Abstract
The goal of this study was to isolate endophytic fungi from Ocimum sanctum L. and to evaluate their biological potential, including antibacterial, antioxidant, and enzymatic activities and further characterization of the bioactive compounds. Nine endophytic fungi were isolated from the leaves and stem tissues of O. sanctum collected from the botanical garden of Banaras Hindu University, Varanasi, India. All isolates were identified based on their microscopic structures and molecular sequencing of the ITS rDNA. Aspergillus clavatonanicus (SS7) and Cochliobolus hawaiiensis (SL3) showed the highest colonization frequencies in the stem and leaves (16 and 14%, respectively). All fungal isolates were tested for extracellular enzymatic activities of amylase, cellulase, and pectinase. Of the nine fungal isolates, 60% tested positive for amylase and cellulase, whereas 50% showed pectinase activity. Using a disc diffusion assay, the extracted secondary metabolites were checked for antibacterial activity against three human pathogenic bacteria. Two isolates, SL2 and SS7, exhibited the highest antibacterial activity against all pathogens, including Enterococcus faecalis, Klebsiella pneumoniae, and methicillin-resistant Staphylococcus aureus (MRSA). Crude extracts of the six fungal isolates showed positive antioxidant activity. The crude extract of Aspergillus allahabadii (isolate SL2) showed strong antibacterial and antioxidant activities and crystallized during purification. X-ray crystallography confirmed the identity of the crystal as citrinin, which also exhibited strong anticancer activity against Dalton’s lymphoma cells. The results of this study suggest that endophytic fungi isolated from the leaf and stem tissues of Ocimum sanctum are potential sources of antibacterial, antioxidant, and anticancer compounds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The emergence of new diseases and the development of drug-resistant pathogenic microorganisms pose a serious problem to humankind. To resolve this issue, search for efficient antimicrobial agents is required. For thousands of years, natural products of plant origin have been used in traditional medicine to cure diseases in humans (Mahajan et al., 2013). Endophytes are considered potential sources of bioactive natural products (Hagag et al., 2022). Fungal endophytes, the microorganisms that reside in the internal tissues of plants without causing any apparent disease, are known to produce useful bioactive compounds, including antibacterial, antiviral, antioxidant, and anticancer ones, as well as industrially important hydrolytic enzymes (Wilson, 1995; Verma et al., 2014; Kharwar et al., 2014; Gupta and Chaturvedi, 2019). Endophytic fungi that living inside healthy plant tissues effectively protect their hosts from biotic and abiotic stresses (Omacini et al., 2001; Redman et al., 2002). Since endophytes occupy unique biological niches in plants growing in diverse environments, they may be considered a source of novel bioactive natural compounds (Strobel et al., 2003; Verma et al., 2009). Recent findings also suggest that endophytic fungi may mimic the synthesis of host-origin metabolites (Singh et al., 2021). Since the discovery of penicillin from Penicillium notatum, fungi have been a great source of bioactive metabolites (Berdy, 2005), with even higher chances of finding novel compounds in endophytic fungi, since they colonize a special niche (Gupta and Chaturvedi, 2019). This prediction led to the discovery of Paclitaxel (Taxol) from Taxus brevifolia (Stierle et al., 1993). Taxol is the world’s first billion-dollar anticancer drug, the main source of which is Taxus spp. Successful identification of the fungal taxol has led to discovery of other bioactive compounds, such as vincristine, vinblastine, piperine, and azadirachtin from the endophytic fungi of their original hosts (Kusari et al., 2013; Singh et al., 2021). Bioactive secondary metabolites of endophytic origin are a good alternative to existing synthetic drugs, which provides more resistance to pathogens. The plant targeted for the endophyte isolation in this study was Ocimum sanctum L. (holy basil, member of the family Lamiaceae), a well-known medicinal plant. It is native to the Indian subcontinent, is widely cultivated throughout the Southeast Asian tropics, and is known for its religious and versatile medicinal properties (Mahajan et al., 2013). The leaves of O. sanctum are well-known for their antibacterial, antifungal, antiulcerogenic, antistress, anticancer, analgesic, antipyretic, anti-inflammatory, antihypertensive, radioprotective, and antitumor activities (Mahajan et al., 2013; Cohen, 2014). In India, the leaves of this plant are generally used for the treatment of fever and allergies. Keeping in mind the medicinal importance of this plant, the goal of the present work was to isolate the endophytic fungi from O. sanctum leaf and stem tissues and to characterize their biological activities, including antibacterial, antioxidant, and anticancer ones, as well as extracellular enzyme production and to further characterize their bioactive molecules.
MATERIALS AND METHODS
Plant selection site. Young and healthy leaves and stems were collected from disease-free plants in the botanical garden of the Banaras Hindu University (BHU), Varanasi, U.P., India, in a sterile polyethylene bag and immediately transported to the laboratory in an icebox. The samples were stored at 4°C and processed for isolation within 24 h. A total of 100 explants (50 from the leaves and 50 from the stem segments) were randomly cut and surface-sterilized for the isolation of endophytic fungi.
Surface treatment. The collected plant parts were rinsed in running tap water for 15‒20 min an then in double distilled water to remove soil particles from the surface of the plant segments. Surface sterilization was performed as described previously (Petrini et al., 1993; Sahu et al., 2022), with minor modifications. For that purpose, a sterile pinch cutter was used to cut the stems into 0.25-cm thick sections and the leaves into the segments of 0.5 × 0.5 cm2 using. The segments were immersed in 70% ethanol for 2–3 min, followed by sterilization with 4% aqueous sodium hypochlorite (NaOCl) for 2 min; the segments were then rinsed several times with sterile distilled water, in 70% ethanol for 1‒2 min and finally the tissues were rinsed 5 times in sterile double distilled water and allowed to surface dry under aseptic conditions.
