Abstract—The importance of root nodule bacteria in biotechnology is determined by their distinctive feature: symbiotic nitrogen fixation resulting in the production of organic nitrogen-containing compounds. While interacting with host legume plants, the cells of these bacteria undergo global changes at all levels of expression of genetic information leading to the formation in root nodules of so-called bacteroids functioning as nitrogen fixation factories. The molecular mechanisms underlying plant-microbial symbiosis are actively investigated, and one of the most interesting and poorly studied aspects of this problem is the species-specificity of interaction between root nodule bacteria and host plants. In this work we have performed the proteomic analysis of the Sinorhizobium meliloti bacteroids isolated from two legume species: alfalfa (Medicago sativa L.) and yellow sweet clover (Melilotus officinalis L.). It has been shown that the S. meliloti bacteroids produce a lot of proteins (many of them associated with symbiosis) in a host-specific manner, i.e., only in certain host plant species. It has been demonstrated for the first time that the levels of expression in bacteroids of the genes encoding the ExoZ and MscL proteins responsible for the synthesis of surface lipopolysaccharides and formation of a large conductance mechanosensitive channel, respectively, depend on a host plant species that confirms the results of proteomic analysis. Overall, our data show that the regulation of bacteroid development by the host plant has species-specific features.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The root nodule bacteria of the order Rhizobiales, class Alphaproteobacteria, are a unique group of microorganisms capable of fixing atmospheric nitrogen in symbiosis with legumes [1, 2]. During the formation of the symbiotic interactions, in plant root nodules these bacteria form bacteroids, large immobile branched cells that have lost reproduction ability and are specialized for atmospheric nitrogen fixation [3, 4]. The formation of bacteroids is accompanied by the rearrangement of gene expression profiles leading to global changes at the proteomic level [5, 6].
It is known that the symbiotic interactions between root nodule bacteria and plants are under complex genetic control [2]. Symbiosis is induced by a flavonoid complex secreted by a leguminous plant [5], root nodule bacteria respond to it by producing LysR-type transcriptional regulators, which are encoded by the nod operon [2], and mediate the synthesis of lipochitooligosaccharides, so-called Nod factors (from nodulation) [7]. The proteins encoded by the nod operon, play the key role in host selection by root nodule bacteria [8–10]. It has also been shown that the levels of production of some other proteins by the Bradyrhizobium japonicum bacteroids, such as ABC transporters, dehydrogenases and transketolases, significantly depend on a host plant species [11]. Nevertheless, diversity of proteins and molecular mechanisms determining the species specificity of interactions between bacteria and host plants remain to be poorly studied [1, 2], in spite of significant progress in the transcriptomics [12–14] and proteomics [15] of root nodule bacteria in recent years.
In this work we attempted to reveal the key participants and potential mechanisms of bacteroid regulation by a host plant. To identify bacterial proteins that are host plant-specific, we compared the proteomes of bacteroids of the root nodule bacteria Sinorhizobium meliloti isolated from the root nodules of legumes from two different genera: Medicago (alfalfa, M. sativa L.) and Melilotus (yellow sweet clover, M. officinalis L.).
EXPERIMENTALS
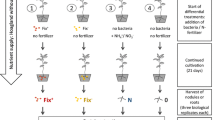
Cultivation of microorganisms. The S. meliloti bacteroids were obtained as follows: the plants of alfalfa (M. sativa L. no. 45 969, the Catalog of Vavilov Research Institute of Plant Industry (VRIPI)) and yellow sweet clover (M. officinalis L. no. 44 565, the Catalog of VRIPI) were inoculated with the efficient nitrogen-fixing strain RCAM04491 of S. meliloti and grown in vessels with sterile vermiculite for five weeks in a green house under standard lighting conditions [16]. The bacteroids were isolated from plant root nodules according to the previously published protocol [17]. The free-living culture of S. meliloti used as a control was grown on a solid TY medium for two days at 28°С.
Protein assay. Total proteins were isolated from the bacteria with the Complete protease inhibitor (Roche, Switzerland); the proteins were denatured through boiling in sodium phosphate buffer with 1% sodium dodecyl sulfate; salts and detergents were removed in HiPPR and Zeba Spin Desalting Columns (ThermoScientific, United States) according to the manufacturer’s protocol. The concentrations of isolated proteins were aligned using the Qubit (Thermo Scientific) fluorescent assay.
