Abstract
A spectrum of differentially expressed microRNAs was determined by the massively parallel sequencing method in normal healthy prostate tissues, in hormone-dependent prostate cancer samples, and in the LNCaP and DU145 cell lines. A set of microRNAs in tumors and prostate cancer (PCa) cell lines was identified on the basis of the changes in expression compared with that in normal prostate tissues. Twenty-seven aberrantly expressed microRNAs were detected in tumor tissues and ten of them showed significant changes in expression in LNCaP and DU145 cells. Seven of them demonstrated the change of the expression in the same direction in all the tumor samples as well as in the PCa cell lines. The expression of miR-148a changed in DU145 cells in the opposite direction compared with that in LNCaP cells and tumors. The expression of let-7c, let-7b, miR-99a, miR-125b-2, miR-100, miR-10a, and miR-31 was reversed in DU145 cells compared with LNCaP cells. However, these microRNAs exhibited no significant changes in expression in tumors. It turns out that the target of miR-148a, let-7b, and microRNAs, included in the miR-99a/let-7c/miR-125b-2 cluster, the expression of which increased in LNCaP cells and decreased in DU145 cells, is the insulin-like growth factor receptor gene 1 (IGF1R). The obtained data make it possible to assume that the differences in the effect of microRNAs in cell lines are connected with their repressive influence on IGF1R expression in hormone-sensitive LNCaP cells and an absence of such influence in the hormone-independent DU145 cell line.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
MicroRNAs are short, 19–23 nucleotides in size, RNA molecules, which, associating with Argonaute proteins (AGO1–4), form a RISC complex (RNA-induced silencing complex). The function of the mature microRNA is to recognize the target messenger RNAs by the RISC complex. The interaction of this complex with mRNA leads either to its degradation or to the suppression of translation [1, 2]. According to different estimates, from 30 to 60% of human genes are under the direct control of microRNAs [3]. MicroRNAs play a key role in the regulation of major genetic processes, such as cell division [4], apoptosis [5], DNA repair [6], differentiation [7, 8], and the cell’s response to stress [9].
Mutations and, as shown in the last 15–20 years, epigenetic disorders of oncogenes and tumor suppressor genes lead to malignant degeneration of cells of various organs, including the prostate. In developed countries, prostate cancer is the second most common cancer among men. It is well known that normal tissue cells, like prostate tumors, need androgens in the early stages of development [10]. However, during development, a malignant tumor of the prostate often acquires the capability of hormone-independent growth and progression. This limits the possibilities of hormone therapy, which is widely used in the treatment of prostate cancer.
Despite intensive research, the mechanism of the origin of hormone-independent prostate tumors remains unclear, and today the role of microRNAs in this process is not clear either. Earlier, microRNAs were detected the expression of which decreases in cells of hormone-independent prostate cancer lines as compared to cells of hormone-dependent lines. These include the miR families miR-99 (miR-99a-99b-100) [11], miR-148a [12], let-7c [13], and others, such as miR-34a,c, miR-146a, miR-124, miR-31, miR-200b, and miR-185 [14].
It is known that the IGF1R signaling pathway plays a significant role in the development of prostate cancer. The expression level of the IGF1R gene in cells of the hormone-independent DU145 line is significantly higher than that in the cells of the hormone-dependent LNCaP line [15, 16]. However, the role of microRNAs in overexpression of IGF1R in DU145 cells remains unclear. This paper is devoted to the analysis of the possible role of microRNA-mediated regulation of IGF1R in changing the dependence of prostate cancer cells on hormones.
MATERIALS AND METHODS
Tissue samples. Samples of hormone-dependent tumors of local prostate cancer (PCa) and normal prostate tissues were provided by the Research Institute of Urology and Nephrology of Rostov State Medical University. Normal prostate tissues used as a control were obtained during radical cystectomy. None of the patients received hormone therapy or chemotherapy before surgery. Tissue samples were treated with stabilizing solution of RNAlater (Ambion, United States) immediately after surgery for 24 h and then crushed and frozen for subsequent isolation of RNA. All tumor samples were morphologically characterized according to the TNM system [17] and the Gleason scale [18] (Table 1). The study was approved by the ethics committee of Rostov State Medical University.