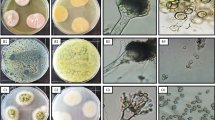
Isolation and morphological identification of the endophytic fungal isolates. The segments of sterilized leaf and stem tissues were placed on potato dextrose agar (PDA) amended with streptomycin sulfate (100 μg mL–1). A total of 100 segments (50 leaves and 50 stems) were used to observe the emergence of endophytic fungi. The plates were sealed with parafilm to avoid contamination and incubated at 27 ± 2°C for 15 days. The plates were observed regularly at alternate days for the emergence of endophytic fungi. The emerging fungal hyphae around the tissues were transferred to fresh PDA plates and purified. The endophytic fungi were grouped and identified based on morphological characteristics, including colony color, morphology, and microscopic observations of fruiting bodies and spore/conidia structures. The standard taxonomic manuals were used to identify the fungal genera and species (Ellis, 1971; Barnett and Hunter, 1972; Ainsworth et al., 1973; Von and Schipper, 1978). All isolated endophytic fungi were maintained in cryovials layered with 20% glycerol at –20°C in a deep freezer (Blue Star) in our laboratory in the Department of Botany, Banaras Hindu University, India.
Molecular identification and phylogeny of the endophytic fungal isolates. For molecular identification, genomic DNA was extracted from pure cultures of endophytic fungi following the protocol described by Sim et al. (2010). The universal primers ITS1: 5′TCCGTAGGTGAACCTGCGG3′ and ITS4: 5′TCCTCCGCTTATTGATAGC3′ (GeNei) (White et al., 1990) were used to amplify the 5.8S rDNA and two ITS regions between the 18S and 28S rDNA. The PCR mixture (25 µL) contained 1 µL DNA template, 1 µL each primer, 0.33 µL (3-unit µL–1) Taq polymerase, 1.5 µL of 25 mM MgCl2, 0.25 µL dNTPs, buffer (10×) 2.5 µL and 17.42 µL MQ water. PCR reactions were performed in a My-cycler (Bio-Rad, Hercules, CA, United States) under the following conditions: pre-denaturation at 94°C for 4 min, 35 cycles at 94°C (denaturation) for 1 min, 55°C (annealing) for 1 min, 72°C (extension) for 1 min, and final extension for 5 min at 72°C. Amplified PCR products were resolved by electrophoresis in a 1.5% (wt/vol) agarose gel followed by staining with ethidium bromide (0.5 µg mL–1) for visual examination. PCR products were sequenced by AgriGenome Labs Pvt. Ltd. (Cochin, Kerala, India). The amplified rDNA sequences obtained were used to retrieve similar sequences from the NCBI GenBank sequence database using the NCBI BLAST program. The sequences were submitted to the NCBI GenBank database and accession numbers were obtained for each isolate. MEGA version 7 software was used to align the sequence with the ClustalW function, and the phylogenetic tree was constructed using the maximum-likelihood method with closely related sequences of all nine isolates, retrieved from the NCBI database (two or three sequences for each i.e., a total of 32 and 1 outgroup).
Colonization frequency (%CF) of endophytic fungi. The collective colonization frequency (%CF) of the endophytic fungi was calculated using the formula given by Hata and Futai (1995): %CF = (Ncol/Nt) × 100, where Ncol is the number of segments of plant tissue colonized by each fungus and Nt is the total number of segments of plant tissue studied.
Extracellular enzyme production activity. A qualitative assay for the extracellular production of enzymes by the fungal endophytes was performed by digestion of suspended substrate in basal agar medium on petri plates. For that, mycelial plugs of each isolate (5 mm in diameter) were inoculated in the center of already prepared plates and incubated for 7 days at 27 ± 2°C in a BOD incubator. The clear zone around the fungal colony was measured as positive a test for the enzyme activity. In brief, an amylase assay was performed on glucose yeast extract peptone (GYP) agar medium (Hankin and Anagnostakis, 1975) composed of 1 g glucose, 0.1 g yeast extract, 0.5 g peptone, 15 g agar, and 1 L distilled water, pH 6 and supplemented with 2% soluble starch. After seven days of incubation, the plates were flooded with 1% iodine in 2% potassium iodide. Clear zones surrounding the colonies were measured. Cellulase activity was observed on yeast extract peptone agar medium (Lingappa, 1962) composed of 0.1 g yeast extract, 0.5 g peptone, 15 g agar, 1 L distilled water, pH 6 and supplemented with 0.5% Na-carboxymethyl cellulose (CMC). After seven days of incubation, the plates were flooded with 0.2 aqueous solutions of Congo red for 10 min and destained with 1 M NaCl for 15 min. The clear zones surrounding the fungal colonies were measured. Pectinase activity was determined using a slightly modified protocol described by Aguilar and Huitron (1990). For that, pectin agar was prepared containing 5.0 g yeast extract, 5.0 g pectin, and 15 g agar in 1 L distilled water with pH 6. The endophytic fungi were inoculated onto pectin agar plates and incubated for 7 days. After incubation, the plates were flooded with a freshly prepared 1% aqueous cetyltrimethyl ammonium bromide (CTAB) solution. The clear zone around the fungal colony was measured.
Production and extraction of secondary metabolites from the fungal endophytes. For the production of secondary metabolites, the actively growing mycelial discs of each pure fungal endophyte were cut and inoculated in 500 mL of potato dextrose broth (PDB). After 21 days of incubation at 27 ± 2°C, the mycelium was removed by filtration through four layers of muslin cloth, and the filtrate was poured into a separating funnel. For the extraction of secondary metabolites, the filtrate was mixed well with an equal volume of ethyl acetate. The solvent layer was collected separately, and the filtrate was subjected to extraction twice with an equal volume of ethyl acetate. The extracted secondary metabolites were concentrated and evaporated in a rotary vacuum evaporator (IKA, Germany). The concentrated secondary metabolites obtained from each isolate were transferred in glass vials separately for further evaporation at room temperature. After complete dryness, metabolites were stored in a refrigerator at 4°C for further study.