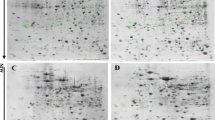
The trypsinolysis of proteins was performed according to [18]. The peptide mixtures (1 µL) were applied to an Acclaim™ PepMap 300 HPLC column for reversed-phase chromatography (150 mm × 75 µm; particle size, 5 µm (Thermo Scientific)) and separated according to [18] in the UltiMate 3000 UHPLC+ RSLC nano high-performance liquid chromatography system (Dionex, United States). The proteins were identified with the Ultraflextrime tandem time-of-flight mass spectrometer (BrukerDaltonics, United States). The mass spectra of each fraction were determined. The spectra were analyzed with the WARP-LC software, taking into account their production as a result of liquid chromatography. The MS/MS analysis of unique peptides was performed in the fractions with maximum peak intensity of these peptides. The correspondence of spectra to proteins was determined by Mascot version 2.4.2 (MatrixScience, http://www.matrixscience.com) in the UniProt database (http://www.uniprot.org/) with the limitation for S. meliloti. The α-cyano-4-hydroxycinnamic acid was used as a matrix. Mass tolerance settings were as follows: precursor mass tolerance, 100 ppm; fragment mass tolerance, 0.9 Da. Carboxymethylation of cysteine, partical oxidation of methionine and one missed site of trypsinolysis were indicated as permissible modifications.
Real-time polymerase chain reaction (qPCR). The total RNA for qPCR was isolated with Trizol (Invitrogen, United States). Reverse transcription was performed with the SuperScript III reverse transcriptase (Invitrogen) according to the producer’s protocol. The amounts of DNA templates were controlled by the reference SMc00128 gene with the previously published primers [19]. The levels of expression of the exoZ and mscL genes were analyzed using primers F—5'-ATC-ATGTGGGTCATCAGCGA and R—5'-TCCAGTGTCAGCACCAGATT; F—5'-ATCATGTGGGTCA-TCAGCGA and R—5'-TCCAGTGTCAGCACCAG-ATT, respectively. qPCR was performed with SYBR GreenMasterMix (BioRad, United States). The results were processed by the method of Livak and Schmittgen [20]. Statistical significance of the differences was assessed by the Mann–Whitney U test in Statistica version 6.0 (StatSoft).
RESULTS AND DISCUSSION
The Proteomic Profiles of the S. meliloti Bacteroids Isolated from Plants of the Genera Medicago and Melilotus Are Substantially Different
We have identified 417 proteins of the free-living culture, 684 proteins of the bacteroids isolated from the M. sativa L. nodules, and 634 proteins of the bacteroids isolated from the M. officinalis L. nodules (Fig. 1). This result might be an evidence in favor of a slightly limited resolving power of proteomic methodology compared to transcriptomics, because according to the data of transcriptomic analysis, more genes are expressed in free-living cells compared to bacteroids [13].
The Venn diagram (a) and column histogram (b) illustrating the number of identified proteins of S. meliloti. The numbers of identified proteins are shown for the free-living culture, the bacteroids isolated from Medicago sativa L. nodules, and the bacteroids isolated from Melilotus officinalis L. nodules. The number of proteins in each of the overlapping samples is indicated.
The genome of S. meliloti contains a ring chromosome and two megaplasmids [21]. The comparison of localizations of the genes encoding the identified proteins showed in bacteroids the activation of production of the proteins encoded by the genes located on megaplasmids (Fig. 2), which is in agreement with the ideas of significance of the products of such genes in plant-microbial interactions [21].
