Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that function in transcriptional and post-transcriptional regulation of gene expression. Several miRNAs have been implicated in regulating prostate cancer (PCa) progression. Deregulations of miRNA regulatory networks have been reported in ERG positive PCa, which accounts for ~50 % of PCa and have been suggested to affect tumor aggressiveness. The function of miR338-3p, its prognostic significance, and its association with ERG positive PCa has not been fully investigated. Using microarray expression profiling, we identified miRNA338-3p as among the top deregulated miRNAs associated with ERG status in PCa. We investigated miR338-3p function using in vitro and in vivo experimental models and its expression was assessed and validated in clinical samples and a public cohort of localized and metastatic prostate cancer. miR338-3p was significantly down-regulated with disease progression from benign prostate tissue to primary and metastatic lesions. In localized disease, patients with lower miR338-3p expression levels showed increased association to biochemical recurrence and several adverse pathological parameters compared to patients with higher miRNA338-3p tissue expression levels. Using in vitro PCa cell models, overexpression of miR338-3p resulted in a decrease in cell invasion and expression of chemokine signalling genes CXCL12, CXCR4, and CXCR7. In vivo, orthotropic implantation of PC3 cells stably expressing miR338-3p was associated with a significant decrease in tumor weights compared to control cells. miR338-3p has anti-proliferative and anti-invasive properties. It affects CXCR4 axis, and its down-regulation is associated with adverse clinical outcomes in PCa patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer is the most common cancer diagnosed in North American men, excluding skin cancers. It is estimated that in 2015, approximately 220,800 new cases and 27,540 prostate cancer-related deaths will occur in the United States [1], primarily as a result of metastatic dissemination of the primary tumor. Although androgen ablation therapy represents a significant milestone in the management of advanced PCa disease, tumors eventually become resistant in castration-resistant PCa (CRPC), which usually manifest by rapid disease progression and shorter survival time [2]. ERG gene rearrangements are the most common genetic aberration described in PCa (affecting approximately 50 % of localized PCa cases), with TMPRSS2 representing the most common ERG partner [3]. Earlier studies suggested that ERG gene rearrangements define a specific molecular subtype of PCa with potential prognostic and therapeutic implications [4]. However, although the current prognostic significance of ERG gene rearrangements remains debated, several reports have shown that TMPRSS2-ERG gene fusions promote cancer progression and invasion [5, 6] and induce epithelial-mesenchymal transition in PCa cellular models [7]. Moreover, in vivo TMPRSS2-driven ERG expression was reported to increase cancer stem cells (CSC) self-renewal in a castration resistant subpopulation [8].
The transcript abundances of miRNAs are subject to regulatory control by many more loci than previously observed for mRNA expression particularly in TMPRSS2-ERG positive prostate cancer cells [9]. As well, a possible relationship between ERG and microRNAs (miRNAs), commonly deregulated during PCa progression [10, 11], has been proposed although the connection to ERG gene rearrangements has not yet been fully established. For instance, one study reported that miR221 is down-regulated in association with TMPRSS2-ERG gene fusion [12], and another reported the regulation of ERG expression by miR-145 [13].
miRNAs are an evolutionarily conserved class of endogenous, small, noncoding RNAs (19–25 nucleotides), involved in the regulation of expression of several target genes. They exert their function by binding to the 3′-untranslated region of a subset of mRNAs, resulting in their degradation and/or repression of their translation [14, 15]. About 30 % of the proteins coding the human transcripts are predicted to be regulated by miRNAs [16, 17] and evidence points to the controversial role of miRNAs in promoting either tumor suppressor or oncogenic activity [18–20].
The precursor miR338-3p sequence is intronically encoded within the Apoptosis-associated Tyrosine Kinase (AATK, also known as AATYK) host gene [21]. Both AATK and miR338-3p are highly conserved genes that are prominently expressed in the vertebrate central nervous system [22–24]. A recent study reported a possible mechanism by which miR338-3p participates in the regulation of its host genes via modulation of the levels of AATK mRNA, a kinase that plays a role in differentiation, apoptosis and possibly neuronal degeneration [24]. miR338-3p also contributes to the formation of basolateral polarity in epithelial cells [25]. Of equal relevance to this study, among potential functional target gene for ERG transcription factor in PCa is CXCR4, an alpha-chemokine receptor specific for CXCL12 [26, 27]. CXCR-4 plays a crucial role in invasion and metastasis of PCa cells along with CXCL12 and CXCR7 [28–30]. Moreover, CXCR4 overexpression in PCa cells accelerated tumor metastasis and growth in vivo [31] and has been shown to be directly affected by ERG, which directly binds to and promotes CXCR4 expression [32].
In this study, we performed a miRNA expression profiling comparing PCa tumors harbouring ERG gene rearrangements versus those without. We identified miR338-3p among the top differentially expressed miRNAs between the two groups. As the role of miR338-3p in PCa has never been investigated before, the current study investigates and characterizes the role of miR338-3p in PCa progression and in relation to ERG gene rearrangements and CXCR axis.
Materials and methods
Clinical samples and identification of miRNA signature in relation to ERG rearrangement
We used seventeen samples representing locally advanced castration resistant prostate cancer (CRPC) [9 ERG positive (ERG1) and 8 ERG negative (ERG0)], for miRNA differential gene expression analysis. Differential expression analysis using mean fold difference (MFD) and statistical student t test were conducted. All patients’ information and samples were collected with appropriate ethical approval from the local institutional review board at the University of Calgary, Calgary, Alberta.
Assessment of miR338-3p levels in clinical samples
miR338-3p expression levels was assessed in a cohort of 25 matched benign and localized PCa samples collected with appropriate ethical approval from the Institutional Review Board at University of Calgary. To assess and validate the clinical and prognostic significance of miR338-3p expression in PCa, we used the Memorial Sloan Kettering Cancer Center (MSKCC) cohort [33] (n = 139), where we investigated the relationship between miR338-3p expression and ERG status, aggressive or non-aggressive cancer, pathological stage, PSA at diagnosis and Gleason score as well as the association of miRNA338-3p expression to patients’ clinical outcome.
