Abstract
In Drosophila, a large group of actively transcribed genes is located in pericentromeric heterochromatin. It is assumed that heterochromatic proteins recruit transcription factors to gene promoters. Two proteins, Ouib and Nom, were previously shown to bind to the promoters of the heterochromatic genes nvd and spok. Interestingly, Ouib and Nom are paralogs of the M1BP protein, which binds to the promoters of euchromatic genes. We have shown that, like M1BP, the Quib and Nom proteins bind to CP190, which is involved in the recruitment of transcription complexes to promoters. Unlike heterochromatic proteins, Ouib and Nom do not interact with the major heterochromatic protein HP1a and bind to euchromatic promoters on polytene chromosomes from the larval salivary glands. The results suggest a new mechanism for the recruitment of transcription factors into the heterochromatic compartment of the nucleus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
In Drosophila, euchromatin and heterochromatin are two independent compartments that differ in the set of chromatin proteins and nucleosomal modifications [1, 2]. Recent studies have shown that heterochromatin is actively transcribed and comprises many housekeeping genes with a high level of expression [2]. The promoters of genes located in euchromatin and heterochromatin can effectively initiate transcription only in their own compartment [1, 2]. Unlike prokaryotes, the majority of promoters of higher eukaryotes, including Drosophila and humans, do not have pronounced motifs to which general transcription factors can bind [3]. According to one model, the key role in the organization of promoters is played by proteins containing C2H2-type zinc finger clusters (C2H2 proteins), which bind to specific relatively long (12–15 bp) motifs in gene promoters, which leads to the formation of open chromatin and the recruitment of the TFIID complex [3, 4]. In Drosophila, there is a large class of proteins that contain a cluster at the C terminus consisting of 5 C2H2 domains, and at the N terminus there is a ZAD domain (Zinc-finger Associated Domain), which ensures protein homodimerization [5, 6]. The ZAD domain and C2H2 clusters are connected by an unstructured linker. In Drosophila, the best described protein is M1BP, which binds to a motif located in approximately 2000 promoters that determine the activity of euchromatic genes [7, 8]. The CP190 protein, which is involved in the recruitment of transcription-stimulating complexes, binds to the unstructured linker of the M1BP protein [9, 10].
The m1bp gene is located in a cluster (85A9 on chromosomal arm 3R) that includes four more genes encoding ZAD-C2H2 proteins—CG8159, ranshi, ouib, and nom (Fig. 1b). All these genes are paralogous and are presumably the result of tandem duplication during evolution. Unexpectedly, it was found that the ouib and nom genes encode proteins that bind to gene promoters located in heterochromatin [11–13]. Earlier, two other ZAD-C2H2 proteins, Odj and Nnk, were found, which effectively bind to regulatory sequences in heterochromatin [14]. These proteins were shown to interact with the main heterochromatin protein HP1a. This is believed to determine the preferential localization of these proteins in the heterochromatin compartment.
(a) Domain structure of proteins Ouib and Nom. The ordinal numbers to the amino acid residues corresponding to domains boundaries are indicated. (b) Genomic location of the m1bp, ouib, and nom genes in the cluster in D. melanogaster and D. virilis. The same colors are used to indicate orthologs. (c) Study of the interaction between ZAD domains in yeast two-hybrid system. ZAD fragments were fused to the GAL4 DNA-binding domain (DBD), and their interaction with fragments fused to the GAL4 activation domain (AD) was examined. The results are presented in columns where signs + and – indicate the presence or absence of the interaction, respectively. As a positive control, the ability of ZAD domains to dimerize was tested; as a negative control, the interaction only with the activation domain (AD) or DNA-binding domain (DBD) of the GAL4 protein was tested. (d) Study of the interaction of the full-length proteins Ouib and Nom fused with the GAL4 AD with the full-length proteins CP190 and HP1a fused with the GAL4 DBD. The remaining notations are the same as in (c).
The purpose of this work was to study how the Ouib and Nom proteins acquire the ability to bind to promoters of genes located in heterochromatin regions. It was previously shown that the ZAD domains of even close C2H2 paralogs predominantly form only homodimers [5, 6]. Since the ZAD domains of the M1BP, Ouib, and Nom proteins have a high degree of sequence similarity, the ability of the ZAD domains to form heterodimers was tested. For this purpose, we used the yeast two-hybrid (Y2H) system. We created constructs in which the sequences of the ZAD domains of the Ouib, Nom, and M1BP proteins were cloned into the same reading frame with the activation domain (AD) or DNA-binding domain (DBD) of the GAL4 protein. A search for orthologs in Drosophila species showed that the ouib orthologue is present in all species, whereas its duplication, the nom gene, occurred later. For example, nom orthologs are absent in D. virilis (Fig. 1b). We tested the interaction of the ZAD domains of the Ouib and Nom proteins with the Ouib ortholog of D. virilis (GJ22678) in Y2H. Interaction analysis showed that the ZAD domains of M1BP, Ouib, and Nom homodimerize but do not form heterodimers. Both paralogous proteins Ouib and Nom interact with the D. virilis ortholog GJ22678 (Fig. 1c), which is additional evidence that they originated from the same gene.