The localization of genes encoding the identified proteins on the chromosome and the pSymA and pSymB megaplasmids (a) and the results of comparative analysis of the exoZ and mscL gene expression in the free-living culture of S. meliloti, the bacteroids from Medicago and the bacteroids from Melilotus by qPCR (b). (a) The genes encoding the proteins identified only in the free-living culture (blue); in both variants of bacteroids isolated from the nodules of Medicago sativa L. and Melilotus officinalis L. (yellow); in bacteroids from Medicago or from Melilotus (species-specific) (red); identified both in the free culture and in bacteroids isolated from both plant species (house-keeping proteins) (green). The distances on the genetic map are given in kilobases (kb). (b) The cDNA amount of the respective gene in the control (free-living culture) is set as 1. The observed differences were assessed by the nonparameteric Mann–Whitney test. p is the level of significance.
The sets of proteins identified in the two variants of bacteroids from the Medicago and Melilotus root nodules demonstrate a high similarity: 597 common proteins, 223 of them being identified only in the bacteroids but not in the free-living culture (Fig. 1). As expected, the highest mass-spectrometric score (Mascot Score) in the protein samples of the bacteroids was shown for nitrogenase (NifH), chaperones and chaperonins (Tig, DnaK, GroEL), and the proteins encoded by the fix operon that play an important role in symbiosis (Table 1). The comparison of the proteomes of bacteroids showed 87 proteins that were present in the bacteroids from Medicago nodules and not in the bacteroids from Melilotus nodules (Fig. 1b, Table 1) and 38 proteins identified in the bacteroids from Melilotus but not from Medicago (Fig. 1b, Table 1). All of these proteins were detected with a low score (no more than 114), which might reflect the minor level of their production compared to the major bacteroid proteins with a score up to several thousands (Table 1). Most of these host-specific bacterial proteins are metabolic enzymes, transcriptional regulators, components of the translational machinery, and factors responsible for the synthesis and transport of polysaccharides and siderophores.
It is noteworthy that some operons had a high content of the genes encoding the host-specific proteins of S. meliloti bacteroids (Fig. 2). For example, the exo operon encoding the machinery for surface lipopolysaccharide production, which plays a key role in plant-microbial interactions [22], carries the genes encoding six proteins identified in our work as host-specific (Table 1) and is located on the pSymB plasmid in the region of 1169–1185 kbp. (Fig. 2). The rhb operon responsible for the siderophore transport system and probably involved in the regulation of nitrogen fixation efficiency [23], carries genes of three host-specific proteins (Table 1) and is located on the pSymA plasmid in the region of 1306–1318 kbp. (Fig. 2). Generally, we may conclude that many of the host-specific proteins of S. meliloti bacteroids are involved in the regulation of plant–microbial interactions.
The Levels of Expression of the exoZ and mscL Genes Significantly Vary in Bacteroids Isolated from Different Plant Species
qPCR was used for the quantitative analysis of expression levels of the genes encoding some host-specific proteins of S. meliloti. The statistically significant changes in expression levels were revealed for two (exoZ and mscL) out of five (exoZ, mscL, rnc, tam, tal) genes under study, which partially correspond to the proteomic data (Fig. 2b).
The similarity of modifications at the mRNA and protein levels is substantial evidence for the dependence of the gene expression regulation in bacteroids on a host plant organism. Thus, the level of expression of the exoZ gene encoding acetyltransferase, which is involved in the synthesis of surface lipopolysaccharides and detected only in the proteome of Medicago bacteroids, was higher in the Medicago bacteroids than in the Melilotus bacteroids and free-living culture (p < 0.01) (Fig. 2b). It is known that the strains with mutations in this gene form bacteroids in Medicago root nodules but have the lower ability to form infection threads [24]. These and our data suggest that ExoZ is important for the formation of infection threads during the symbiotic interactions with Medicago but not with Melilotus; therefore, the levels of the exoZ gene expression (Fig. 2b) and ExoZ protein production (Table 1) in bacteroids isolated from Medicago is much higher than in bacteroids isolated from Melilotus.