Cell culture
Human Prostate carcinoma cell lines LNCaP, DU145, VCaP, PC3, and human normal prostate epithelial cell line RWPE-1 were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). Various prostate cell lines with different PTEN and TMPRSS2-ERG status were utilized in our study to evaluate whether miR338-3p overexpression or knockdown would have an effect on proliferation and invasion in these cells. While PC-3 cells have sustained a homozygous deletion of PTEN, LNCaP cells have a deletion of one allele and a mutation of the other PTEN allele. Furthermore, DU145 cells contain one wild-type PTEN allele and a second variant allele while VCaP retained intact PTEN. Cells were maintained in culture medium according to the manufacturers’ instructions and in 5 % CO2 at 37 °C. All cell lines were authenticated by on-site DNA (STR) profiling using an Applied Biosystems 3130 Genetic Analyzer with AmpF/STR Identifiler PCR Amplification Kit from Applied Biosystems (Life Technologies Corp., Carlsbad, CA, USA).
PC3-miR338-3p and transiently silenced VCaP-miR338-3p-siRNA cells were generated in our laboratory. Details of these cell variants are illustrated in the supplementary materials and methods.
RNA extraction
Total RNA was extracted from PCa cell lines using the miRNeasy Mini Kit (Qiagen Sciences, Maryland, USA) according to the manufacturer’s protocol.
RNA was extracted from formaldehyde-fixed, paraffin-embedded (FFPE) tissue samples with the RecoverAll™ Total Nucleic Acid Isolation Kit (Ambion, Austin, TX, USA).
miRNA and RNA analysis
Mature miRNA expression was quantified in tissue samples and in cell lines with the TaqMan® miRNA assay RT kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s protocol. Sample analysis was performed using the ABI StepOne Plus system (Applied Biosystems). Fold changes in miRNA expression between samples and controls were determined by the comparative (ΔΔCt) method. RNU48 was used as endogenous reference gene and experiments were done in triplicates. For different gene expression analyses, 1 µg of total RNA was used as template in the reverse transcription reaction using qScript™ cDNA SuperMix (Quanta BioSciences, Gaithersburg, MD) according to manufacturer’s protocol. Real-time PCR was performed by the ABI StepOne Plus system (Applied Biosystems) using PerfeCTa™ SYBR Green FastMix™ (Quanta BioSciences). Beta-glucoronidase (GUSB) was used as the endogenous control. The relative expression of target genes (CXCR4, CXCR7 and CXCL12) was calculated using the comparative (ΔΔCt) method using the data assist software provided by Applied Biosystems.
Cell viability assay
Cell viability was assessed by XTT Cell Viability Kit (Biotium, Hayward, CA) as recommended by the manufacturer. LNCaP and PC3 cells were seeded in 96-well plates and left overnight to adhere to the surface of the plates. VCaP cells were allowed to adhere for 48 h. Cells were seeded at a density of 2.5 × 103 (PC3), 5 × 103 (LNCaP), and 2.5 × 104 (VCaP) cells/well. Fifty microlitres of the prepared XTT working solution was added to each well and absorbance was read at 490 nm with a reference wavelength of 630–690 nm.
Invasion assay
Cell invasion assay was performed using 8 μm porous BD BioCoat™ Matrigel™ Invasion Chambers (BD Biosciences, NJ, USA) according to the manufacturer’s recommendations. Control cell migration experiments were performed in uncoated transwell chambers (BD Biosciences) under exact conditions as invasion assays. Briefly, cells were serum starved for 24 h and then 105 cells in 500 µl were seeded in serum free media in the upper compartment of the trans-well chambers. The lower compartment contained 750 µl of either serum free media (RPMI-1640) or media supplemented with 100 ng/ml recombinant human SDF-1α (Peprotech, Ricky Hill, NJ) as a chemoattractant. After 24 h incubation, the Matrigel (including non-migrating cells) was removed and cells invading the membrane were fixed and stained with 0.1 % crystal violet in 95 % ethanol, and quantified. Data was expressed as percent invasion which is calculated by dividing the mean of cells invaded through Matrigel insert membrane by the mean of cells migrating through the control inserts membrane multiplied by 100.
In vivo othrotopic tumor implantation in mice
In vivo studies were approved by the McGill Animal Care Committee (Protocol number 4101) and were conducted in accordance with institutional and Canadian Federal Guidelines. SCID mice (male) were purchased from Charles River Laboratories (St. Zotique, Quebec, Canada). Mice (n = 15) were anaesthetized using sodium pentobarbital (given intra-peritoneal) and one million PC3 cells (at 70 % confluence) were implanted in the prostate via a lower midline incision, as previously described [34]. Formation of a bulla indicated a satisfactory injection. The incision was then closed with a single layer of surgical clips. At the end of experiment (larger palpable tumors), animals were sacrificed, lymph nodes examined, and prostate tumors isolated and weighed.
ChIPBase analysis
To find potential transcription factors (TFs) that have binding sites in the region upstream of the miR-338 gene we used CHIPBase database that has a comprehensive annotation of the transcription factors binding maps that were inferred from more than 500 Chip-seq data [35]. We only focused on the 5 kb upstream region of the miR-338-3p gene.
Statistical analysis and bioinformatics
Data were analyzed by the student’s t test (Two-tailed) using p < 0.05 to define statistically significant differences. For differential expression analysis, we used significant analysis of microarray (SAM) to identify differentially expressed miRNAs in ERG positive vs ERG negative prostate samples. Kaplan–Meier survival model, cox proportional hazards regression analysis and the log rank test were used for disease recurrence analysis and for the multivariate analysis. Independent miRNA expression data (GSE21036) was downloaded from public cohort cBio Cancer Genomics Portal (http://cbio.mskcc.org) to validate the significance of association between miR338-3p and other clinical factors. To identify potential targets for miR338-3p, we used microRNA.org, FindTar3 and CHIPbase online bioinformatics tools.