Antibacterial activity of secondary metabolites of the fungal endophytes. The fungal secondary metabolites were evaluated for their antibacterial activity against three strains of human pathogenic bacteria, Enterococcus faecalis (EF), Klebsiella pneumoniae (KP), and methicillin-resistant Staphylococcus aureus (MRSA) procured from the Institute of Medical Sciences (IMS) BHU, Varanasi, India using the disc diffusion assay (Hudzicki and Bauer, 2009). Pre-weighed crude metabolite extracts were dissolved in methanol and diluted to 10 mg mL–1 for the assay. A sterile paper disc (5 mm diameter, Whatman no. 1) was impregnated with 20 µL of the methanol extract using a micropipette and dried under a laminar hood for 20 min. Air-dried paper discs containing 0.2 mg crude metabolite extract from each isolate were placed separately on nutrient agar plates lawn-inoculated with pathogenic bacterial cultures. The disc containing only 20 µL of methanol was used as the control. The plates were incubated at 30 ± 2°C for 24 h. The zones of inhibition around the discs were measured for each extract. All experiments were performed in triplicates.
Antioxidant activity of secondary metabolites of the fungal endophytes. Crude metabolites of endophytic fungi were screened for free radical scavenging activity by the DPPH (2,2-diphenyl 2-picryl hydrazyl) method. DPPH, a violet-colored compound, is a stable free radical that accepts an electron or a hydrogen atom to form a stable diamagnetic molecule, which is yellow in color. This de-colorization is showing a positive test for anti-oxidant activity. Crude extract of endophytic fungi (100 µL) dissolved in methanol (1 mg mL-1) were mixed with 2900 µL DPPH solution (100 µM) in methanol and incubated in the dark for 30 min. A change in the color of DPPH was observed as a positive test.
X-ray crystallography. The crude metabolite extract of the isolate SL2 was fractionated with hexane, and the remaining extract was dissolved in ethyl acetate and slowly evaporated at 20°C in glass vials. After 2‒3 days of evaporation, fine crystals were formed. The crystals were cleaned with ethyl acetate and analyzed by X-ray crystallography. Data collection was performed using an Xcalibur Eos CCD (OXFORD) single-crystal diffractometer at room temperature (20°C). Graphite-monochromated MoKα radiation (γ = 0.71073 Å) was used for diffraction. The structures of the successfully diffracted crystals were analyzed.
NMR spectroscopy. The 1H and 13C NMR spectra were recorded on a JNM-ECZ500R/S1 Spectrometer with an operating field strength of 500 MHz. The 1H NMR spectra of the crystals were analyzed in DMSO (d6). The NMR software was imported into Mest ReNova (version 12.0), and manual baseline correction and automatic phase correction were performed. The tetramethylsilane (TMS) proton signal was calibrated to 0.00 ppm. The peaks of –CH3 (a, b, c) were recorded in the range of 1–3 ppm. The peak appeared at 3.1 ppm may be the (d) proton. Due to deshielding of oxygen atom proton of (e and f) appeared at 4.9 and 8.5 ppm. Phenolic group (–OH) proton appeared at 15.4 ppm and carboxylic acid proton (–COOH) appeared at 16.3 ppm (Fig. 5).
On the other hand, 13C-NMR spectroscopy provides information about the carbon environment in a molecule. This is particularly useful for determining the connectivity and carbon framework of organic compounds. The carbon skeleton and arrangement of functional groups can be identified by analyzing the chemical shifts of the carbon atoms in the crystal. In 13C-NMR of the crystal, the –CH3 carbon peak appeared in the range of 10‒20 ppm. The ketonic (‒C=O) carbon appeared at 187 ppm and carboxylic carbon (–COOH) appeared at 170 ppm. Further, carbon peaks appeared at 76, 131, 137, 110, and 162 ppm respectively (Fig. S1).
Anticancer activity of the pure compound. The anticancer activity of this pure compound was assessed by the MTT (3-[4,5-dimethylthiazol-2yl]-2,5-diphenyl tetrazolium bromide) assay, as described previously by Jaiswara et al. (2021) on the Dalton’s Lymphoma (DL) cell line. DL is a murine T-cell lymphoma used to evaluate the antitumor potential of various synthetic and natural compounds both in vitro and in vivo. DL cells (1 × 106 cells mL–1) were inoculated into 96-well culture plates and treated with the indicated concentrations of the pure compound for 24 and 48 h. After the indicated time periods, 25 µL of the MTT solution (5 mg MTT in 1 mL PBS) was added to each well of the 96 well culture plates containing DL cells and incubated at 37°C for 4 h in a humidified CO2 incubator. Thereafter, the pellet of formazan crystals was collected by centrifugation (3000 rpm for 15 min) and dissolved in 50 µL of acidified isopropanol followed by measurement of absorbance at 540 nm by using an ELISA plate reader (ERBA LISA SCAN, Germany).
RESULTS
Isolation, identification, and phylogenetic analysis of endophytic fungi. Nine morphologically distinct endophytic fungal isolates were recovered from 100 segments of the leaf and stem tissues of Ocimum sanctum (Table 1). Based on microscopic studies and the ITS rDNA sequencing, the nine isolates were identified as Corynespora cassiicola (SS1), Alternaria alternata (SS2), Pleosporales sp. (SS4), Alternaria longipes (SS5), Alternaria tenuissima (SS6), and Aspergillus clavatonanicus (SS7) from the stem, and Penicillium citrinum (SL1), Aspergillus allahabadii (SL2), and Cochliobolus hawaiiensis (SL3) from the leaf samples (Table 1). Sequences of all the isolates were submitted to the NCBI database, and the accession numbers obtained along with the closest match from the NCBI database with their similarity query coverage (%QC) are listed in Table 1. A phylogenetic tree was constructed using the maximum-likelihood method implemented in the Mega 7 software, which provided more accurate descriptions of the patterns of relatedness, and all nine strains clustered at strongly supported and consistent nodes inside their respective clusters. Bootstrap values for all pertinent nodes were consistent and robust. The rest of their related taxa were clustered at individual well-supported nodes as shown in Fig. 1. Out of nine isolates, Aspergillus clavatonanicus (SS7) showed the highest CF% (16%) among the stem isolates, followed by Corynespora cassiicola (SS1) (10%) and Alternaria tenuissima (SS6) (10%). However, the highest CF% in leaf samples were found for Cochliobolus hawaiiensis (SL3) (14%), followed by Penicillium citrinum (SL1) (8%) (Table 1).