The gene encoding the mechanosensitive channel protein (MscL), which we have found in the free-living culture and bacteroids from Medicago but not from Melilotus, shows a higher level of expression in the bacteroids from Medicago nodules compared to the bacteroids from Melilotus (p < 0.01) (Fig. 2b), confirming our proteomic data (Table 1). It should be ascertained that having no MscL in the list of proteins found in the bacteroids from Melilotus does not suggest having no production of this protein but rather demonstrates a drastic decrease in its synthesis, probably due to the reduced expression of mscL that we have revealed by qPCR (Fig. 2b). Mechanosensitive channels are important osmotic regulators responding to the changes in plasma membrane tension [25]. It is notable that the genes encoding the small mechanosensitive channel of root nodule bacteria (MscS) are differentially expressed in symbiosis with legumes [25]. Our data also contribute to the involvement of mechanosensitive channels in the regulation of species specificity of symbiosis. The role of these proteins can be associated with the adaptation of bacteroids to an osmotically aggressive internal medium of root nodules of different legume species.
Overall, the comparative proteomic analysis of the free-living culture of S. meliloti and the bacteroids isolated from two host plant species, Medicago sativa L. and Melilotus officinalis L., has shown that the proteomes of bacteroids formed in the root nodules of different host plant species are substantially different. This result suggests for the specificity of regulation of bacteroids by different plant species. The revealed host-specific proteins of S. meliloti bacteroids are characterized by the low level of production and great diversity of functions (Table 1). These proteins should be considered candidates that require additional verification by targeted methods, including the comparative analysis of the level of expression of the corresponding genes by qPCR. We have used this technique to show host-specific changes in the level of expression of the genes encoding the MscL large conductance mechanosensitive channel and the ExoZ protein responsible for the synthesis of surface lipopolysaccharides, confirming that these genes and their products are associated with the regulation of species-specificity of plant-microbial interactions.
REFERENCES
Andrews M., Andrews M.E. 2017. Specificity in legume–rhizobia symbioses. Int. J. Mol. Sci. 18, e705.
Wang D., Yang S., Tang F., Zhu H. 2012. Symbiosis specificity in the legume: Rhizobial mutualism. Cell. Microbiol. 14, 334–342.
Gourion B., Berrabah F., Ratet P., Stacey G. 2015. Rhizobium–legume symbioses: The crucial role of plant immunity. Trends Plant Sci. 20, 186–194.
Sprent J.I. 2009. Legume Nodulation: A Global Perspective. Ed. Sprent J.I. Chichester UK: Wiley-Blackwell.
Rose C.M., Venkateshwaran M., Volkening J.D., Grimsrud P.A., Maeda J., Bailey D.J., Park K., Howes-Podoll M., den Os D., Yeun L.H., Westphall M.S., Sussman M.R., Ané J.-M., Coon J.J. 2012. Rapid phosphoproteomic and transcriptomic changes in the rhizobia–legume symbiosis. Mol. Cell. Proteomics. 11, 724–744.
Marx H., Minogue C.E., Jayaraman D., Richards A.L., Kwiecien N.W., Siahpirani A.F., Rajasekar S., Maeda J., Garcia K., Del Valle-Echevarria A.R., Volkening J.D., Westphall M.S., Roy S., Sussman M.R., Ané J.-M., Coon J.J. 2016. A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotechnol. 34, 1198–1205.
Oldroyd G.E.D., Murray J.D., Poole P.S., Downie J.A. 2011. The rules of engagement in the legume–rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144.
Debellé F., Maillet F., Vasse J., Rosenberg C., de Billy F., Truchet G., Dénarié J., Ausubel F.M. 1988. Interference between Rhizobium meliloti and Rhizobium trifolii nodulation genes: Genetic basis of R. meliloti dominance. J. Bacteriol. 170, 5718–5727.
Spaink H.P., Weinman J., Djordjevic M.A., Wijffelman C.A., Okker R.J., Lugtenberg B.J. 1989. Genetic analysis and cellular localization of the Rhizobium host specificity-determining NodE protein. EMBO J. 8, 2811–2818.
McIver J., Djordjevic M.A., Weinman J.J., Bender G.L., Rolfe B.G. 1989. Extension of host range of Rhizobium leguminosarum bv. trifolii caused by point mutations in nodD that result in alterations in regulatory function and recognition of inducer molecules. Mol. Plant Microbe Interact. 2, 97–106.
Koch M., Delmotte N., Rehrauer H., Vorholt J.A., Pessi G., Hennecke H. 2010. Rhizobial adaptation to hosts, a new facet in the legume root–nodule symbiosis. Mol. Plant. Microbe. Interact. 23, 784–790.
