Abstract—
The chloroplast genome of the synthetic octaploid Triticum timonovum Heslot et Ferrary k-43065 (France) was sequenced for the first time. Plastome sequencing was carried out on a Genolab M sequencer (GeneMind, China). The genome assembly was carried out using the NOVOwrap program. The size of the chloroplast genome of T. timonovum was 136 158 bp. The length of the inverted repeat region was 21 552 bp, that of the SSC region was 12 795 bp, and the LSC length was 80 257 bp. The chloroplast genomes of T. timonovum and different T. timopheevii accessions from the GenBank database were compared. As for the chloroplast genome, T. timonovum was closer to T. timopheevii (AB976560.1) but differed from it by the presence of one insert A at position 47891.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Triticum timonovum Heslot et Ferrary (Timonovum wheat) is octaploid wheat artificially created by doubling the number of T. timopheevii Zhuk. chromosomes using colchicine. The authors of [1] designated this plant as an aberrant tetraploid. It is known that Timonovum wheat is a synthetic octaploid (2n = 56), which is characterized by such useful traits as high protein content in grain and good baking qualities, as well as the ability to cause cytoplasmic male sterility in interspecific hybrids. Meanwhile, this wheat is characterized by a slow rate of development and lower yield than T. timopheevii, which appears to be associated with the high ploidy of T. timonovum [2].
Taking into account the genotype of the original wheat, the T. timonovum genome should be designated as GGAtAtGGAtAt, but according to E. Badaeva et al. [3, 4], it should be denoted as GGAtAtBBAuAu since several chromosomes of the A genome and one chromosome of the G genome were replaced by homeologs of the A and B genomes from some unidentified ferro. In addition to this, in a karyotype study of T. fungicidum Zhuk., genomes A, B, At, and G of this wheat were identical to the parent species T. timopheevii and T. carthlicum Nevsk., but at the same time the genomes A and B differed from the corresponding T. timonovum genomes. The authors of the study suggested that genomes A and B were transmitted into the genomes of T. fungicidum and T. timonovum by different species of wheat. Synthetic octaploid T. timonovum is a little-studied plant, and its genome has not yet been studied; in addition, possible changes in the chloroplast genome of this wheat under the influence of such a strong mutagen as colchicine were not studied during the creation of T. timonovum by polyploidization of T. timopheevii.
The purpose of this work is to sequence and annotate the chloroplast genome of T. timonovum.
T. timonovum seeds were provided by the Vavilov All-Russian Institute of Plant Genetic Resources (VIR, St. Petersburg). The plants were germinated in a greenhouse at 21°C for three weeks, then about 20 g of green leaves were collected and placed in the cold (4°C) and darkness for 48 h to reduce starch content. Homogenization of the leaves was carried out using a porcelain mortar and pestle in liquid nitrogen. Chloroplast DNA was isolated from green leaves using the sucrose gradient method [5] with some modifications.
DNA libraries were prepared for sequencing by the shotgun method using the SG GM Plus kit (Raissol, Russia). First, enzymatic fragmentation of chloroplast DNA was carried out to a fragment size of 350 bp with simultaneous end restoration. Next, the adapters were ligated, and the reaction product was purified using Smart beads magnetic particles (Raissol, Russia). At the final stage, index PCR and purification of the reaction product were carried out using Smart beads magnetic particles (Raissol, Russia). DNA libraries were pooled and sequenced using the Genolab M Kit V1.0 100 M reads/flow cell on a Genolab M sequencer (GeneMind, China). The sequencing mode was 2 × 150 bp, base files were demultiplexed on a Genolab M instrument to obtain files with the FASTQ extension. A total of 13 million reads were obtained. The adapters of the prepared library were removed from FASTQ files using the program Trimmomatic v. 0.22 [6]. Trimmed reads were assembled into contigs, which were combined and written into a FASTA file containing information about the complete circular chloroplast DNA molecule, which was generated using the NOVOwrap program [7].
The chloroplast genome was used as a reference genome of T. timopheevii with access number KJ614408.1. Complete T. timonovum chloroplast genome was annotated using the Chloroplast Genome Annotation, Visualization, Analysis, and GenBank Submission 2 (CPGAVAS2) resource (http://47.96. 249.172:16019/analyzer/home) [8]. The circular map of the chloroplast genome was visualized using the Chloroplot resource (https://irscope.shinyapps.io/ Chloroplot) [9]. Alignment of nucleotide sequences of chloroplast genomes was carried out using MAFFT v.7 [10]. The phylogenetic tree was built and visualized using Archaeopteryx (bootstrap = 1000) [11]. Secale cereale subsp. segetale (MZ507427.1) was taken from GenBank as the external species for phylogenetic analysis.