Extracellular enzyme production. Of the nine isolates tested, seven of showed positive results in one or more enzyme production tests. Aspergillus allahabadii (SL2) showed positive results in all three enzymatic assays. Cochliobolus hawaiiensis (SL3) showed the highest amylase activity, with a clear zone of >10 mm, whereas SS2 and SS4 did not show any enzymatic activity (Table 2). Cellulase activity was observed in the isolates of Aspergillus allahabadii (SL2), Alternaria tenuissima (SS6), and Aspergillus clavatonanicus (SS7). Three isolates, Penicillium citrinum (SL1), Aspergillus allahabadii (SL2), and Aspergillus clavatonanicus (SS7), also showed pectinase activity (Table 2).
Extraction of crude metabolites and antibacterial activity. Crude metabolites were successfully extracted from 21 day-old broth cultures of all fungal isolates using ethyl acetate. The antibacterial activity of crude metabolite extracts was tested against the three standard bacterial pathogens, Klebsiella pneumoniae (KP), Enterococcus faecalis (EF), and MRSA using the disc diffusion assay. Of the nine isolates, the crude extracts of five isolates showed strong antibacterial activity against one or more of the tested pathogens (Fig. 2 and Table 3). The inhibition zone diameters varied from 6 to >10 mm. The extracts from Aspergillus allahabadii (SL2) and Aspergillus clavatonanicus (SS7) were effective against all tested bacterial pathogens, with an inhibition zone of 10 mm or more (except SL2 against MRSA). Penicillium citrinum (SL1) was effective against two bacterial pathogens. Alternaria longipes (SS5) and Alternaria tenuissima (SS6) were only effective against one bacterial pathogen each. Crude extracts of SL3, SS1, SS2, and SS4 did not show any antibacterial activity against the tested pathogens.
Antibacterial activity of secondary metabolites of endophytic fungi against pathogenic bacteria using disc diffusion assay. (a) Endophytic fungi isolated from leaves where 1—Penicillium citrinum, 2—Aspergillus allahabadii, 3—Cochliobolus hawaiiensis, (b) Endophytic fungi isolated from stem where 1—Corynespora cassiicola, 2—Alternaria alternata, 4—Pleosporales sp., 5—Alternaria longipes, 6—Alternaria tenuissima; 7—Aspergillus clavatonanicus and c—control.
Antioxidant activity. DPPH, a stable free radical, was used to study the radical-scavenging effects of the extracts as antioxidant activity. The scavenging effects of the samples were evaluated based on the extent of decolorization. The crude fungal metabolite extracts of six out of nine isolates showed positive results. The fungal extract of Cochliobolus hawaiiensis (SL3) showed maximum decolorization of the DPPH radical. The extracts from Penicillium citrinum (SL1), Aspergillus allahabadii (SL2), Corynespora cassiicola (SS1), Alternaria alternata (SS2), and Aspergillus clavatonanicus (SS7) also showed significant antioxidant activities. However, SS4, SS5, and SS6 did not exhibit any antioxidant activity (Fig. 3).
Characterization of the pure compound from the endophytic fungus Aspergillus allahabadii (SL2). Fine crystals were formed from partially purified metabolites of A. allahabadii under slow evaporation in ethyl acetate. Microscopic examination confirmed the purity of the crystals. X-ray crystallography confirmed the structure of the compound as citrinin (Fig. 4). Moreover, the 1H-NMR (Fig. 5) and 13C-NMR (Fig. S1) spectra also confirmed that the purified compound was citrinin.
Anticancer activity of the pure compound citrinin. The anticancer activity of citrinin was assessed using the MTT assay, as discussed in materials and methods. The results are shown in Fig. 6. Our results showed that citrinin exhibited a significant antitumor potential in a dose-dependent manner after 48 h with the IC50 value ~150 µM, although low cytotoxic activity was noted after 24 h.
Assessment of antitumor activity of citrinin extracted from Aspergillus allahabadii. Dalton’s lymphoma cells-DL (1 × 106 cells mL–1) were treated with mentioned concentrations of citrinin (0, 50, 100, 150, 200 and 400 μM) for 24 and 48 h followed by estimation of percent cytotoxicity by MTT assay as described in materials and methods. The values shown are the mean ± SD of three independent experiments done in triplicate. * p < 0.05 vs. cells treated with citrinin for 24 h.
DISCUSSION
Since the discovery of penicillin from Penicillium notatum, fungi have been recognized as a great source of a plethora of bioactive compounds. Because of their specialized niche, endophytic fungi of medicinal plants have shown the potential to produce novel bioactive compounds that have greatly attracted the attention of researchers (Gupta and Chaturvedi, 2019; Singh, 2019; Jiang et al., 2022). The selection of medicinal plants for the isolation of endophytes is influenced by such factors as chemo-diversity, the traditional use of the plants, and the region from which the plant is obtained (Verpoorte, 1998). Given the growing need for novel compounds with antimicrobial, antioxidant, and anticancer potential, this study focuses on exploring the diversity of endophytic fungi derived from the O. sanctum plant and investigating their potential as a source of compounds with antibacterial, antioxidant, and anticancer properties.