Immunohistochemistry. Immunohistochemical staining was performed for morphological characterization of tissue samples and assessment of the status of androgen receptors. Five 4 μm consecutive sections were obtained from formalin-paraffin-fixed tissue samples and mounted on prepared glass. Immunostaining was performed using the Ventana HX BenchMark™ system (Ventana Medical Systems, United States) according to the manufacturer’s recommendations. Monoclonal mouse antibodies against the androgen receptor were used in a dilution of 1/50 (clone AR441, DAKO, United States). Diaminobenzidine was used as a chromogenic substrate. Hematoxylin and eosin were used for contrasting staining.
Cell lines. The hormone-dependent prostate cancer LNCaP cells were cultured in RPMI-1460 medium, and the hormone-independent DU145 cells were cultured in EMEM medium. Ten volume percent of fetal bovine serum was added to the basic medium. Cells were cultured at 37°C in the presence of 5% CO2 in 75 cm2 flasks (with a time interval between consecutive subcultures of 3 days).
RNA isolation. To isolate total RNA from prostate tissue samples, a fragment of 0.5 cm3 was crushed in liquid nitrogen and placed in QIAzol lysis buffer (Qiagen, United States). To isolate RNA from cell lines, 2 × 106 cells were used. Total RNA was isolated using the miRNeasy Mini Kit (Qiagen, United States) according to the manufacturer’s protocol. The miRNeasy Mini Kit (Qiagen, United States) and the RNeasy MinElute Cleanup Kit (Qiagen, United States) were used to isolate the small RNA fractions. Quantitative determination of total RNA was measured on a Qubit 3.0 Fluorometer (Life Science Technologies, United States). For qualitative analysis of total RNA, electrophoresis was carried out in a denaturing 1% agarose gel with formaldehyde.
Massively parallel sequencing. The microRNA expression was determined using massively parallel sequencing on the Illumina platform. For the evaluation of the first repeat in DU145 and LNCaP, the MiSeq system (Illumina Inc., United States) was used; for the second replication of these cell lines and samples of tumor and normal tissues, the HiSeq system (Illumina Inc., United States) was used. In the first series of experiments, the MiSeq system (Illumina Inc., United States) was used for DU145 and LNCaP and HiSeq (Illumina Inc., United States) was used for the other cases. Creating a cDNA library and massively parallel sequencing were performed according to the method described by us earlier [19].
Statistical analysis. In identifying microRNAs, cDNA sequences were compared with canonical microRNA sequences (miRBase, v. 21) using Bowtie, v. 1.2.0 [20]. In this case, one nucleotide was allowed to be replaced in the cDNA sequences of mature microRNA. When normalizing the expression level of the compared variants, the TMM method implemented in the edgeR computer program was used [21]. MicroRNAs are included in the analysis, the mature forms of which are represented in the analyzed sample by no less than 25 copies in a sample of a continuous cell line or samples of prostate tissue and which showed a change in expression with a false detection rate (FDR) ≤ 0.05.
RESULTS
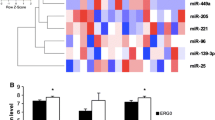
Using the method of massively parallel sequencing, the spectrum of differential expression of microRNAs was obtained in cells of three samples of hormone-dependent prostate cancer and three samples of normal prostate tissues, as well as in cells of the LNCaP and DU145 lines in duplicate. The expression of microRNAs was compared in cells of each tumor sample and in LNCaP and DU145 cells with expression of microRNAs in cells of each sample of normal prostate tissues. As a result, nine comparisons were performed for tumors and six comparisons were performed for each of the LNCaP and DU145 lines. The analyzed microRNAs showed a statistically significant (FDR ≤ 0.05) change in expression with each comparison in the same direction. Twenty-seven microRNAs (Table 2) were identified in tumor cells, while 116 were identified in LNCaP cells and 87 were identified in DU145 cells (Fig. 1).
