that with rs9277534G-linked HLA-DPB1 mismatches. Thomas and colleagues also demonstrated (Thomas et al. 2012) that rs9277534GG genotype, which confers susceptibility to HBV persistence, with significantly higher levels of HLA-DP surface protein and transcription in healthy donors. O’Brien reported that rs9277535AA with higher HLA-DPB1 mRNA expression in the liver was associated with lower risk of chronic HBV infection (O'Brien et al. 2011). We obtained a similar result with HLA-DPB1 expression in peripheral blood mononuclear cells. The HLA-DPB1*05:01 and HLA-DPB1*21:01 alleles in the HBV group showed significantly lower mRNA expression compared with the HCs. Previous research (Cho et al. 2008) showed that decreased expression of HLA-DPA1 and HLA-DPB1 mRNA was correlated with HBV activation after 2 years’ treatment with anti-HBV drugs, and high expression of HLA-DPA1 mRNA was associated with lower HBV viral load (Matsuda et al. 2018). These results suggest that persistence of HBV infection is influenced by differences in HLA-DPB1 expression. Lower HLA-DPB1 expression may lower antigen peptide presentation, resulting in decreased immune response to HBV, and increased risk of persistent infection.
The amino acids at positions 84–87 of the second exon antigen presentation sequence of HLA-DPB1 have a complete linkage with the HLA-DPB1 alleles, and there are two major amino acids at these positions with 84Gly and 84Asp (Zhu et al. 2016), from IPD-IMGT/HLA of EMBL-EBI database, we can see that the DPB1 amino acid with GGPM at position 84–87 were linkage with DPB1*02:01, 02:02, 04:01, 04:02 (r2 = 1), while amino acid with DEAV at position 84–87 were linkage with DPB1* 03:01, 05:01, 09:01, 13:01, 14:01, 17:01 (r2 = 1). Bioinformatics analysis has found that the four amino acids are located at the groove contacting peptide residues pocket-1, which can cause different antigen-presenting functions and immunity to HBV infection. Class II HLA molecules expression on the surface of antigen-presenting cells, to combine with antigenic peptide to CD4 + T helper cells. The T cells play a crucial role in the response of HBV to the host immune response.
Usually, HLA class II molecules present exogenous antigen peptides; however, HLA-DP molecules with beta-chains encoding DP84Gly (Yamashita et al. 2017) do not bind invariant chain via the class-II-associated invariant chain peptide region to constitutively present endogenous peptides. And DP84Gly processed by the proteasome and transported to the endoplasmic reticulum by the transporter associated with antigen processing. Therefore, DP84Gly can uniquely uses both class I and II antigen-processing pathways to present peptides derived from intracellular and extracellular sources, but DP84Asp has no such endogenous antigen-presentation function. Therefore, this polymorphism has different functions in autoimmune, antiviral, and tumor mechanisms through its different functions of antigen presentation (Anczurowski and Hirano 2018).
The DP84Gly genotype not only has a unique antiviral function in adaptive immunity but also acts as a ligand of NKp44 to activate NKT cells to play an antiviral role in natural immune function (Niehrs et al. 2019). Compared with HCs, NK cells in CHB patients show inhibitory phenotypes, in which the expression of activated receptors NKp44 and NKp46 is downregulated (Li et al. 2017). Whether the DP84Gly genotype can promote the spontaneous clearance of HBV by binding NKp44 to activated NK cells after HBV infection has not been studied.
The association of HLA-DPB1 expression with HBV infection has not been verified in the Chinese population. To the best of our knowledge, the present study is the first to investigate HLA-DPB1 mRNA expression in different HLA-DPB1 and SNP alleles, and the association between mRNA expression and HBV infection. Further research is needed to verify the role of NK cells and CD4+ T cells in DP84Gly, including HLA-DPB1*04:01, HLA-DPB1*04:02, HLA-DPB1*02:01, and HLA-DPB1*02:02 in HBV protection and clearance.