In this study, a total of nine endophytic fungi were isolated from leaf and stem tissues of the plant. The colonization frequency data suggest that different endophytic fungal strains colonize different plant tissues (such as stem and leaves) to a different extent. This suggests the varying degree of affinity of the isolated strains towards different tissues of the host plant. Aspergillus clavatonanicus was found to be the most prevalent species in stem tissue, whereas Cochliobolus hawaiiensis predominated in the leaf tissue. These isolates were identified using a combination of morphological and molecular methods (Table 1). A combined approach using microscopic spore structures and molecular ITS rDNA sequencing methods is considered a reliable method for the identification of culturable fungi at the species level (Verma, 2014; Singh et al., 2018). Advancements in sequencing technologies, such as next-generation sequencing, have enabled the exploration of the functional roles of non-culturable microorganisms (Tao et al., 2008; Maghembe et al., 2020). Species of Aspergillus, Cochliobolus, Alternaria, and Corynespora were the most common isolates in our study. These species have also been isolated as endophytes from other medicinal plants, including Aegle marmelos, Azadirachta indica, Adenocalymma alliaceum, and Tectona grandis (Verma et al., 2007; Kharwar et al., 2010, 2011; Singh et al., 2018).
In the antibacterial assay against three human pathogenic bacteria, Enterococcus faecalis, Klebsiella pneumoniae, and MRSA, more than 55% of the isolates significantly inhibited one, two, or all three pathogens tested in the disc diffusion assay (Table 3, Fig. 2). Gong and Guo (2009) reported that only 8.3% of the fungal isolates of Dracaena cambodiana and Aquilaria sinensis showed antibacterial activity. However, in our previous study, 58.33% of Madhuca indica fungal isolates screened positive for antibacterial activity against the tested pathogens (Verma et al., 2014). This suggests that these endophytic fungi have the potential to combat such drug-resistant bacteria as MRSA. Interestingly, two endophytic fungi, Aspergillus allahabadii and Aspergillus clavatonanicus, inhibited the growth of all three human pathogens tested, showing broad-spectrum antibacterial activity (Table 3, Fig. 2). The crude extracts of these two endophytic fungi also showed strong antioxidant activity (Fig. 3). Many studies have reported significant antioxidant activity of many endophytic fungi (Huang et al., 2007; Sadananda et al., 2011; Mishra et al., 2018). Hence, these can be used as a source of natural antioxidants and further efforts in identification and isolation of the bioactive constituents can lead to the development of antioxidant-based drugs or dietary supplements.
Fungi have been a great source of industrially important enzymes, and many endophytic fungi have also been reported to successfully produce hydrolytic enzymes, including amylase, cellulase, protease, and pectinase, which probably help the endophytes to successfully colonize their host plants (Karnchanatat et al., 2007; Caroll and Pertini, 2014; Verma and White, 2018). In this study, out of nine isolates, seven showed positive test results for one or more enzymes, in which A. allahabadii tested positive for all enzymes (amylase, cellulase, and pectinase) (Table 2). The amylase activity can help the fungus to break down the starch into simpler forms of sugar, which it may utilize for growth and development during unfavourable conditions (Sunitha et al., 2012). Furthermore, the high amylase activity in endophytic fungi may also have industrial applications, such as in food and pharmaceutical industries. Similarly, cellulases can play important roles in lignocellulosic biomass degradation, bioremediation, and industrial applications (Robl et al., 2013). Finally, pectinases play a crucial role in the degradation of pectin, the polysaccharide present in plant cell walls, and hence can act as an important tool in the decomposition of host tissue and the establishment of symbiosis between endophytic fungi and their plant host (Rahul et al., 2015; Hawar, 2022).
The crude metabolite extract of one isolate, A. allahabadii, showed strong antioxidant and antibacterial activities. During the purification process, fine crystals were formed and X-ray crystallography confirmed the identity of the pure crystal as citrinin (Fig. 4). Citrinin is a low molecular weight (250.25 g mol−1) polyketide compound produced by many fungi, including the Penicillium and Aspergillus species, a known mycotoxin contaminant of several food grains (Dunn, 1994). Despite its toxicity, citrinin exhibits a range of biological activities, including antibacterial, antifungal, and cytotoxic ones (Mazumder et al., 2002; Park et al., 2008, Filho et al., 2017; Wang et al., 2019). Previous studies reported that citrinin extracted from Penicillium citrinum inhibited the growth of many bacterial pathogens including gram-positive (Staphylococcus aureus, Bacillus pumilus, Klebsiella pneumoniae, and Streptococcus pneumoniae) and gram-negative ones (Escherichia coli, Shigella boydii, and Salmonella typhimurium) (Mazumder et al., 2002; Wang et al., 2019). Recently, a few studies have shown the cytotoxic action of citrinin against different cancers, such as breast and colon cancer, and reported the antitumor activity of citrinin possibly due to induction of apoptosis (Filho et al., 2021). A study also reported that citrinin produced by the fungus P. citrinum increased the motility of the rhizospheric bacteria Paenibacillus polymyxa (Park et al., 2008).
In the present study, we found that the crude extract of A. allahabadii containing citrinin showed strong antibacterial activity against all tested pathogens (Fig. 2 and Table 3). Interestingly, recent studies have demonstrated the anticancer potential of citrinin against colon and breast cancers (Salah et al., 2017; Filho et al., 2021; Menezes et al., 2023). However, to date, the antitumor potential of citrinin has not yet been examined in hematological malignancies such as T-cell lymphoma, which is considered a highly complex neoplastic disorder. Moreover, to establish citrinin as a promising anticancer drug, it is essential to test its tumoricidal activity against a wide range of cancers. In our study, citrinin exerted a significant cytotoxic effect in a dose-dependent manner after 48 h, although a low cytotoxic activity was noted after 24 h. The results of the present study suggest that fungal endophytes are a great source of bioactive molecules having great therapeutic potential (Kharwar et al., 2011; Singh et al., 2018). Further studies are required to characterize the pure compounds isolated from other active fungal isolates. Moreover, investigations are also warranted to explore the mechanisms underlying these bioactivities and their potential applications in drug development and plant-protection strategies.