Results
miRNA expression profiling in ERG-positive and ERG-negative PCa
To identify miRNAs associated with ERG gene rearrangements, we assessed global miRNA gene expression profiles in 17 samples of CRPC (9 ERG-positive and 8 ERG-negative. As shown in Fig. 1A, miR338-3p was identified to be among the top de-regulated miRNAs between the two groups with an average 1.5 fold changes in expression. To validate miR338-3p expression levels relative to ERG in a larger clinical cohort (Fig. 1B), we used the MSKCC miRNA data [33], which confirmed miR338-3p to be significantly upregulated in ERG-positive compared to ERG-negative samples (p = 0.009). After segregating samples based on tumor localization, there was still significant up-regulation of miRNA338-3p levels in ERG-positive primary tumors (p = 0.03) but not in ERG-positive metastatic samples (p = 0.36) as compared to their corresponding ERG-negative samples (Fig. 1B).
Identification and expression of miR338-3p in relation to ERG expression. A Heatmap of miRNA Gene expression profiling of seventeen cancer prostate clinical samples with known ERG status. Samples with no ERG expression (ERG0) versus samples with ERG expression (ERG1). Shades of red represent increased miRNA gene expression while shades of blue represent decreased expression. B MSKCC data showing ERG1 (positive) primary tumors having higher expression of miR338-3p compared to ERG0 (negative) primary tumors (p = 0.03), however, no significant difference between metastatic ERG fusion positive versus metastatic ERG fusion negative tumors (p = 0.36). Comparing the miR338-3p expression in ERG1 with ERG0 regardless of tumor type, results demonstrated a significant up-regulation of miR338-3p in ERG1 subset (p = 0.009). (Color figure online)
Characterization of miR338-3p in clinical samples and the MSKCC cohort
Using a different cohort from the University of Calgary, 25 patients’ samples with localized PCa and adjacent benign prostate tissue were utilized. miR338-3p expression levels were determined by quantitative RT-PCR as described in materials and methods. As shown in Fig. 2A, miR338-3p levels were significantly down regulated in prostate cancer tissues compared to their matched benign prostate tissue (p = 0.037). Total results of the 25 cases are shown in Supplementary Fig. S1 including any outliers. To validate miR338-3p expression levels in relation to disease progression and patients’ prognosis, we used the MSKCC cohort [33] (n = 139), which is composed of 98 primary PCa, 13 metastatic and 28 benign prostate tissues. miR338-3p expression levels were significantly down-regulated in primary and metastatic samples compared to benign tissues and between metastasis compared to primary cancer (p = 1.6E−4, p = 4.8E−7 and p = 0.001, respectively; Fig. 2B). In addition, miR338-3p expression levels were significantly lower in the aggressive cancer group, as defined by the original Taylor et al. study [33], compared to non-aggressive cancer group (p = 0.016; Fig. 2C). Regarding miR338-3p association to other clinical and pathological parameters, miRNA338-3p levels were lower in PCa patients presenting with serum PSA (>4 ng/ml) compared to patients with serum PSA ≤ 4 (p = 0.04), and lower in Gleason score (GS) > 7 compared to GS = 7 and < 7 (p = −0.01 and p = 0.04, respectively) (Fig. 2D, E, respectively). However, there was no statistical difference between patients with pT3 stage disease compared to those with pT2 (p = 0.3) (Fig. 2F). To further assess the prognostic significance of miR338-3p in clinical progression, analysis of the MSKCC cohort of patients with lower miR338-3p expression levels revealed decreased survival probability compared to patients with higher miR338-3p levels as assessed by time from radical prostatectomy to PSA recurrence (p = 0.02; Fig. 2G).
Analysis of miR338-3p expression in prostate clinical samples. A Twenty-five pairs of patient matched benign versus cancer specimen were studied for the expression of miR338-3p by qPCR and results are shown by grouping benign versus cancer for all patients’ samples (p = 0.037). RNU48 was used as an endogenous control. Results are presented as relative expression as calculated by the comparative CT method (ΔΔCT) and error bars represent the SEM. p < 0.05 is considered significant. B MSKCC data showing that miR338-3p is significantly down-regulated in primary tumors (p = 1.6E−4) compared to its level in benign prostate tissues. miR338-3p expression is significantly low in metastatic samples compared to primary tumors (p = 0.001). There is also a significant decrease of miR338-3p expression in metastatic compared to benign samples (p = 4.8E−7). C MSKCC data showing that mir338-3p is significantly lower in aggressive tumors (p = 0.016) compared to its levels in non-aggressive ones. D MSKCC data showing that mir338-3p is significantly lower in patients with PSA levels (>4) compared to patients with PSA levels (≤4) (p = 0.04). E MSKCC data showing that mir338-3p is significantly lower in tumors with higher Gleason score (GS > 7) compared to low Gleason score (GS < 7) (p = 0.04). There is also significant decrease of miR338-3p in GS = 7 compared to its levels in GS > 7 (p = 0.01). F MSKCC data showing that mir338-3p is not significantly different in (pT3) stage compared to (pT2) (p = 0.3). G MSKCC data showing that mir338-3p is significantly lower (p = 0.02) in tumors with higher risk of recurrence
Biological significance of miR338-3p in prostate cancer
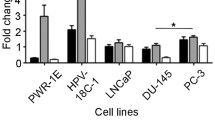
Clinical results described above suggest a tumor suppressive function of miRNA338-3p in PCa. To confirm this hypothesis, we investigated the impact of miR338-3p in vitro and in vivo using established PCa cell models. Initially, we assessed miR338-3p expression levels in the immortalized normal prostate epithelial cell line RWPE-1 and the PCa cell lines LNCaP, VCaP, DU145 and PC3 by quantitative RT-PCR. miR338-3p expression was significantly higher in most PCa cell lines compared to RWPE-1, and highest in the fusion-positive VCaP cell (p < 0.004), followed by PC3 (p = 0.003) and DU145 (p = 0.017). However, it was almost absent in RWPE-1 and LNCaP cells (Fig. 3A). Based on this data, we chose LNCaP, and PC3 to overexpress miR338-3p by creating stable cell lines, and VCaP cells to knock-down miR338-3p transiently. We tested the over expression and knockdown by qPCR in each of those cells (Fig. 3B, C, respectively).