Ampe F., Kiss E., Sabourdy F., Batut J. 2003. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 4, R15.
Becker A., Berges H., Krol E., Bruand C., Ruberg S., Capela D., Lauber E., Meilhoc E., Ampe F., de Bruijn F.J., Fourment J., Francez-Charlot A., Kahn D., Kuster H., Liebe C., et al. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant. Microbe Interact. 17, 292–303.
Li Y., Tian C.F., Chen W.F., Wang L., Sui X.H., Chen W.X. 2013. High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PLoS One. 8, e0070531.
Larrainzar E., Wienkoop S. 2017. A proteomic view on the role of legume symbiotic interactions. Front. Plant Sci. 8, e1267.
Rumyantseva M.L., Simarov B.V., Onishchuk O.P., Andronov E.E., Chizhevskaya E.P., Belova V.S., Kurchak O.N., Muntyan A.N., Rumyantseva T.B. 2011. Biologicheskoe raznoobrazie kluben’kovykh bakterii v ekosistemakh i agrotsenozakh. Teoreticheskie osnovy i metody (Biological Diversity of Rhizobia in Ecosystems and Agrocenoses: Theoretical Foundations and Methods). Rumyantseva M.L., Simarov B.V., Eds. St. Petersburg: VNIISKhM.
Vedam V., Kannenberg E., Datta A., Brown D., Haynes-Gann J.G., Sherrier D.J., Carlson R.W. 2006. The pea nodule environment restores the ability of a Rhizobium leguminosarum lipopolysaccharide acpXL mutant to add 27-hydroxyoctacosanoic acid to its lipid A. J. Bacteriol. 188, 2126–2133.
Antonets K.S., Volkov K.V., Maltseva A.L., Arshakian L.M., Galkin A.P., Nizhnikov A.A. 2016. Proteomic analysis of Escherichia coli protein fractions resistant to solubilization by ionic detergents. Biochemistry (Moscow). 81 (1), 34–46.
Rinaudi-Marron L.V., González J.E. 2013. Role of quorum sensing in the Sinorhizobium meliloti–alfalfa symbiosis. Mol. Microbial. Ecol. Rhizosphere. 1, 535–540.
Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25, 402–408.
Blanca-Ordóñez H., Oliva-García J.J., Pérez-Mendoza D., Soto M.J., Olivares J., Sanjuán J., Nogales J. 2010. pSymA-dependent mobilization of the Sinorhizobium meliloti pSymB megaplasmid. J. Bacteriol. 192, 6309–6312.
Mendrygal K.E., González J.E. 2000. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 182, 599–606.
Lynch D., O’Brien J., Welch T., Clarke P., Cuív P.O., Crosa J.H., O’Connell M. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183, 2576–2585.
Skorupska A., Janczarek M., Marczak M., Mazur A., Król J. 2006. Rhizobial exopolysaccharides: Genetic control and symbiotic functions. Microb. Cell Fact. 5, e7.
Booth I.R. 2014. Bacterial mechanosensitive channels: Progress towards an understanding of their roles in cell physiology. Curr. Opin. Microbiol. 18, 16–22.
ACKNOWLEDGMENTS
The study was carried out with equipment of the Research Resource Center for Molecular and Cell Technologies, St. Petersburg State University, and the Collective Use Center for Genome Technologies, Proteomics, and Cell Biology, Federal State Budgetary Scientific Institution, All-Russia Research Institute of Agricultiral Microbiology.
The comparative proteomic analysis of the S. meliloti bacteroids was supported by the Russian Science Foundation, Grant no. 14-26-00094. The development of visualization tools for gene expression on circular replicons was supported by the Grant of the President of the Russian Federation, MK-3240.2017.4.
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by E. Makeeva
Rights and permissions
About this article
Cite this article
Antonets, K.S., Onishchuk, O.P., Kurchak, O.N. et al. Proteomic Profile of the Bacterium Sinorhizobium meliloti Depends on Its Life Form and Host Plant Species. Mol Biol 52, 779–785 (2018). https://doi.org/10.1134/S0026893318050035
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893318050035