MicroRNAs expressed in the same direction in the studied lines and prostate cancer. (a) Spectra of microRNAs with expression increased in the same direction in LNCaP and DU145 cells and prostate cancer; (b) spectra of microRNAs with expression reduced in the same direction in LNCaP and DU145 cells and prostate cancer.
Comparison of microRNAs expressed in the same direction in hormone-dependent tumor cells and the LNCaP showed that expression of only 16 microRNAs changed at the same time. And in seven of them, expression changed in the same direction in DU145 cells (Table 3). The expression of these microRNAs reflects changes in metabolism in malignant cells that are not related to their dependence on hormones.
For four of them—miR-150-5p [22, 23], miR-223-3p [24], miR-378a-3p [25], and miR-486-5p [26]—participation in the control of prostate cancer was previously shown. Direct data on the participation in the regulation of prostate carcinogenesis of three other microRNAs included in this group were not found. However, it is known that miR-451a is involved in the control of colorectal cancer and breast cancer [27, 28], miR-126-3p is involved in the control of breast cancer [29] and thyroid cancer [30], and miR-144-5p is involved in the control of bladder cancer [31].
In eight microRNAs of cell lines, the direction of change in expression was reversed when moving from LNCaP cells to DU145 cells, while the miR-148a expression level was also changed in tumor cells (Table 4). For all five microRNAs that showed an increase in expression in LNCaP cells and, accordingly, its decrease in DU145 cells, the target is the IGF1R gene.
DISCUSSION
The key role in the development of prostate cancer is played by the androgen receptor (AR), whose interaction with androgens leads to its activation [32, 33]. Activated AR acquires the ability to move from the cytoplasm to the cell nucleus [34]. Its interaction with AREs (androgen response elements)—canonical DNA sequences in promoter regions of genes—leads to the suppression or activation of their transcriptional activity [35]. However, activation of AR and its movement into the cell nucleus can occur in the absence of androgens when the IGF-1 signaling pathway is activated [36]. Over 400 genes are androgen receptor targets [37]. These include the miR-148a gene [38], the direction of expression of which changes to the opposite when moving from LNCaP cells to DU145 (Table 4). The direct targets of this microRNA are DNA methyltransferases DNMT3b [39, 40] and DNMT1 [41], which are responsible for de novo methylation and methylation maintenance, respectively (Fig. 2).
MicroRNA dependent regulation of IGF1R expression in cells of the LNCaP and DU145 lines. (a) Regulation scheme in LNCaP cells; (b) regulation scheme in DU145 cells. ( ) Increased gene expression or protein activation; (
) Increased gene expression or protein activation; ( ) suppression of expression; (
) suppression of expression; ( ) absence (weakening) of regulatory action; (
) absence (weakening) of regulatory action; ( ) increased gene expression; (
) increased gene expression; ( ) reduced gene expression; (
) reduced gene expression; ( ) predicted regulation; (
) predicted regulation; ( ) verified regulation.
) verified regulation.
In addition, according to miRDB (http://mirdb.org) and TargetScan (http://www.targetscan.org), the KDM2B gene encoding lysine-specific demethylase 2B also belongs to the predicted miR-148a targets. KDM2B is part of the polycomb complex 1 (PRC1). Its binding to completely unmethylated DNA of CpG islands leads to lysine monoubiquitinating at position 119 of histone 2A (H2AK119 Ub1) and recruitment to the binding site of polycomb complex 2 (PRC2), of which histone methyltransferase 2 (EZH2) is a component. This leads to trimethylation of lysine in the 27th position of histone 3 (H3K27me3) and, accordingly, to histone-dependent suppression of the transcriptional activity of genes whose promoters contain CpG islands. In addition, H3K27me3 is a molecular label for de novo DNA methylation [42].