In-vitro experiments and relation of miR338-3p and ERG in various cell lines. A miR338-3p expression in different prostate cell lines. Levels of expression of miR338-3p in different prostate cell lines (RWPE-1, LNCaP, VCaP, DU145 and PC3) as measured by qPCR. miR338-3p expression was significantly higher in VCaP (p = 0.004), DU145 (p = 0.017) and PC3 (p = 0.003) compared to its level in RWPE-1 cell line. RNU48 was used as an endogenous control. Results are presented as relative expression as calculated by the comparative CT method (ΔΔCT) and error bars represent SEM. p < 0.05 is considered significant. B Overexpression of miR338-3p in different prostate cell lines. Overexpression of miR338-3p in LNCaP (p < 0.001) and PC3 (p = 0.0005) cell lines as measured by qPCR compared to their control. RNU 48 was included as an endogenous control. Results are presented as relative expression as calculated by the comparative CT method (ΔΔCT) and errors bars represent the SEM. C Knockdown of miR338-3p in VCaP cell line. VCaP cell line was transiently transfected different with 100 nM of MIRIDIAN Hairpin inhibitor specific for miR338-3p or with hairpin inhibitor negative control. RNA was isolated at different time points (day 2, 3, 6, 9 and 12), and subjected to qPCR analysis of miR338-3p. miR338-3p was significantly knockdown at day 2 (p = 0.0003), day 3 (p = 0.001), day 6 (p = 0.0005), day 9 (p = 1.1E−05), and at day 12 (p = 5.2E−05). miR338-3p expression was normalized to RNU48 as an endogenous control. Results are presented as relative expression as calculated by the comparative CT method (ΔΔCT) and errors bars represent SEM. p < 0.05 is considered significant. D Effect of Knocking down miR338-3p in VCaP cell line on TMPRSS2-ERG expression. qPCR results showing that knocking down miR338-3p in VCaP cell line has no significant effect on TMPRSS2-ERG expression (p = 0.99). GUSB was included as an endogenous control. Results are presented as relative expression as calculated by the comparative CT method (ΔΔCT) and errors bars represent the SEM. p < 0.05 is considered significant
Effect of ERG on miR338-3p in PCa cells
To fully understand ERG-miR338-3p functional relationship and to test any reciprocal regulation between them, we investigated the influence of miR338-3p on ERG. In VCaP cells, silencing miR338-3p did not have any effect on TMPRSS2-ERG mRNA levels (p = 0.99) (Fig. 3D). Based on the CHIPBase database, regulatory network of miR-338 showed that ERG and Androgen Receptor (AR) are potential transcription factors (TFs) that regulate miR-338 gene (Supplementary Fig. S2). Our clinical data suggested a functional link between ERG gene rearrangements and miR338-3p expression levels.
Effect of miR338-3p on chemokine targets
Considering the significant impact of miR338-3p expression seen on PCa tumorigenicity, demonstrated both by in vitro analysis and its association with PCa progression and prognosis in PCa patients, we further investigated potential downstream mechanism(s) involved. Using available online tools to identify potential miRNA targets (microRNA.org and FINDTar3), the chemokine receptor, CXCR4, was identified as a candidate target for miR338-3p. Since ERG gene rearrangements have been shown to be associated with increased PCa invasiveness and progression to metastasis [3, 6], we focused on chemokine targets which have been involved in these processes in prostate cancer and are potentially linked to ERG [6]. In addition, FindTar3 showed that CXCR7, and CXCL12 could be additional potential targets. The bioinformatic analysis reveals a putative target sites for miR338-3p in the 3′-UTR of these chemokine genes, as shown in (Supplementary Fig. S3). These data suggest that miR338-3p regulates cancer molecular modulators, such as chemokines, and act as a tumor suppressor for prostate cancer.
In support of the involvement of the CXCR axis, miR338-3p significantly reduced the transcript levels of CXCL12 (p = 0.0001), CXCR4 (p = 0.0003) and CXCR7 (p = 0.001) in PC3 cells. LNCaP cells shows similar trend of decreased chemokine expression but was not significant (Fig. 4A), whereas no significant changes in the protein levels could be seen in these chemokines (data not shown) However, silencing miR338-3p significantly increased the expression levels of CXCR4 (p = 0.006) but did not affect CXCR7 or CXCL12 in VCaP cells (Fig. 4B).