It is known that many proteins associated with heterochromatin interact with the HP1a protein through the amino acid motif PxVxL (where x is any amino acid residue). However, the analysis of the Ouib and Nom sequences did not reveal this motif. To test the direct interaction of the proteins with HP1a in Y2H, we obtained constructs in which the cDNA sequences of the Ouib and Nom proteins were cloned into the same reading frame with the GAL4 AD and a construct in which the cDNA of the HP1a protein was cloned in the same reading frame with the GAL4 DBD. As a result, no direct interaction of Ouib and Nom with HP1a was detected in Y2H (Fig. 1d).
Many ZAD-C2H2 proteins that have binding sites in gene promoters interact with the CP190 protein. In particular, the recruitment of CP190 is important for the functional activity of M1BP [9]. We tested the direct interaction of the Ouib and Nom paralogs with CP190 in Y2H (Fig. 1c). It was found that Ouib and Nom also interact with CP190, which binds exclusively to euchromatic promoters. This suggests that Ouib and Nom are involved in the organization of euchromatic promoters.
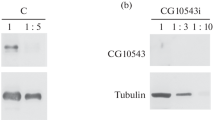
To study the genome-wide distribution of binding sites for the Ouib and Nom proteins, we used a model of polytene chromosomes from the salivary gland nuclei of third instar larvae. Polytene chromosomes consist of interbands, which are promoters that control the expression of housekeeping genes, and bands, which correspond to predominantly inactive genes [15]. The constitutive heterochromatin regions are clearly visible in the pericentromeric heterochromatin region and in telomeres at the ends of chromosomes. Since Ouib and Nom are not expressed in the salivary glands, we used the inducible UAS/GAL4 system to express these proteins. For this purpose, we created genetic constructs in which cDNAs of the Nom and Ouib proteins in the same reading frame with the HA epitope were cloned into the pUAS vector, which contains the UAS sequence (Upstream activating sequence) and an attB site for integration into the Drosophila genome (Fig. 2a). The obtained constructs were inserted into the same genomic position at the attP 38D site using the phiC31 integration system. For ectopic gene expression, we crossed flies of a strain containing the pUAS-OuibxHA or pUAS-NomxHA construct with flies of a strain expressing GAL4 under the control of a strong tubulin promoter (tub:GAL4), which ensures a high level of transcription in all tissues. Protein expression was confirmed by Western blot analysis of protein lysate from adult flies using anti-HA antibodies (Fig. 2b).
(a) Genetic construction scheme used to create transgenic flies expressing a protein fused to an HA epitope. (b) Western blot analysis of protein extracts from 2–3-day-old males expressing OuibxHA and NomxHA stained with antibodies to the HA epitope. (c) Polytene chromosomes from larval salivary glands of flies expressing OuibxHA and NomxHA were examined. The chromosomes were stained with antibodies to the HA epitope, and DNA was stained with DAPI. For comparison, immunostaining of polytene chromosomes from the Oregon-R strain with rabbit antibodies to the heterochromatin protein Odj is shown. The arrows indicate the position of the chromocenter in different preparations of polytene chromosomes. Scale bar: 10 μm.
Immunostaining of polytene chromosomes gave unexpected results. It was found that the Nom and Ouib proteins have many binding sites in the interbands of polytene chromosomes; however, no binding in the pericentromeric heterochromatic regions was observed. Thus, the ectopically expressed proteins are not recruited themselves into the heterochromatic compartment (Fig. 2c). This distinguishes the Nom and Ouib proteins from the Odj protein, which actively binds to the heterochromatin of polytene chromosomes (Fig. 2c).
It is most likely that the gene encoding M1BP is the founder of the gene cluster (85A9), because its expression is much higher and the protein has binding motifs conserved among Drosophila in the promoters of a large group of genes. The remaining genes of the cluster, except ranshi, show a low level of expression and are probably associated with the promoters of a small number of genes. For example, it was shown that the Nom and Ouib proteins determine the activity of the promoters of the nvd and spok genes, located in heterochromatin [11, 12]. Activation of these promoters is the main function of the Nom and Ouib proteins. Similarly to M1BP, the Nom and Ouib proteins bind to CP190, whose recruitment to promoters determines the formation of an open chromatin zone and transcription activation. Thus, the Nom and Ouib proteins are similar in structure and function to M1BP. Moreover, when overexpressed, the Nom and Ouib proteins bind to the promoters (interbands) of housekeeping genes, which are located in euchromatin, and their association with pericentromeric heterochromatin is not observed. The ZAD domains of three close paralogs M1BP, Nom, and Ouib form only homodimers. Thus, ZAD domains determine the functional independence of the newly emerged C2H2 proteins and block their interaction with M1BP. The results obtained indicate that the mechanism of recruitment of Ouib and Nom to heterochromatic regions is fundamentally different from that described previously for Odj and Nnk, for which interaction through a consensus motif with HP1a was shown. The Nom and Ouib proteins do not interact directly with the major heterochromatic protein HP1a.