It is known that the promoter region of the let-7b microRNA gene, the target of which is histone methyltransferase EZH2 [43], contains CpG islands and its transcription depends on the level of DNA methylation [44]. Using the example of epigenetic regulation of cell division of mouse embryonic fibroblasts [43] and myeloid cell lines [45], it was shown that activation of KDM2B leads to activation of EZH2 and a decrease in let-7b expression, and, on the contrary, a decrease in expression of KDM2B is accompanied by a decrease in expression of EZH2 and an increase in expression of let-7b. In turn, San et al. [46] showed that EZH2 activity inhibits the activity of the MIR99AHG gene. The transcript of this gene contains a miR-99a/let-7c/miR-125b-2 microRNA cluster. The target of microRNAs included in this cluster, as well as miR-148a and let-7b, is the IGF1R gene.
For miR-148a, which showed an increase in expression in LNCaP cells, we also observe an increase in expression in hormone-dependent tumor cells. Differences in microRNA expression in LNCaP cells and tumor cells affect let-7c, let-7b, miR-99a, and miR-125b. The nature of these differences remains unclear, but it can be assumed that it is associated with changes in the expression of genes whose proteins are involved in the differential maturation of microRNA, for example, LIN-28 [13, 47]. It has now been shown that IGF1R plays a significant role in the development of hormonal independence of prostate cancer cells [48]. This may, in particular, be associated with androgen-independent activation of androgen receptors [36]. However, Alimir et al. [49] showed that, in the hormone-independent DU145 and PC3 cells, the activity of the AR gene is reduced compared with that in hormone-dependent LNCaP cells. In this regard, it is possible that the bypass signaling pathway, dependent on IGF1R, but not on AR, lies in the basis of the division and survival of hormone-independent cells cultured in vitro [50, 51].
Our results suggest that the differences in the effect of microRNAs in the two cell lines studied are mainly related to their repressive effect on the expression of IGF1R in cells of the hormone-dependent LNCaP and the lack of this effect in cells of the hormone-independent DU145.
REFERENCES
Lin, S. and Gregory, R.I., MicroRNA biogenesis pathways in cancer, Nat. Rev. Cancer, 2015, vol. 15, no. 6, pp. 321—333. https://doi.org/10.1038/nrc3932
Kim, Y.K., Kim, B., and Kim, V.N., Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis, Proc. Natl. Acad. Sci. U.S.A., 2016, vol. 113, no. 13, pp. E1881—E1889. https://doi.org/10.1073/pnas.1602532113
Friedman, R.C., Farh, K.K.H., Burge, C.B., and Bartel, D.P., Most mammalian mRNAs are conserved targets of microRNAs, Genome Res., 2009, vol. 19, no. 1, pp. 92—105. https://doi.org/10.1101/gr.082701.108
Shenoy, A. and Blelloch, R.H., Regulation of microRNA function in somatic stem cell proliferation and differentiation, Nat. Rev. Mol. Cell Biol., 2014, vol. 15, no. 9, pp. 565—576. https://doi.org/10.1038/nrm3854
Lima, R.T., Busacca, S., Almeida, G.M., et al., MicroRNA regulation of core apoptosis pathways in cancer, Eur. J. Cancer, 2011, vol. 47, no. 2, pp. 163—174. https://doi.org/10.1016/j.ejca.2010.11.005
Landau, D.A. and Slack, F.J., MicroRNAs in mutagenesis, genomic instability, and DNA repair, Semin. Oncol., 2011, vol. 38, no. 6, pp. 743—751. https://doi.org/10.1053/j.seminoncol.2011.08.003