Effects of miR338-3p on CXCR axis, cell viability and invasions in various cell lines. A Effect of miR338-3p over-expression on CXCl12, CXCR4 and CXCR4 mRNA in different prostate cell lines. qPCR results showing no significant effect of miR338-3p over-expression on CXCl12 (p = 0.18), CXCR4 (p = 0.21) and CXCR7 (p = 0.43) mRNA transcripts in LNCaP cell line. In PC3 cells, miR338-3p over-expression significantly down-regulated CXCL12 (p = 0.0001), CXCR4 (p = 0.003) and CXCR7 (p = 0.001) mRNA transcripts. B Effect of knocking down miR338-3p in VCaP cell line on CXCL12, CXCR4 and CXCR7 transcript levels. qPCR results showing that knocking down of miR-338-3p in VCaP cells is associated with a significant increase in CXCR4 (p = 0.006) mRNA levels, but did not affect CXCR7 (p = 0.84) or CXCL12 (p = 0.51). GUSB was included as an endogenous control. Results are presented as relative expression as calculated by the comparative CT method (ΔΔCT) and errors bars represent the SEM. C Effect of over-expression of miR338-3p on cell viability of LNCaP cell lines. Over-expression of miR338-3p significantly decreased cell viability in LNCaP Cell line at day 3 (p = 0.014) and day 4 (p = 0.003) after adding the XTT reagents to the cells. Data are represented as mean ± SEM. Data are from three independent experiments done in triplicate. p < 0.05 is considered significant. D Effect of over-expression of miR338-3p on cell viability of PC3 cell lines. Over-expression of miR338-3p in PC3 cells did not have any significant effect on cell proliferation. Data are represented as mean ± SEM. Data are from three independent experiments done in triplicate. p < 0.05 is considered significant. E Influence of miR338-3p over-expression on CXCR4-mediated migration and invasion of PC3 cell line. PC3 cells were serum starved for 24 h and then 105 cells were seeded in the upper Transwell chamber and allowed to migrate and invade towards either vehicle (no SDF 1∝) or SDF 1∝ in the lower wells for 24 h at 37 °C. Five fields from each Transwell were randomly selected and counted for migrated or invaded cells at 10X magnification using light microscope. Percent invasion was calculated by dividing the mean of cells invaded through Matrigel insert membrane by the mean of cells migrating through the control inserts membrane multiplied by 100. miR338-3p over-expression significantly decreased CXCR4—mediated invasion in PC3 (p = 0.03). Data are represented as mean ± SEM. Data are from three independent experiments done in triplicate. p < 0.05 is considered significant. F Anti-tumoral activity of miR338-3p in PC3 preclinical model. PC3 cells were implanted into the prostate of male SCID mice. Tumor size is significantly smaller in PC3-338-3p compared to PC3-control miRNA (p = 0.007). The number of macroscopic lymph nodes was not significantly affected (please refer to results)
Effects of miR338-3p on cell viability and invasiveness
We investigated whether miR338-3p expression influence cell proliferation in prostate cancer cells using in vitro cell viability assay. Over-expression of miR338-3p significantly inhibited the proliferation of LNCaP (p = 0.014 day 3 and p = 0.003 day 4; Fig. 4C). However, over-expression of miR338-3p in PC3 cells (Fig. 4D) and silencing miR338-3p in VCaP cells did not have any significant effect on cell proliferation (Data not shown). To assess miR338-3p effects on cell invasion, we used PC3 cell lines stably expressing miR338-3p and their corresponding control cells. In the absence of chemotactic agents, over expression of miR338-3p had no significant effect on cell migration or invasion compared to control PC3 cells. However, in the presence of the CXCR4 ligand, SDF-1α (CXCL12), used as a chemo-attractant in the lower chamber of the Boyden chamber, over-expression of miR338-3p significantly reduced invasion of PC3-miR338-3p cells (p = 0.03; Fig. 4E).
miR338-3p reduces PCa progression in an in vivo orthotopic PC3 mouse model
To assess the effect of miR338-3p on prostate cancer progression in in vivo, we implanted PC3 cells stably expressing miR338-3p orthotopically into mouse prostates (n = 7). Matched PC3 cells expressing control miRNA were used as a control (n = 8). As shown in Fig. 4F, miR338-3p-induced a significant antitumor effect in comparison to controls based on tumor weights at sacrifice (p = 0.007 compared to controls). Examination of macroscopic lymph nodes revealed a slight reduction in the number of lymph nodes but this difference did not reach statistical significance (7.4 ± 1.47 vs. 4.3 ± 1.35; p = 0.14).
Discussion
Down-regulation of miR338-3p is a frequent event seen in various cancer cell types including gastric cancer [36] and colorectal carcinoma [37]. Up to our best knowledge, very few literature works was associating miR-338 to prostate cancer. In this study, we selected miR-338-3p for further investigation as it showed to be upregulated in Taylor’s data comparing ERG1 vs ERG0 and we hypothesized that it might play a novel role in PCa progression. In this study, we investigated the role and prognostic significance of miR338-3p expression in PCa using clinical samples from a cohort of patients with localized PCa and the MSKCC cohort [33]. Parallel studies were conducted in PCa preclinical models, both in vitro and in vivo, to examine the impact on prostate cancer progression. miR338-3p expression was significantly decreased in localized PCa compared to benign prostate tissues, and analysis of miR338-3p expression levels in the MSKCC miRNA expression data (GSE21036) supported our results by confirming that miR338-3p is significantly down-regulated in localized and metastatic PCa, and also in the subgroup of aggressive tumors, defined by Taylor et al., in addition to being associated with patient prognosis [33]. Our data support a potential prognostic role and effect on disease progression for miR338-3p in PCa, possibly through a tumor suppressor activity. This is also supported by results from our preclinical models, where a decrease in cell invasion in vitro and tumor growth in vivo was observed following enforced expression of miR338-3p. These results are in concordance with previous reports in hepatocellular carcinoma (HCC) documenting lower miR338-3p expression levels in comparison to non-cancerous liver tissues, where those levels were also inversely related to increased stage [38].
Our miRNA expression profiling data indicated that ERG potentially effects miR-338-3p expression. To confirm and validate the relation between ERG and miR338-3p, we used ChIPBase [35] analysis to identify transcription factors (TFs) that have binding sites within 5 kb upstream of the miR338-3p gene. ChIPBase is a comprehensive collection of TF maps and transcriptional regulatory relationships of TFs and genes from CHIP-Seq data. ChIPBase analysis identified ERG and AR binding sites upstream from miR-338-3p, in addition to other TFs. Using the deepView genome browser within ChIPBase, ERG and AR were shown to bind to the 5 kb upstream region of miR338-3p in multiple experiments. In support of the notion that ERG regulate miR-338-3p, our in vitro gene expression analysis demonstrated that PC3, DU145, and LNCaP cells, which do not harbor TMPRSS2-ERG fusion gene, show significantly lower miR338-3p expression than the fusion-positive VCaP cell line. One striking feature of miRNA species is their ability to participate in negative auto-regulatory feedback loops to control their own expression [39]. A recent report showed that Paired-Like Homeodomain Transcription Factor 3 (PITX3) transcription factor regulates miR133b, which consecutively can silence PITX3 [40]. Based on this notion, we investigated if ERG is a putative target for miR-338-3p. The only Evidence on the association between miR338-3p was based on our study from analysis of MSKCC miRNA data. However, this was not evident from our experiments on VCaP cell lines which harbor TMPRSS2-ERG mRNA rearrangement. Thus, we concluded that there is no reciprocal relationship between ERG and miR338-3p.