ABBREVIATIONS AND NOTATION
ITS | internal transcribed spacer |
PDA | potato dextrose agar |
CF | colonization frequency |
DMSO | dimethyl sulfoxide |
MRSA | methicillin-resistant Staphylococcus aureus |
CTAB | cetyltrimethyl ammonium bromide |
TMS | tetramethyl silane |
DPPH | (2,2-diphenyl 2-picryl hydrazyl) |
MTT | (3-[4,5-dimethylthiazol-2yl]-2,5-diphenyl tetrazolium bromide) |
DL | Dalton’s lymphoma |
PBS | phosphate buffer saline |
IC50 | half maximum inhibitory concentration |
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
Aguilar, G. and Huitrón, C., Constitutive exo-pectinase produced by Aspergillus sp. CH-Y-1043 on different carbon source, Biotechnol. Lett., 1990, vol. 12, pp. 655–660.
Ainsworth, G.C. and Austwick, P.K.C., Fungal Diseases of Animals, 1973. https://doi.org/10.2307/3758441
Barnett, H.L. and Hunter, B.B., Illustrated Genera of Imperfect Fungi, 1972, 3rd ed.
Berdy, J., Bioactive microbial metabolites, The J. Antibiot., 2005, vol. 58, no. 1, pp. 1–26. https://doi.org/10.1038/ja.2005.1
Carroll, G. and Petrini, O., Patterns of substrate utilization by some fungal endophytes from coniferous foliage, Mycologia, 1983, vol. 75, no. 1, pp. 53–63. https://doi.org/10.1080/00275514.1983.12021637
Cohen, M.M. Tulsi-Ocimum sanctum: A herb for all reasons. Journal of Ayurveda and Integrative Medicine, 2014, vol. 5, no. 4, p. 251. https://doi.org/10.4103/0975-9476.146554
de Menezes, A.A.P., Aguiar, R.P., Santos, J.V., Sarkar, C., Islam, M.T., Braga, A.L., ... and Sousa, J.M., Citrinin as a potential anti-cancer therapy: a comprehensive review, Chem.-Biol. Interact., 2023, p. 110561. https://doi.org/10.1016/j.cbi.2023.110561
de Oliveira Filho, J.W.G., Andrade, T.D.J.A.D.S., de Lima, R.M.T., Dos Reis, A.C., Silva, D.H.S., Santos, J.V.D.O., ... and Melo Cavalcante, A.A.D.C., Citrinin against breast cancer: a cytogenotoxicological study, Phytother. Res., 2021, vol. 35, no. 1, pp. 504–516. https://doi.org/10.1002/ptr.6830
de Oliveira Filho, J.W.G., Islam, M.T., Ali, E.S., Ud-din, S.J., de Oliveira Santos, J.V., de Alencar, M.V.O.B., ... and de Carvalho Melo-Cavalcante, A.A., A comprehensive review on biological properties of citrinin, Food Chem. Toxicol., 2017, vol. 110, pp. 130–141. https://doi.org/10.1016/j.fct.2017.10.002
Dunn, B.B. and Friedman, L., Distribution and metabolism of citrinin: a review, in Mycotoxins, Wood Decay, Plant Stress, Biocorrosion, and General Biodeterioration, 1994, pp. 27−40.
Ellis, M.B., Dematiaceous Hyphomycetes, 1971.
Gong, L. and Guo, S., Endophytic fungi from Dracaena cambodiana and Aquilaria sinensis and their antimicrobial activity, Afr. J. Biotechnol., 2009, vol. 8, no. 5.
Gupta, S. and Chaturvedi, P., Enhancing secondary metabolite production in medicinal plants using endophytic elicitors: a case study of Centella asiatica (Apiaceae) and asiaticoside, in Endophytes for a Growing World, 2019, pp. 310−323.
Hagag, A., Abdelwahab, M.F., Abd El-kader, A.M., and Fouad, M.A., The endophytic Aspergillus strains: a bountiful source of natural products, J. Appl. Microbiol., 2022, vol. 132, no. 6, pp. 4150–4169. https://doi.org/10.1111/jam.15489
Hankin, L. and Anagnostakis, S.L., The use of solid media for detection of enzyme production by fungi, Mycologia, 1975, vol. 67, no. 3, pp. 597–607. https://doi.org/10.1080/00275514.1975.12019782
Hata, K. and Futai, K., Endophytic fungi associated with healthy pine needles and needles infested by the pine needle gall midge, Thecodiplosis japonensis, Can. J. Bot., 1995, vol. 73, no. 3, pp. 384–390. https://doi.org/10.1139/b95-040
Hawar, S.N., Extracellular enzyme of endophytic fungi isolated from Ziziphus spina leaves as medicinal plant, Int. J. Biomater., 2022. https://doi.org/10.1155/2022/2135927
Huang, W.Y., Cai, Y.Z., Xing, J., Corke, H., and Sun, M., A potential antioxidant resource: endophytic fungi from medicinal plants, Econ. Bot., 2007, vol. 61, no. 1, pp. 14–30.
Hudzicki, J., Kirby-Bauer disk diffusion susceptibility test protocol, Amer. Soc. Microbiol., 2009, vol. 15, pp. 55–63.