Nucleotide sequence of the chloroplast genome of T. timonovum has been deposited in GenBank (accession number OR936056). Annotation of the chloroplast genome was carried out using the resource CPGAVAS 2; the size of the T. timonovum chloroplast genome amounted to 136 158 bp. The length of the inverted repeat region is 21 552 bp; SSC, 12 795 bp, and LSC, 80 257 bp. The content of GC pairs in the complete plastid genome is 38.3%. In the SSC area, the GC content is 32.17, and in the LSC region, 36.25%. In the region of IR T. timonovum, GC content reaches 43.92%.
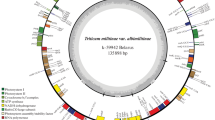
In Timonovum wheat, 132 structural genes have been annotated, of which 85 are protein-coding genes, 31 are tRNA genes, and 4 are rRNA genes. Seven protein-coding genes (rps19, rpl2, rpl23, ndhB, rps7, rps12, rps15), 8 tRNA genes (trnH-GUG, trnM-CAU, trnL-CAA, trnV-GAC, trnT-CGU, trnA-UGC, trnR-ACG, trnN-GUU), and 4 rRNA genes (rRNA4.5, rRNA23, rRNA16 and rRNA5) were duplicated due to being in the IR repeat region. At the same time, 10 protein-coding genes and 1 tRNA gene were found in the SSC region, and 68 protein-coding genes and 22 tRNA genes were found in the LSC region. In addition, 16 of the 132 genes have one intron each (atpF, ndhB, pet В, petD, rpl2, trnl-GAU, ndhA, rpl16, rps16, trnG-UCC, trnK-UUU, trnS-CGA, trnL-UAA, trnV-UAC, trnT-CGU, trnA-UGC) and 1 gene has two introns each (ycf3). The largest intron (2559 bp) is located in the trnk-UUU gene, inside which another gene is located, matK. All obtained results are presented in Fig. 1a in the form of a ring structure obtained using an online resource Chloroplot. Various color blocks on the outer circle of the ring reflect the belonging of genes to certain functional groups.
(a) Visual representation in the form of a ring of a sequenced chloroplast genome T. timonovum k-43065. Genes are shown in different colors; the blue circle in the middle shows the GC level. IRA is inverted repeat region A, IRB is inverted repeat region B. Genes located outside the outer circle are transcribed clockwise, and genes located inside are transcribed counterclockwise. (b) Phylogenetic tree constructed based on alignment of nucleotide sequences of chloroplast genomes of various samples of T. timopheevii from GenBank, T. turgidum MG958546.1 and T. timonovum k-43065. Secale cereale subsp. segetale (MZ507427.1) is presented as an external species.
Based on the analysis of nucleotide sequences of complete chloroplast genomes of various samples of T. timopheevii from GenBank, T. turgidum MG958546.1 and T. timonovum k-43065 (sequenced by us), a phylogenetic tree was constructed, which showed that T. timonovum k-43065 is closest to T. timopheevii (AB976560.1) (Fig. 1b), while other samples of T. timopheevii are located on the tree a little further. T. turgidum MG958546.1 and Secale cereale subsp. segetale (MZ507427.1), as expected, turned out to be external species in relation to the studied wheats. Chloroplast genomes of T. timonovum and T. timopheevii (AB976560.1) differed only in one insert A at position 47891, while other samples of T. timopheevii differed from these wheat samples, similar in chloroplast genome, in various deletions and insertions.
Previously, Japanese authors [12], based on microsatellite analysis of chloroplast DNA of a large number of T. timopheevii samples, identified three subgroups from the wild flora, which can also be seen on the tree we constructed (Fig. 1b). Only sample no. KJ614407.1 was located separately from these three groups.
Unfortunately, the French authors [1] did not indicate in their article which specific sample of tetraploid wheat T. timopheevii with the GA genome they used to create an octaploid species T. timonovum with the GGAA genome. However, according to the nucleotide sequence of the plastid genome, this octaploid turned out to be closest to the Timofeev wheat sample (AB976560.1), sequenced by Georgian authors [13]; however, their paper also does not report the catalog number or the exact geographic origin of the sequenced sample of this wheat.