miRNAs are capable to modulate cancer driver genes by complementary binding to their 3′-UTR and subsequently repress their translation [41]. Our bioinformatic analysis showed that 3′-UTR region of CXCL12, CXCR4, and CXCR7 has a putative binding site for miR338-3p. It has been shown that chemokines play important roles in tumor-associated angiogenesis, cancer stem cell motility, and tumor metastasis [42]. In particular, CXCR4 was reported as the most common over-expressed chemokine receptor in human cancer [43]. It has been reported that CXCR4 upregulation positively correlates with metastatic epithelial cancer cell types including non-small cell lung cancer [44] and breast cancer [45]. In addition, previous studies have alluded to potential interaction linking ERG and the CXCR axis [46]. In the study by Cai et al., it was demonstrated that ERG binds to the CXCR4 gene promoter, providing a potential link between ERG gene rearrangements and enhanced metastasis of tumor cells through CXCR4 function in prostate cells [46]. More recently, it was demonstrated that ERG factor activates CXCR4 expression by binding to the specific ERG/ETS responsive elements and intracellular kinases phosphorylate at ERG to induce CXCR4 expression [32]. We observed that overexpression of miR338-3p in parental PC3 cell line shows significantly lower transcript levels of these chemokines. We have performed a loss of function experiment to silence miR-338-3p expression in VCaP and observed significant up-regulation of CXCR4 mRNA levels. These data support a functional link between miR338-3p and the CXCR axis (CXCl12-CXCR4-CXCR7).
A drawback in our results is that, although miR338-3p decreased CXCL12, CXCR4 and CXCR7 at the transcriptional level, it did not affect the protein level in any of the cell lines investigated (data not shown). This difference could be extrapolated to the differential kinetics between CXCR4 mRNA and protein. In neuroblastoma cells, it was shown that 55 kDa surface expressed CXCR4 isoform is slowly ubiquitinated in comparison to 87, 67 kDa isoforms [47]. Clearly, additional studies are warranted to further investigate the heterogeneity of CXCR4 protein kinetics in comparison to its mRNA half-life, in the context of PCa cell lines. Moreover, we were unable to demonstrate that CXCR4 is a direct target of miR338-3p by luciferase reporter assay (data not shown), implying that the role of miR338-3p on this axis may be indirect. It was shown that CXCR4 increases pancreatic cancer invasion and Hh pathway activation by increasing the expression of Smoothened (SMO) [48], which was found to be a validated target of miR338-3p, at least in several cancer types [37]. These data is suggesting that miR338-3p decrease expression of CXCR4, hence this could lead to targeting of SMO protein [49, 50], as shown in our suggested model (Fig. 5).
The observation that miR338-3p overexpression significantly decreased cell viability in LNCaP but not in PC3 cells, despite significantly decreasing invasiveness of the latter cells, indicate a critical role for miR338-3p in PCa metastasis. It is possible that alternative mechanisms are being operative in different PCa lines. These cells have different genetic makeup that influences the anti proliferative effects of miR338-3p. Perhaps LNCaP cells are most suitable for miR338-3p to carry out its anti-proliferative effects, primarily under in vitro conditions. Furthermore, it should be kept in mind that since PC3 cell line has a very high growth rate, the anti-proliferative effect of miR338-3p in vitro might be subtle.
It is noteworthy that the high mortality rate of prostate cancer is mostly attributed to the invasion of malignant cells to distant organs [51]. It is intriguing that miR338-3p inhibited CXCL12, CXCR4 and CXCR7, at least at the transcriptional level with no negative effects on proliferation in the PC3 cells. Although the down-regulation of CXCL12 and CXCR7 was clear in PC3 cell lines when miR338-3p was overexpressed, only CXCR4 change in VCaP cells was noticed by knocking down miR338-3p. Our conclusions will be therefore focused on CXCR4 in this article.
Herein, we also demonstrate that that overexpression of miR338-3p decreases CXCR4 mediated invasion in vitro where SDF-1α /CXCL12 is used as chemoattractant, supporting that the decrease of CXCR4 at the transcriptional level have influenced the invasiveness of PC3 cell line. These results are in line with several previous publications where targeting CXCR4 led to decreased invasion and metastasis [51, 52]. Essentially, CXCL12/CXCR4 axis has been found to regulate bone metastasis in PCa [31]. Consistent with our results, Uygur and Wu’s study showed that knocking-down CXCR4 ligand, in PC3 cell lines inhibits invasion but not proliferation [30]. On the other hand, the mechanism(s) through which miR338-3p-induced a significant antitumor effect in vivo based on tumor weights is not yet clear. However, there is an increasing evidence that cells implanted in vivo, but not those growing in vitro, are able to release growth factors and recruit stroma cells [53]. For instance, it was reported that myofibroblasts facilitate angiogenesis due to their ability to secrete stroma-derived factor-1 (SDF-1/CXCL12) [53]. Since our data propose CXCL12 as a potential target for miR338, we speculate that it might be that the inhibitory effect is seen more clearly in vivo. However, the mechanisms, through which miR-338-3p inhibit tumor growth in vivo, warrant further investigations.
In summary, although the exact mechanism is not yet clear, our study clearly supports and demonstrates anti-proliferative and anti-invasive properties, and a tumor suppressive role of miRNA338, as documented by our clinical studies and in vitro and in vivo experimental models. There is clearly a potential association between miR338-3p and ERG but its exact nature needs further investigations. The inhibitory effects of miR338-3p could be potentially mediated-at least in part- by the CXCR axis. Future work on miR338-3p to better characterize the regulation of miR338-3p expression and its direct targets may lead to better understanding of pathways associated with PCa progression and may impact the diagnosis and prognosis of prostate cancer patients.