Jaiswara, P.K., Gupta, V.K., Sonker, P., Rawat, S.G., Tiwari, R.K., Pathak, C., ... and Kumar, A., Nimbolide induces cell death in T lymphoma cells: Implication of altered apoptosis and glucose metabolism. Environ. Toxicol., 2021, vol. 36, no. 4, pp. 628−641. https://doi.org/10.1002/tox.23067
Jiang, H., Cai, R., Zang, Z., Yang, W., Wang, B., Zhu, G., ... and She, Z., Azaphilone derivatives with anti-inflammatory activity from the mangrove endophytic fungus Penicillium sclerotiorum ZJHJJ-18., Bioorg. Chem., 2022, vol. 122, p. 105721. https://doi.org/10.1016/j.bioorg.2022.105721
Karnchanatat, A., Petsom, A., Sangvanich, P., Piaphukiew, J., Whalley, A.J., Reynolds, C. D., and Sihanonth, P., Purification and biochemical characterization of an extracellular β-glucosidase from the wood-decaying fungus Daldinia eschscholzii (Ehrenb.: Fr.) Rehm, FEMS Microbiol. Lett., 2007, vol. 270, no. 1, pp. 162−170. https://doi.org/10.1111/j.1574-6968.2007.00662.x
Kharwar, R.N., Gond, S.K., Kumar, A., and Mishra, A., A comparative study of endophytic and epiphytic fungal association with leaf of Eucalyptus citriodora Hook., and their antimicrobial activity, World J. Microbiol. Biotechnol., 2010, vol. 26, pp. 1941–1948. https://doi.org/10.1007/s11274-010-0374-y
Kharwar, R.N., Sharma, V.K., Mishra, A., Kumar, J., Singh, D.K., Verma, S.K., ... and Kusari, S., Harnessing the phytotherapeutic treasure troves of the ancient medicinal plant Azadirachta indica (Neem) and associated endophytic microorganisms, Planta Medica, 2020, vol. 86, nos. 13/14, pp. 906–940. https://doi.org/10.1055/a-1107-9370
Kharwar, R.N., Verma, S.K., Mishra, A., Gond, S.K., Sharma, V.K., Afreen, T., and Kumar, A., Assessment of diversity, distribution and antibacterial activity of endophytic fungi isolated from a medicinal plant Adenocalymma alliaceum Miers, Symbiosis, 2011, vol. 55, pp. 39–46. https://doi.org/10.1007/s13199-011-01422
Kusari, S., Pandey, S.P., and Spiteller, M., Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites, Phytochemistry, 2013, vol. 91, pp. 81–87. https://doi.org/10.1016/j.phytochem.2012.07.021
Lingappa, B.T. and Lockwood, J.L., Relationship of soil microbes to the widespread soil fungistasis, Abstr. Phytopathol., 1962, vol. 52, p. 739.
Maghembe, R., Damian, D., Makaranga, A., Nyandoro, S.S., Lyantagaye, S.L., Kusari, S., and Hatti-Kaul, R., Omics for bioprospecting and drug discovery from bacteria and microalgae, Antibiotics, 2020, vol. 9, no. 5, p. 229. https://doi.org/10.3390/antibiotics9050229
Mahajan, N., Rawal, S., Verma, M., Poddar, M., and Alok, S., A phytopharmacological overview on Ocimum species with special emphasis on Ocimum sanctum, Biomed. Prevent. Nutrit., 2013, vol. 3, no. 2, pp. 185–192. https://doi.org/10.1016/j.bionut.2012.08.002
Mazumder, P.M., Mazumder, R., Mazumder, A., and Sasmal, D.S., Antimicrobial activity of the mycotoxin citrinin obtained from the fungus Penicillium citrinum, Ancient Sci. Life, 2002, vol. 21, no. 3, p. 191.
Mishra, J., Rajput, R., Singh, K., Puri, S., Goyal, M., Bansal, A., and Misra, K., Antibacterial natural peptide fractions from Indian Ganoderma lucidum, Int. J. Peptide Res. Therap., 2018, vol. 24, pp. 543–554. https://doi.org/10.1007/s10989-017-9643-z
Omacini, M., Chaneton, E.J., Ghersa, C.M., and Müller, C.B., Symbiotic fungal endophytes control insect host–parasite interaction webs, Nature, 2001, vol. 409, no. 6816, pp. 78–81. https://doi.org/10.1038/35051070
Park, S.Y., Kim, R., Ryu, C.M., Choi, S.K., Lee, C.H., Kim, J.G., and Park, S.H., Citrinin, a mycotoxin from Penicillium citrinum, plays a role in inducing motility of Paenibacillus polymyxa, FEMS Microbiol. Ecol., 2008, vol. 65, no. 2, pp. 229–237. https://doi.org/10.1111/j.1574-6941.2008.00492.x
Petrini, O., Sieber, T.N., Toti, L., and Viret, O., Ecology, metabolite production, and substrate utilization in endophytic fungi, Natural Toxins, 1993, vol. 1, no. 3, pp. 185–196. https://doi.org/10.1002/nt.2620010306
Rahul, Y., Ajay, V.S., Samiksha, J., and Manish, K., Antifungal and enzyme activity of endophytic fungi isolated from Ocimum sanctum and Aloe vera, Afr. J. Microbiol. Res., 2015, vol. 9, no. 29, pp. 1783–1788. https://doi.org/10.5897/AJMR2015.7451
Redman, R.S., Sheehan, K.B., Stout, R.G., Rodriguez, R.J., and Henson, J.M., Thermotolerance generated by plant/fungal symbiosis, Science, 2002, vol. 298, no. 5598, p. 1581. https://doi.org/10.1126/science.1078055
Robl, D., Delabona, P.D.S., Mergel, C.M., Rojas, J.D., Costa, P.D.S., Pimentel, I. C., ... and Padilla, G., The capability of endophytic fungi for production of hemicellulases and related enzymes, BMC Biotechnol., 2013, vol. 13, no. 1, pp. 1–12. https://doi.org/10.1186/1472-6750-13-94
Sadananda, T.S., Nirupama, R., Chaithra, K., Govindappa, M., Chandrappa, C.P., and Vinay Raghavendra, B., Antimicrobial and antioxidant activities of endophytes from Tabebuia argentea and identification of anticancer agent (lapachol), J. Med. Plants Res., 2011, vol. 5, no. 16, pp. 3643−3652.