Ivey, K.N. and Srivastava, D., MicroRNAs as regulators of differentiation and cell fate decisions, Cell Stem Cell, 2010, vol. 7, no. 1, pp. 36—41. https://doi.org/10.1016/j.stem.2010.06.012
Turner, M.L., Schnorfeil, F.M., and Brocker, T., MicroRNAs regulate dendritic cell differentiation and function, J. Immunol., 2011, vol. 187, no. 8, pp. 3911—3917. https://doi.org/10.4049/jimmunol.1101137
Leung, A.K. and Sharp, P.A., MicroRNA functions in stress responses, Mol. Cell, 2010, vol. 40, no. 2, pp. 205—215. https://doi.org/10.1016/j.molcel.2010.09.027
Cucchiara, V., Yang, J.C., Mirone, V., et al., Epigenomic regulation of androgen receptor signaling: potential role in prostate cancer therapy, Cancers, 2017, vol. 9, no. 1, p. 9. https://doi.org/10.3390/cancers9010009
Sun, D., Lee, Y.S., Malhotra, A., et al., miR-99 family of microRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation, Cancer Res., 2011, vol. 71, no. 4, pp. 1313—1324. https://doi.org/10.1158/0008-5472.can-10-1031
Fujita, Y., Kojima, K., Ohhashi, R., et al., miR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression, J. Biol. Chem., 2010, vol. 285, no. 25, pp. 19076—19084. https://doi.org/10.1074/jbc.M109.079525
Nadiminty, N., Tummala, R., Lou, W., et al., MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells, J. Biol. Chem., 2012, vol. 287, no. 2, pp. 1527—1537. https://doi.org/10.1074/jbc.M111.278705
Li, F. and Mahato, R.I., MicroRNAs and drug resistance in prostate cancers, Mol. Pharm., 2014, vol. 11, no. 8, pp. 2539—2552. https://doi.org/10.1021/mp500099g
Iwamura, M., Sluss, P.M., Casamento, J.B., and Cockett, A.T., Insulin-like growth factor I: action and receptor characterization in human prostate cancer cell lines, Prostate, 1993, vol. 22, no. 3, pp. 243—252. https://doi.org/10.1002/pros.2990220307
Rochester, M.A., Riedemann, J., Hellawell, G.O., et al., Silencing of the IGF1R gene enhances sensitivity to DNA-damaging agents in both PTEN wild-type and mutant human prostate cancer, Cancer Gene Ther., 2005, vol. 12, no. 1, pp. 90—100. https://doi.org/10.1038/sj.cgt
Schröder, F.H., Hermanek, P., Denis, L., et al., The TNM classification of prostate cancer, Prostate Suppl., 1992, vol. 21, no. 4, pp. 129—138. https://doi.org/10.1002/pros.2990210521
Mellinger, G.T., Gleason, D., and Bailar, J., The histology and prognosis of prostatic cancer, J. Urol., 1967, vol. 97, no. 2, pp. 331—337. https://doi.org/10.1016/S0022-5347(17)63039-8
Tarasov, V.A., Boiko, N.V., Makhotkin, M.A., et al., The miRNA aberrant expression dependence on DNA methylation in HeLa cells treated with mitomycin C, Russ. J. Genet., 2016, vol. 52, no. 11, pp. 1117—1123. https://doi.org/10.1134/S1022795416110156
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S.L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol., 2009, vol. 10, no. 3. R25. https://doi.org/10.1186/gb-2009-10-3-r25
Robinson, M.D., McCarthy, D.J., and Smyth, G.K., edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics, 2010, vol. 26, no. 1, pp. 139—140. https://doi.org/10.1093/bioinformatics/btp616
Dezhong, L., Xiaoyi, Z., Xianlian, L., et al., miR-150 is a factor of survival in prostate cancer patients, J. BUON, 2015, vol. 20, no. 1, pp. 173—179.
Liu, D.Z., Zhang, H.Y., Long, X.L., et al., MIR-150 promotes prostate cancer stem cell development via suppressing p27Kip1, Eur. Rev. Med. Pharmacol. Sci., 2015, vol. 19, no. 22, pp. 4344—4352.