Abbreviations
- AR:
-
Androgen receptor
- CRPC:
-
Castration resistant prostate cancer
- GS:
-
Gleason score
- miRNA:
-
microRNA
- PCa:
-
Prostate cancer
- PSA:
-
Prostate specific antigen
- MSKCC:
-
Memorial Sloan-Kettering Cancer Center
- SCID:
-
Severe combined immunodeficiency
- TFs:
-
Transcription factors
References
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65(1):5–29. doi:10.3322/caac.21254
Fong MK, Hare R, Jarkowski A (2012) A new era for castrate resistant prostate cancer: a treatment review and update. J Oncol Pharm Pract. doi:10.1177/1078155212437599
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW et al (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310(5748):644–648. doi:10.1126/science.1117679
Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC et al (2012) The TMPRSS2: ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarker Prev 21(9):1497–1509. doi:10.1158/1055-9965.EPI-12-0042
Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S et al (2008) The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell 13(6):519–528. doi:10.1016/j.ccr.2008.04.016
Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A et al (2009) Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 41(5):619–624. doi:10.1038/ng.370
Leshem O, Madar S, Kogan-Sakin I, Kamer I, Goldstein I, Brosh R et al (2011) TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PLoS ONE 6(7):e21650. doi:10.1371/journal.pone.0021650
Casey OM, Fang L, Hynes PG, Abou-Kheir WG, Martin PL, Tillman HS et al (2012) TMPRSS2- driven ERG expression in vivo increases self-renewal and maintains expression in a castration resistant subpopulation. PLoS ONE 7(7):e41668. doi:10.1371/journal.pone.0041668
Fayyaz S, Farooqi AA (2013) miRNA and TMPRSS2-ERG do not mind their own business in prostate cancer cells. Immunogenetics 65(5):315–332. doi:10.1007/s00251-012-0677-2
Wang G, Wang Y, Feng W, Wang X, Yang JY, Zhao Y et al (2008) Transcription factor and microRNA regulation in androgen-dependent and -independent prostate cancer cells. BMC Genomics 9(Suppl 2):S22. doi:10.1186/1471-2164-9-S2-S22
Selcuklu SD, Donoghue MT, Spillane C (2009) miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans 37(Pt 4):918–925. doi:10.1042/BST0370918
Gordanpour A, Stanimirovic A, Nam RK, Moreno CS, Sherman C, Sugar L et al (2011) miR-221 Is down-regulated in TMPRSS2:ERG fusion-positive prostate cancer. Anticancer Res 31(2):403–410
Hart M, Wach S, Nolte E, Szczyrba J, Menon R, Taubert H et al (2013) The proto-oncogene ERG is a target of microRNA miR-145 in prostate cancer. FEBS J 280(9):2105–2116. doi:10.1111/febs.12236
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Lai EC (2002) Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 30(4):363–364. doi:10.1038/ng865
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20. doi:10.1016/j.cell.2004.12.035
Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K et al (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434(7031):338–345. doi:10.1038/nature03441
Chen X, Gong J, Zeng H, Chen N, Huang R, Huang Y et al (2010) MicroRNA145 targets BNIP3 and suppresses prostate cancer progression. Cancer Res 70(7):2728–2738. doi:10.1158/0008-5472.CAN-09-3718
Mezzanzanica D, Bagnoli M, De Cecco L, Valeri B, Canevari S (2010) Role of microRNAs in ovarian cancer pathogenesis and potential clinical implications. Int J Biochem Cell Biol 42(8):1262–1272. doi:10.1016/j.biocel.2009.12.017
Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F et al (2010) MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS ONE 5(4):e10147. doi:10.1371/journal.pone.0010147
Barik S (2008) An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res 36(16):5232–5241. doi:10.1093/nar/gkn513
Tomomura M, Fernandez-Gonzales A, Yano R, Yuzaki M (2001) Characterization of the apoptosis-associated tyrosine kinase (AATYK) expressed in the CNS. Oncogene 20(9):1022–1032. doi:10.1038/sj.onc.1204210
Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E et al (2005) MicroRNA expression in zebrafish embryonic development. Science 309(5732):310–311. doi:10.1126/science.1114519
Kos A, Olde Loohuis NF, Wieczorek ML, Glennon JC, Martens GJ, Kolk SM et al (2012) A potential regulatory role for intronic microRNA-338-3p for its host gene encoding apoptosis-associated tyrosine kinase. PLoS ONE 7(2):e31022. doi:10.1371/journal.pone.0031022
Tsuchiya S, Oku M, Imanaka Y, Kunimoto R, Okuno Y, Terasawa K et al (2009) MicroRNA-338-3p and microRNA-451 contribute to the formation of basolateral polarity in epithelial cells. Nucleic Acids Res 37(11):3821–3827. doi:10.1093/nar/gkp255
Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K et al (2000) International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev 52(1):145–176
Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB (2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 63(6):1256–1272. doi:10.1124/mol.63.6.1256
Zlotnik A (2006) Involvement of chemokine receptors in organ-specific metastasis. Contrib Microbiol 13:191–199. doi:10.1159/000092973
Begley LA, MacDonald JW, Day ML, Macoska JA (2007) CXCL12 activates a robust transcriptional response in human prostate epithelial cells. J Biol Chem 282(37):26767–26774. doi:10.1074/jbc.M700440200
Uygur B, Wu WS (2011) SLUG promotes prostate cancer cell migration and invasion via CXCR4/CXCL12 axis. Mol Cancer 10:139. doi:10.1186/1476-4598-10-139
Sun YX, Wang J, Shelburne CE, Lopatin DE, Chinnaiyan AM, Rubin MA et al (2003) Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers (PCa) in vivo. J Cell Biochem 89(3):462–473. doi:10.1002/jcb.10522
Singareddy R, Semaan L, Conley-Lacomb MK, St John J, Powell K, Iyer M et al (2013) Transcriptional regulation of CXCR4 in prostate cancer: significance of TMPRSS2-ERG fusions. Mol Cancer Res 11(11):1349–1361. doi:10.1158/1541-7786.MCR-12-0705