Conclusion
HLA-DPB1 gene has a major effect on the risk of HBV infection. rs9277535A, HLA-DPB1*02:01, HLA-DPB1*02:02, HLA-DPB1*04:01, and HLA-DPB1*04:02 protect against HBV infection, while rs9277535G, HLA-DPB1*05:01, HLA-DPB1*09:01, and HLA-DPB1*13:01 are associated with susceptibility to HBV infection. HLA-DPB1*02:01, HLA-DPB1*02:02, and HLA-DPB1*04:01 are associated with HBV spontaneous clearance, while HLA-DPB1*05:01 is associated with chronic HBV infection. rs9277535GG has significantly higher HLA-DPB1 mRNA expression in patients with HBV infection compared with HCs. Alleles associated with susceptibility to HBV infection appear to have decreased HLA-DPB1 mRNA expression.
Data availability
All the data and materials supporting the conclusions were included in the main paper.
Abbreviations
- HBV:
-
Hepatitis B virus
- HLA:
-
Human leukocyte antigens
- APCs:
-
Antigen-presenting cells
- CHB:
-
Chronic HBV carriers
- HC:
-
Healthy controls
- HBsAg:
-
Hepatitis B surface antigen
- HBsAb:
-
Hepatitis B surface antibody
- HBeAg:
-
Hepatitis Be antigen
- HBeAb:
-
Hepatitis Be antibody
- HBcAb:
-
Hepatitis core antibody
- HCV:
-
Hepatitis C virus
- HIV:
-
Human immunodeficiency virus
- HWE:
-
Hardy–Weinberg equilibrium
References
Anczurowski M, Hirano N (2018) Mechanisms of HLA-DP antigen processing and presentation revisited. Trends Immunol 39:960–964
Cho SW, Cheong JY, Ju YS, Oh DH, Suh YJ, Lee KW (2008) Human leukocyte antigen class II association with spontaneous recovery from hepatitis B virus infection in Koreans: analysis at the haplotype level. J Korean Med Sci 23:838–844
Cui Y, Jia J (2013) Update on epidemiology of hepatitis B and C in China. J GastroenterolHepatol 28(Suppl 1):7–10
Doganay L, Fejzullahu A, Katrinli S, Yilmaz Enc F, Ozturk O, Colak Y, Ulasoglu C, Tuncer I, DinlerDoganay G (2014) Association of human leukocyte antigen DQB1 and DRB1 alleles with chronic hepatitis B. World J Gastroenterol 20:8179–8186
Donaldson PT, Ho S, Williams R, Johnson PJ (2001) HLA class II alleles in Chinese patients with hepatocellular carcinoma. Liver 21:143–148
Hiramatsu K, Matsuda H, Nemoto T, Nosaka T, Saito Y, Naito T, Takahashi K, Ofuji K, Ohtani M, Suto H et al (2017) Identification of novel variants in HLA class II region related to HLA DPB1 expression and disease progression in patients with chronic hepatitis C. J Med Virol
Hu L, Zhai X, Liu J, Chu M, Pan S, Jiang J, Zhang Y, Wang H, Chen J, Shen H, Hu Z (2012) Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology 55:1426–1431
Huang YH, Liao SF, Khor SS, Lin YJ, Chen HY, Chang YH, Huang YH, Lu SN, Lee HW, Ko WY et al (2020) Large-scale genome-wide association study identifies HLA class II variants associated with chronic HBV infection: a study from Taiwan Biobank. Aliment PharmacolTher 52:682–691
Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H et al (2009) A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 41:591–595
Katrinli S (2017) NilayKaratasErkut G, Ozdil K, Yilmaz Enc F, Ozturk O, Kahraman R, Tuncer I, DinlerDoganay G, Doganay L: HLA DPB1 15:01 allele predicts spontaneus hepatitis B surface antigen seroconversion. Acta Gastroenterol Belg 80:351–355
Li X, Zhou L, Gu L, Gu Y, Chen L, Lian Y, Huang Y (2017) Veritable antiviral capacity of natural killer cells in chronic HBV infection: an argument for an earlier anti-virus treatment. J Transl Med 15:220