Kurozumi, A., Goto, Y., Matsushita, R., et al., Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer, Cancer Sci., 2016, vol. 107, no. 1, pp. 84—94. https://doi.org/10.1111/cas.12842
Avgeris, M., Stravodimos, K., and Scorilas, A., Loss of miR-378 in prostate cancer, a common regulator of KLK2 and KLK4, correlates with aggressive disease phenotype and predicts the short-term relapse of the patients, Biol. Chem., 2014, vol. 395, no. 9, pp. 1095—1104. https://doi.org/10.1515/hsz-2014-0150
Zhang, X., Zhang, T., Yang, K., et al., miR-486-5p suppresses prostate cancer metastasis by targeting snail and regulating epithelial–mesenchymal transition, Onco. Targets Ther., 2016, vol. 9, pp. 6909—6914. https://doi.org/10.2147/ott.s117338
Bitarte, N., Bandres, E., Boni, V., et al., MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells, Stem Cells, 2011, vol. 29, no. 11, pp. 1661—1671. https://doi.org/10.1002/stem.741
Kovalchuk, O., Filkowski, J., Meservy, J., et al., Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin, Mol. Cancer Ther., 2008, vol. 7, no. 7, pp. 2152—2159. https://doi.org/10.1158/1535-7163.mct-08-0021
Zhu, N., Zhang, D., Xie, H., et al., Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2, Mol. Cell. Biochem., 2011, vol. 351, nos. 1—2, pp. 157—164. https://doi.org/10.1007/s11010-011-0723-7
Xiong, Y., Kotian, S., Zeiger, M.A., et al., miR-126-3p inhibits thyroid cancer cell growth and metastasis, and is associated with aggressive thyroid cancer, PLoS One, 2015, vol. 10, no. 8. e0130496. https://doi.org/10.1371/journal.pone.0130496
Matsushita, R., Seki, N., Chiyomaru, T., et al., Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer, Br. J. Cancer, 2015, vol. 113, no. 2, pp. 282—289. https://doi.org/10.1038/bjc.2015.195
Liu, L.L., Xie, N., Sun, S., et al., Mechanisms of the androgen receptor splicing in prostate cancer cells, Oncogene, 2014, vol. 33, no. 24, pp. 3140—3150. https://doi.org/10.1038/onc.2013.284
Liao, R.S., Ma, S., Miao, L., et al., Androgen receptor-mediated non-genomic regulation of prostate cancer cell proliferation, Transl. Androl. Urol., 2013, vol. 2, no. 3, pp. 187—196. https://doi.org/10.3978/j.issn.2223-4683.2013.09.07
Watson, P.A., Arora, V.K., and Sawyers, C.L., Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer, Nat. Rev. Cancer, 2015, vol. 15, no. 12, pp. 701—711. https://doi.org/10.1038/nrc4016
Denayer, S., Helsen, C., Thorrez, L., et al., The rules of DNA recognition by the androgen receptor, Mol. Endocrinol., 2010, vol. 24, no. 5, pp. 898—913. https://doi.org/10.1210/me.2009-0310
Wu, J.D., Haugk, K., Woodke, L., et al., Interaction of IGF signaling and the androgen receptor in prostate cancer progression, J. Cell. Biochem., 2006, vol. 99, no. 2, pp. 392—401. https://doi.org/10.1002/jcb.20929
Liu, S., Kumari, S., Hu, Q., et al., A comprehensive analysis of coregulator recruitment, androgen receptor function and gene expression in prostate cancer, eLife, 2017, vol. 6. e28482. https://doi.org/10.7554/eLife.28482
Hamilton, M.P., Rajapakshe, K.I., Bader, D.A., et al., The landscape of microRNA targeting in prostate cancer defined by AGO-PAR-CLIP, Neoplasia, 2016, vol. 18, no. 6, pp. 356—370. https://doi.org/10.1016/j.neo.2016.04.008
Li, Y., Deng, X., Zeng, X., and Peng, X., The role of mir-148a in cancer, J. Cancer, 2016, vol. 7, no. 10, pp. 1233—1241. https://doi.org/10.7150/jca.14616