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al (2010) Integrative genomic profiling of human prostate cancer. Cancer Cell 18(1):11–22. doi:10.1016/j.ccr.2010.05.026
Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ et al (1996) Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res 2(9):1627–1636
Yang JH, Li JH, Jiang S, Zhou H, Qu LH (2013). ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res 41(Database issue):D177–D187. doi:10.1093/nar/gks1060
Guo B, Liu L, Yao J, Ma R, Chang D, Li Z et al (2014) miR-338-3p suppresses gastric cancer progression through a PTEN-AKT axis by targeting P-REX2a. Mol Cancer Res 12(3):313–321. doi:10.1158/1541-7786.MCR-13-0507
Xue Q, Sun K, Deng HJ, Lei ST, Dong JQ, Li GX (2014) MicroRNA-338-3p inhibits colorectal carcinoma cell invasion and migration by targeting smoothened. Jpn J Clin Oncol 44(1):13–21. doi:10.1093/jjco/hyt181
Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ et al (2009) Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res 39(8):786–794. doi:10.1111/j.1872-034X.2009.00502.x
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11(9):597–610. doi:10.1038/nrg2843
Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E et al (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317(5842):1220–1224. doi:10.1126/science.1140481
Walter BA, Valera VA, Pinto PA, Merino MJ (2013) Comprehensive microRNA Profiling of Prostate Cancer. J Cancer 4(5):350–357. doi:10.7150/jca.6394
Keeley EC, Mehrad B, Strieter RM (2010) CXC chemokines in cancer angiogenesis and metastases. Adv Cancer Res 106:91–111. doi:10.1016/S0065-230X(10)06003-3
Koizumi K, Hojo S, Akashi T, Yasumoto K, Saiki I (2007) Chemokine receptors in cancer metastasis and cancer cell-derived chemokines in host immune response. Cancer Sci 98(11):1652–1658. doi:10.1111/j.1349-7006.2007.00606.x
Wagner PL, Hyjek E, Vazquez MF, Meherally D, Liu YF, Chadwick PA et al (2009) CXCL12 and CXCR4 in adenocarcinoma of the lung: association with metastasis and survival. J Thorac Cardiovasc Surg 137(3):615–621. doi:10.1016/j.jtcvs.2008.07.039
Smith MC, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D et al (2004) CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res 64(23):8604–8612. doi:10.1158/0008-5472.CAN-04-1844
Cai J, Kandagatla P, Singareddy R, Kropinski A, Sheng S, Cher ML et al (2010) Androgens Induce Functional CXCR4 through ERG Factor Expression in TMPRSS2-ERG Fusion-Positive Prostate Cancer Cells. Transl Oncol 3(3):195–203
Carlisle AJ, Lyttle CA, Carlisle RY, Maris JM (2009) CXCR4 expression heterogeneity in neuroblastoma cells due to ligand-independent regulation. Mol Cancer 8:126. doi:10.1186/1476-4598-8-126
Li X, Ma Q, Xu Q, Liu H, Lei J, Duan W et al (2012) SDF-1/CXCR4 signaling induces pancreatic cancer cell invasion and epithelial-mesenchymal transition in vitro through non-canonical activation of Hedgehog pathway. Cancer Lett 322(2):169–176. doi:10.1016/j.canlet.2012.02.035
Huang XH, Chen JS, Wang Q, Chen XL, Wen L, Chen LZ et al (2011) miR-338-3p suppresses invasion of liver cancer cell by targeting smoothened. J Pathol 225(3):463–472. doi:10.1002/path.2877
Sun K, Deng HJ, Lei ST, Dong JQ, Li GX (2013) miRNA-338-3p suppresses cell growth of human colorectal carcinoma by targeting smoothened. World J Gastroenterol 19(14):2197–2207. doi:10.3748/wjg.v19.i14.2197
Ok S, Kim SM, Kim C, Nam D, Shim BS, Kim SH et al (2012) Emodin inhibits invasion and migration of prostate and lung cancer cells by downregulating the expression of chemokine receptor CXCR4. Immunopharmacol Immunotoxicol 34(5):768–778. doi:10.3109/08923973.2012.654494
Shanmugam MK, Manu KA, Ong TH, Ramachandran L, Surana R, Bist P et al (2011) Inhibition of CXCR4/CXCL12 signaling axis by ursolic acid leads to suppression of metastasis in transgenic adenocarcinoma of mouse prostate model. Int J Cancer 129(7):1552–1563. doi:10.1002/ijc.26120
Weinberg RA (2014) The Biology of Cancer Garland Science, 2nd edn. Taylor & Francis Group, LLC, New York
Acknowledgments
The authors would like to thank Shuhong Liu, Sabrina Daniela Silva, Samir Al Bashir, Tingting Wang and Liang Hong Teng for technical assistance in this study.
Author contribution
AB; executed and planned in vitro experiments and drafted the manuscript. MA and RA; carried out all bioinformatics work in this study and assisted in drafting manuscript. LP; carried out ERG cloning experiments and participated in the preparation of figures; HA helped in performing qPCR experiments, writing the manuscript and preparing of figures; and AA and TW performed western blots, SH performed Q-PCR and contributed to manuscript writing, KB carried out in vitro experiments. MAJ designed and supervised in vitro and in vivo experiments of the study and revised manuscript. TAB; carried out pathological analysis, and contributed to manuscript drafting.
Funding
This work was supported by the Prostate Cancer Foundation Young Investigator Award (T.A.B) and in part by the Canadian Institutes for Health Research, the Canadian Cancer Society, and Quebec Breast Cancer Foundation (MAAJ). This work was also supported by Prostate cancer Canada and is proudly funded by the Movember Foundation-Grant #B2013-01.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare in this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bakkar, A., Alshalalfa, M., Petersen, L.F. et al. microRNA 338-3p exhibits tumor suppressor role and its down-regulation is associated with adverse clinical outcome in prostate cancer patients. Mol Biol Rep 43, 229–240 (2016). https://doi.org/10.1007/s11033-016-3948-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-3948-4