Liao Y, Cai B, Li Y, Chen J, Tao C, Huang H, Wang L (2014) Association of HLA-DP/DQ and STAT4 polymorphisms with HBV infection outcomes and a mini meta-analysis. PLoS One 9:e111677
Mardian Y, Yano Y, Wasityastuti W, Ratnasari N, Liang Y, Putri WA, Triyono T, Hayashi Y (2017) Genetic polymorphisms of HLA-DP and isolated anti-HBc are important subsets of occult hepatitis B infection in Indonesian blood donors: a case-control study. Virol J 14:201
Matsuda H, Hiramatsu K, Akazawa Y, Nosaka T, Saito Y, Ozaki Y, Hayama R, Takahashi K, Naito T, Ofuji K et al (2018) Genetic polymorphism and decreased expression of HLA class II DP genes are associated with HBV reactivation in patients treated with immunomodulatory agents. J Med Virol 90:712–720
Niehrs A, Altfeld M (2020) Regulation of NK-cell function by HLA class II. Front Cell Infect Microbiol 10:55
Niehrs A, Garcia-Beltran WF, Norman PJ, Watson GM, Holzemer A, Chapel A, Richert L, Pommerening-Roser A, Korner C, Ozawa M et al (2019) A subset of HLA-DP molecules serve as ligands for the natural cytotoxicity receptor NKp44. Nat Immunol 20:1129–1137
Nishida N, Ohashi J, Khor SS, Sugiyama M, Tsuchiura T, Sawai H, Hino K, Honda M, Kaneko S, Yatsuhashi H et al (2016) Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis. Sci Rep 6:24767
Nishida N, Ohashi J, Sugiyama M, Tsuchiura T, Yamamoto K, Hino K, Honda M, Kaneko S, Yatsuhashi H, Koike K et al (2015) Effects of HLA-DPB1 genotypes on chronic hepatitis B infection in Japanese individuals. Tissue Antigens 86:406–412
Nishida N, Sawai H, Kashiwase K, Minami M, Sugiyama M, Seto WK, Yuen MF, Posuwan N, Poovorawan Y, Ahn SH et al (2014) New susceptibility and resistance HLA-DP alleles to HBV-related diseases identified by a trans-ethnic association study in Asia. PLoS One 9:e86449
Nishida N, Sugiyama M, Sawai H, Nishina S, Sakai A, Ohashi J, Khor SS, Kakisaka K, Tsuchiura T, Hino K et al (2018) Key HLA-DRB1-DQB1 haplotypes and role of the BTNL2 gene for response to a hepatitis B vaccine. Hepatology 68:848–858
O’Brien TR, Kohaar I, Pfeiffer RM, Maeder D, Yeager M, Schadt EE, Prokunina-Olsson L (2011) Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 12:428–433
Ou G, Xu H, Yu H, Liu X, Yang L, Ji X, Wang J, Liu Z (2018) The roles of HLA-DQB1 gene polymorphisms in hepatitis B virus infection. J Transl Med 16:362
Ou G, Liu X, Yang L, Yu H, Ji X, Liu F, Xu H, Qian L, Wang J, Liu Z (2019) Relationship between HLA-DPA1 mRNA expression and susceptibility to hepatitis B. J Viral Hepat 26:155–161
Paximadis M, Mathebula TY, Gentle NL, Vardas E, Colvin M, Gray CM, Tiemessen CT, Puren A (2012) Human leukocyte antigen class I (A, B, C) and II (DRB1) diversity in the black and Caucasian South African population. Hum Immunol 73:80–92
Petersdorf EW, Malkki M (2015) O’HUigin C, Carrington M, Gooley T, Haagenson MD, Horowitz MM, Spellman SR, Wang T, Stevenson P: High HLA-DP expression and graft-versus-host disease. N Engl J Med 373:599–609
Schone B, Bergmann S, Lang K, Wagner I, Schmidt AH, Petersdorf EW, Lange V (2018) Predicting an HLA-DPB1 expression marker based on standard DPB1 genotyping: linkage analysis of over 32,000 samples. Hum Immunol 79:20–27
Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ (2015) Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386:1546–1555
Shin HR, Oh JK, Masuyer E, Curado MP, Bouvard V, Fang YY, Wiangnon S, Sripa B, Hong ST (2010) Epidemiology of cholangiocarcinoma: an update focusing on risk factors. Cancer Sci 101:579–585