Sahu, P.K., Tilgam, J., Mishra, S., Hamid, S., Gupta, A.K.J., ... and Kharwar, R.N., Surface sterilization for isolation of endophytes: ensuring what (not) to grow, J. Bas. Microbiol., 2022, vol. 62, no. 6, pp. 647–668. https://doi.org/10.1002/jobm.202100462
Salah, A., Bouaziz, C., Prola, A., Pires Da Silva, J., Bacha, H., Abid-Essefi, S., and Lemaire, C., Citrinin induces apoptosis in human HCT116 colon cancer cells through endoplasmic reticulum stress, J. Toxicol. Environ. Health, Part A, 2017, vol. 80, nos. 23/24, pp. 1230–1241. https://doi.org/10.1080/15287394.2017.1359127
Sim, J.H., Khoo, C.H., Lee, L.H., and Cheah, Y.K., Molecular diversity of fungal endophytes isolated from Garcinia mangostana and Garcinia parvifolia, J. Microbiol. Biotechnol., 2010, vol. 20, no. 4, pp. 651–658. https://doi.org/10.4014/jmb.0909.09030
Singh, A., Singh, D.K., Kharwar, R.N., White, J.F., and Gond, S.K., Fungal endophytes as efficient sources of plant-derived bioactive compounds and their prospective applications in natural product drug discovery: Insights, avenues, and challenges, Microorganisms, 2021, vol. 9, no. 1, p. 197. https://doi.org/10.3390/microorganisms9010197
Singh, B.P., Ed., Advances in Endophytic Fungal Research: Present Status and Future Challenges, Springer, 2019.
Singh, D.K., Kumar, J., Sharma, V.K., Verma, S.K., Singh, A., Kumari, P., and Kharwar, R.N., Mycosynthesis of bactericidal silver and polymorphic gold nanoparticles: physicochemical variation effects and mechanism, Nanomedicine, 2018, vol. 13, no. 2, pp. 191–207. https://doi.org/10.2217/nnm-2017-0235
Stierle, A., Strobel, G., and Stierle, D., Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew, Science, 1993, vol. 260, no. 5105, pp. 214–216. https://doi.org/10.1126/science.8097061
Strobel, G. and Daisy, B., Bioprospecting for microbial endophytes and their natural products, Microbiol. Mol. Biol. Rev., 2003, vol. 67, no. 4, pp. 491–502. https://doi.org/10.1128/mmbr.67.4.491-502.2003
Sunitha, V.H., Ramesha, A., Savitha, J., and Srinivas, C., Amylase production by endophytic fungi Cylindrocephalum sp. isolated from medicinal plant Alpinia calcarata (Haw.) Roscoe, Brazil. J. Microbiol., 2012, vol. 43, no. 3, p. 1213. https://doi.org/10.1590/S1517-838220120003000049
Tao, G., Liu, Z.Y., Hyde, K.D., Lui, X.Z., and Yu, Z.N., Whole rDNA analysis reveals novel and endophytic fungi in Bletilla ochracea (Orchidaceae), Fungal Divers., 2008, vol. 33, no. 1, pp. 101–112. https://doi.org/10.1186/s12866-017-0961-2
Verma, S.K. and White, J.F., Indigenous endophytic seed bacteria promote seedling development and defend against fungal disease in browntop millet (Urochloa ramosa L.), J. Appl. Microbiol., 2018, vol. 124, no. 3, pp. 764–778. https://doi.org/10.1111/jam.13673
Verma, S.K., Gond, S.K., Mishra, A., Sharma, V.K., Kumar, J., Singh, D.K., ... and Kharwar, R.N., Impact of environmental variables on the isolation, diversity and antibacterial activity of endophytic fungal communities from Madhuca indica Gmel. at different locations in India, Ann. Microbiol., 2014, vol. 64, no. 2, pp. 721–734. https://doi.org/10.2307/3545919
Verma, V.C., Gond, S.K., Kumar, A., Kharwar, R.N., and Strobel, G., The endophytic mycoflora of bark, leaf, and stem tissues of Azadirachta indica A. Juss (Neem) from Varanasi (India), Microb. Ecol., 2007, vol. 54, pp. 119–125. https://doi.org/10.1007/s00248-006-9179-9
Verma, V.C., Kharwar, R.N., and Strobel, G.A., Chemical and functional diversity of natural products from plant associated endophytic fungi, Nat. Prod. Commun., 2009, vol. 4, no. 11. https://doi.org/10.1177/1934578X0900401114
Verpoorte, R., Exploration of nature’s chemodiversity: the role of secondary metabolites as leads in drug development, Drug Discovery Today, 1998, vol. 3, no. 5, pp. 232–238. https://doi.org/10.1016/S1359-6446(97)01167-7
Von Arx, J.A. and Schipper, M.A.A., The CBS fungus collection, Advances in Аpplied Microbiology, Academic Press, 1978, vol. 24, pp. 215–236. https://doi.org/10.1016/S0065-2164(08)70641-5
Wang, W., Liao, Y., Zhang, B., Gao, M., Ke, W., Li, F., and Shao, Z., Citrinin monomer and dimer derivatives with antibacterial and cytotoxic activities isolated from the deep sea-derived fungus Penicillium citrinum NLG-S01-P1, Marine Drugs, 2019, vol. 17, no. 1, p. 46. https://doi.org/10.3390/md17010046
White, T.J., Bruns, T., Lee, S.J.W.T., and Taylor, J., Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, in PCR Protocols: A Guide to Methods and Applications, 1990, vol. 18, no. 1, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Wilson, D., Endophyte: the evolution of a term, and clarification of its use and definition, Oikos, 1995, pp. 274-276. https://doi.org/10.2307/3545919
ACKNOWLEDGMENTS
The authors are highly thankful to the Head and Coordinator, CAS, DST-FIST in Botany, BHU, Varanasi, for providing the facilities to carry out research work. The authors acknowledge IoE-BHU, CSIR, and UGC for financial support. SKV thanks DBT for the financial support as project (P07/1265). Authors express thanks to Prof. Gopal Nath (Institute of Medical Sciences, BHU) for providing human pathogens and antibacterial activity. Authors also acknowledge the help of Dr. M.K. Bharty and Dr. K. Kumar, Department of Chemistry, BHU for solving the structure of compound.
Funding
This work was supported by ongoing institutional funding. No additional grants to carry out or direct this particular research were obtained.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human and animal subjects.
Additional information
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Verma, A., Kumar, K., Talukdar, U. et al. Assessment of Bioactive Potential and Characterization of an Anticancer Compound from the Endophytic Fungi of Ocimum sanctum. Microbiology 93, 459–471 (2024). https://doi.org/10.1134/S002626172360252X
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S002626172360252X