It is most likely that Nom and Ouib are recruited to the heterochromatic compartment with the help of a partner protein that is also involved in the formation of active promoters of the nvd and spok genes. One of the potential partner proteins may be the large (approximately 2000 aa) ZAD-C2H2 protein Molting Defective (Mld), which also binds to the nvd and spok promoters [12]. The Mld protein is efficiently expressed at all stages of development and apparently has many various functions in gene expression regulation. Further studies are required to answer the question of whether the Ouib and Nom proteins can be recruited to the heterochromatic compartment as a result of interaction with other ZAD-C2H2 proteins that are associated with the main heterochromatic proteins.
REFERENCES
Marsano, R.M., Giordano, E., Messina, G., et al., A new portrait of constitutive heterochromatin: lessons from drosophila melanogaster, Trends Genet., 2019, vol. 35, no. 9, pp. 615–631.
Saha, P., Sowpati, D.T., Soujanya, M., et al., Interplay of pericentromeric genome organization and chromatin landscape regulates the expression of Drosophila melanogaster heterochromatic genes, Epigenetics Chromatin, 2020, vol. 13, no. 1, p. 41.
Vo Ngoc, L., Kassavetis, G.A., and Kadonaga, J.T., The RNA polymerase II core promoter in Drosophila, Genetics, 2019, vol. 212, no. 1, pp. 13–24.
Kyrchanova, O.V., Bylino, O.V., and Georgiev, P.G., Mechanisms of enhancer–promoter communication and chromosomal architecture in mammals and Drosophila, Front. Genet., 2022, vol. 13, p. 1081088.
Bonchuk, A.N., Boyko, K., Nikilaeva, A.Y., et al., Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity, Structure (London, England), 2022, vol. 30, no. 7, pp. 1004–1015.
Bonchuk, A.N., Boyko, K., Fedotova, A.A., et al., Structural basis of diversity and homodimerization specificity of zinc-finger-associated domains in Drosophila, Nucleic Acids Res., 2021, vol. 49, no. 4, pp. 2375–2389.
Baumann, D.G. and Gilmor, D.S., A sequence-specific core promoter-binding transcription factor recruits TRF2 to coordinately transcribe ribosomal protein genes, Nucleic Acids Res., 2017, vol. 45, no. 18, pp. 10481–10491.
Li, J. and Gilmour, D.S., Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor, EMBO J., 2013, vol. 32, no. 13, pp. 1829–1841.
Bag, I., Shue, C., and Rosin, L., M1BP cooperates with CP190 to activate transcription at TAD borders and promote chromatin insulator activity, Nat. Commun., 2021, vol. 12, no. 1, p. 4170.
Sabirov, M., Popovich, A., Boyko, K., et al., Mechanisms of CP190 interaction with architectural proteins in Drosophila melanogaster, Int. J. Mol. Sci., 2021, vol. 22, no. 22, p. 12400.
Komura-Kawa, T., Hirota, K., Shimada-Niwa, Y., et al., The Drosophila zinc finger transcription factor Ouija board controls ecdysteroid biosynthesis through specific regulation of spookier, PLoS Genet., 2015, vol. 11, no. 12, p. e1005712.
Uryu, O., Komura-Kawa, T., Kamiyama, T., et al., Cooperative control of ecdysone biosynthesis in Drosophila by transcription factors Séance, Ouija Board, and Molting Defective, Genetics, 2018, vol. 208, no. 2, pp. 605–622.
Niwa, Y.S. and Niwa, R., Ouija board: a transcription factor evolved for only one target in steroid hormone biosynthesis in the fruit fly Drosophila melanogaster, Transcription, 2016, vol. 7, no. 5, pp. 196–202.
Kasinathan, B., Colmenares, S.U., McConnell, H., et al., Innovation of heterochromatin functions drives rapid evolution of essential ZAD-ZNF genes in Drosophila, eLife, 2020, vol. 9, p. e63368.
Demakova, O.V., Demakov, S.A., Boldyreva, L.V., et al., Faint gray bands in Drosophila melanogaster polytene chromosomes are formed by coding sequences of housekeeping genes, Chromosoma, 2020, vol. 129, no. 1, pp. 25–44.
Funding
The study was supported by the Russian Science Foundation (project no. 22-24-00894).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
In accordance with Directive 2010/63/EU of September 22, 2010 on the protection of animals used for scientific purposes, chapter 1, paragraph 3, the requirements of bioethics do not apply to the object of this study.
Additional information
Translated by M. Batrukova
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pekina, Y.V., Babosha, V.A., Georgiev, P.G. et al. Study of the Association of Ouib and Nom with Heterochromatin in Drosophila melanogaster. Dokl Biochem Biophys 513 (Suppl 1), S26–S29 (2023). https://doi.org/10.1134/S1607672924700741
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1607672924700741