However, based on the fact that the chloroplast genome of Timonovum wheat is distinguished from other T. timopheevii representatives by the presence of a number of indels, it can be suggested that it is the sample T. timopheevii (AB976560.1) or close to it that was original in obtaining T. timonovum; at the same time, the chloroplast genome of the resulting octaploid did not undergo significant changes under the influence of colchicine.
REFERENCES
Heslot, H. and Raymond, R., Obtention experimentale d’un autotetraploide aberrant (Triticum timonovum) a partir de Triticum timopheevi Zhuk., C. R. Hedb. Séanc. Acad. Sci., 1959, vol. 248, pp. 452—455.
Murashov, V.V. and Morozova, Z.A., Comparative morphogenesis of Triticum timopheevii (Zhyk.) and a synthetic octoploid species T. timonovum Heslot et Ferrary, Moscow Univ. Biol. Sci. Bull., 2008, vol. 63, no. 3, pp. 127—133. https://doi.org/10.3103/S0096392508030073
Badaeva, E.D., Badaev, N.S., Filatenko, A.A., et al., Cytological investigation of cereal hexa- and octoploid species containing G genome, Genetika (Moscow), 1990, vol. 26, no. 4, pp. 708—716.
Badaeva, E.D., Filatenko, A.A., and Badaev, N.S., Cytogenetic investigation of Triticum timopheevii (Zhuk.) Zhuk. and related species using the C-banding technique, Theor. Appl. Genet., 1994, vol. 89, pp. 622—628.
Shi, C., Hu, N., Huang, H., et al., An improved chloroplast DNA extraction procedure for whole plastid genome sequencing, PLoS One, 2012, vol. 7, no. 2. https://doi.org/10.1371/journal.pone.0031468
Bolger, A.M., Lohse, M., and Usadel, B., Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 2014, vol. 30, pp. 2114—2120. https://doi.org/10.1093/bioinformatics/btu170
Wu, P., Xu, C., Chen, H., et al., NOVOWrap: an automated solution for plastid genome assembly and structure standardization, Mol. Ecol. Resour., 2021, vol. 21, no. 6, pp. 2177—2186. https://doi.org/10.1111/1755-0998.13410
Shi, L., Chen, H., Jiang, M., et al., CPGAVAS2, an integrated plastome sequence annotator and analyzer, Nucleic Acids Res., 2019, vol. 47, pp. W65—W73. https://doi.org/10.1093/nar/gkz345
Zheng, S., Poczai, P., Hyvönen, J., et al., Chloroplot: an online program for the versatile plotting of organelle genomes, Front Genet., 2020, vol. 11, p. 576124. https://doi.org/10.3389/fgene.2020.576124
Katoh, K. and Standley, D.M., MAFFT multiple sequence alignment software version 7: improvements in performance and usability, Mol. Biol. Evol., 2013, vol. 30, no. 4, pp. 772—780. https://doi.org/10.1093/molbev/mst010
Han, M.V. and Zmasek, C.M., phyloXML: XML for evolutionary biology and comparative genomics, BMC Bioinf., 2009, vol. 10, pp. 1—6. https://doi.org/10.1186/1471-2105-10-356
Mori, N., Kondo, Y., Ishii, T., et al., Genetic diversity and origin of timopheevi wheat inferred by chloroplast DNA fingerprinting, Bred. Sci., 2009, vol. 59, pp. 571—578. https://doi.org/10.1270/jsbbs.59.571
Gogniashvili, M., Naskidashvili, P., Bedoshvili, D., et al., Complete chloroplast DNA sequences of Zanduri wheat (Triticum spp.), Genet. Resour. Crop. Evol., 2015, vol. 62, pp. 1269—1277. https://doi.org/10.1007/s10722-015-0230-x
ACKNOWLEDGMENTS
We thank the team of the Department of Wheat Genetic Resources of the Vavilov All-Russian Institute of Plant Genetic Resources (VIR, St. Petersburg) for providing seed material for research.
Funding
This work was supported within the framework of the State contract under the Russian Science Foundation, project no. 23-24-00275 “Phylogenetic relationships of individual species of the wheat–aegilops complex of different ploidy levels through the prism of their complete chloroplast genomes with an eye to the origin of the B- and G‑subgenomes of polyploid forms of wheat turgidum-aestivum and timopheevii-zhukovskyi lines.”
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human and animal subjects.
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
Additional information
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kuluev, A.R., Matniyazov, R.T., Kuluev, B.R. et al. Sequencing and Annotation of the Chloroplast Genome of Triticum timonovum Heslot et Ferrary. Russ J Genet 60, 992–995 (2024). https://doi.org/10.1134/S1022795424700455
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795424700455