Duursma, A.M., Kedde, M., Schrier, M., et al., miR-148 targets human DNMT3b protein coding region, RNA, 2008, vol. 14, no. 5, pp. 872—877. https://doi.org/10.1261/rna.972008
Braconi, C., Huang, N., and Patel, T., MicroRNA dependent regulation of DNMT-1 and tumor suppressor gene expression by Interleukin-6 in human malignant cholangiocytes, Hepatology, 2010, vol. 51, no. 3, pp. 881—890. https://doi.org/10.1002/hep.23381
Wang, W., Qin, J.J., Voruganti, S., et al., Polycomb group (PcG) proteins and human cancers: multifaceted functions and therapeutic implications, Med. Res. Rev., 2015, vol. 35, no. 6, pp. 1220—1267. https://doi.org/10.1002/med.21358
Tzatsos, A., Paskaleva, P., Lymperi, S., et al., Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells, J. Biol. Chem., 2011, vol. 286, no. 38, pp. 33061—33069. https://doi.org/10.1074/jbc.M111.257667
Vrba, L., Muñoz-Rodríguez, J.L., Stampfer, M.R., and Futscher, B.W., miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer, PLoS One, 2013, vol. 8, no. 1. e54398. https://doi.org/10.1371/journal.pone.0054398
Karoopongse, E., Yeung, C., Byon, J., et al., The KDM2B-let-7b-EZH2 axis in myelodysplastic syndromes as a target for combined epigenetic therapy, PLoS One, 2014, vol. 9, no. 9. e107817. https://doi.org/10.1371/journal.pone.0107817
Sun, D., Layer, R., Mueller, A.C., et al., Regulation of several androgen-induced genes through the repression of the miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells, Oncogene, 2014, vol. 33, no. 11, pp. 1448—1457. https://doi.org/10.1038/onc.2013.77
Nadiminty, N., Tummala, R., Lou, W., et al., MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth, PLoS One, 2012, vol. 7, no. 3. e32832. https://doi.org/10.1371/journal.pone.0032832
Denduluri, S.K., Idowu, O., Wang, Z., et al., Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance, Genes Dis., 2015, vol. 2, no. 1, pp. 13—25. https://doi.org/10.1016/j.gendis.2014.10.004
Alimirah, F., Chen, J., Basrawala, Z., et al., DU-145 and PC-3 human prostate cancer cell lines express androgen receptor: implications for the androgen receptor functions and regulation, FEBS Lett., 2006, vol. 580, no. 9, pp. 2294—2300. https://doi.org/10.1016/j.febslet.2006.03.041
Ferraldeschi, R., Welti, J., Luo, J., et al., Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects, Oncogene, 2015, vol. 34, no. 14, pp. 1745—1757. https://doi.org/10.1038/onc.2014.115
Edwards, J. and Bartlett, J., The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer: 2. Androgen-receptor cofactors and bypass pathways, BJU Int., 2005, vol. 95, no. 9, pp. 1327—1335. https://doi.org/10.1111/j.1464-410X.2005.05527.x
Funding
This work was supported by the state task for the Southern Scientific Center, Russian Academy of Sciences, state registration number 01201363192.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflict of interest.
Statement of compliance with standards of research involving humans as subjects. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants involved in the study.
Additional information
Translated by A. Kashevarova
Rights and permissions
About this article
Cite this article
Tarasov, V.A., Naboka, A.V., Makhotkin, M.A. et al. The Influence of microRNAs in Regulation of Hormone Dependence in Prostate Cancer Cells. Russ J Genet 55, 720–727 (2019). https://doi.org/10.1134/S1022795419050132
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795419050132