Thomas R, Thio CL, Apps R, Qi Y, Gao X, Marti D, Stein JL, Soderberg KA, Moody MA, Goedert JJ et al (2012) A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol 86:6979–6985
Wang FS, Fan JG, Zhang Z, Gao B, Wang HY (2014) The global burden of liver disease: the major impact of China. Hepatology 60:2099–2108
Wang LY, Chen CF, Wu TW, Lai SK, Chu CC, Lin HH (2019) Response to hepatitis B vaccination is co-determined by HLA-DPA1 and -DPB1. Vaccine 37:6435–6440
Yamashita Y, Anczurowski M, Nakatsugawa M, Tanaka M, Kagoya Y, Sinha A, Chamoto K, Ochi T, Guo T, Saso K et al (2017) HLA-DP(84Gly) constitutively presents endogenous peptides generated by the class I antigen processing pathway. Nat Commun 8:15244
Yan ZH, Fan Y, Wang XH, Mao Q, Deng GH, Wang YM (2012) Relationship between HLA-DR gene polymorphisms and outcomes of hepatitis B viral infections: a meta-analysis. World J Gastroenterol 18:3119–3128
Yu L, Cheng YJ, Cheng ML, Yao YM, Zhang Q, Zhao XK, Liu HJ, Hu YX, Mu M, Wang B et al (2015) Quantitative assessment of common genetic variations in HLA-DP with hepatitis B virus infection, clearance and hepatocellular carcinoma development. Sci Rep 5:14933
Zhang Q, Yin J, Zhang Y, Deng Y, Ji X, Du Y, Pu R, Han Y, Zhao J, Han X et al (2013) HLA-DP polymorphisms affect the outcomes of chronic hepatitis B virus infections, possibly through interacting with viral mutations. J Virol 87:12176–12186
Zhu M, Dai J, Wang C, Wang Y, Qin N, Ma H, Song C, Zhai X, Yang Y, Liu J et al (2016) Fine mapping the MHC region identified four independent variants modifying susceptibility to chronic hepatitis B in Han Chinese. Hum Mol Genet 25:1225–1232
Acknowledgments
The authors thank the participants for generously providing the venous blood samples.
Funding
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) under contract 2016-I2M-3–024, Funding of Sichuan Science and Technology Department under contract 2017RZ0047 and 2018SZ0395 and Ministry of Science and Technology of China, Grant/Award number: 2014EG150133. The CIFMS program provided us blood samples and relevant data, funding of Sichuan Science and Technology Department project provide us the cytokine detect methods and methods of HLA-DPB1 typing, program of Ministry of Science and Technology of China provide us HBV relevant detect methods.
Author information
Authors and Affiliations
Contributions
Ou GJ, Xu HX, and Ji X performed the experiments. Liu X collected and evaluated the samples. Ou GJ and Liu XJ wrote the original draft of the manuscript. Wang J and Liu XJ designed the experiments and performed the data analysis, discussed the results, and substantially revised the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethic committees of the Institution of Blood Transfusion, CAMS&PUMC, and was conducted according to the principles of the Declaration of Helsinki. All participants provided written informed consent before enrolment, and the study’s protocol was approved by the ethic committees of the Institution of Blood Transfusion, CAMS&PUMC.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ou, G., Liu, X., Xu, H. et al. Variation and expression of HLA-DPB1 gene in HBV infection. Immunogenetics 73, 253–261 (2021). https://doi.org/10.1007/s00251-021-01213-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-021-01213-w