Abstract—
A facultative methylotroph, strain F5Т, which uses methylamine and a broad spectrum of polycarbon substrates as carbon and energy sources, was isolated from silt of a freshwater lake in the southern Moscow region. The cells were gram-negative, coccoid, non-spore-forming, nonmotile, colorless, reproducing by binary fission and possessing a capsule. The organism was mesophilic, neutrophilic, not halophilic, oxidase- and catalase-positive, and capable of nitrate reduction to nitrite. Methylamine was oxidized by amine dehydrogenase and via the icl- serine pathway of С1 metabolism, as was indicated by activities of hydroxypyruvate reductase and serine-glyoxylate aminotransferase and by the absence of hexulosephosphate synthase and ribulose bisphosphate carboxylase. Predominant fatty acids were C18:1ω7c (72.3%) and C16:0 (11.6%). Phosphatidylcholine, phosphatidylglycerol, diphosphatidylglycerol, and phosphatidylethanolamine were the dominant phospholipids. The G + C DNA content was 65.8 mol % (Tm). Q10 was the dominant ubiquinone. Strain F5T exhibited high similarity of the 16S rRNA gene sequences with Paracoccus strains: P. aminovorans JCM7685T = VKM B-2140Т (98.0%), P. huijuniae FLN-7Т (97.9%), and P. limosus NB88T (97.5%). However, the level of DNA-DNA relatedness between the strains F5Т and P. aminovoransТ was only 21 ± 3%. Based on the data obtained, strain F5Т was identified as a new Paracoccus species with proposed name Paracoccus simplex sp. nov. (= VKM B-3226T = CCUG 71989T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The genus Paracoccus belongs to the family Rhodobacteraceae, the order Rhodobacterales, the class Alp-haproteobacteria, and was originally described by Davis et al. (1969); the description was subsequently emended (Katayama et al., 1995; Liu et al., 2008). The new information was generalized and presented in the chapters of the 3rd and 4th editions of “Prokaryotes” (Kelly et al., 2006a; 2006b; Pujalte et al., 2014). At present, the genus includes 48 species. Paracocci are gram-negative cocci or short rods, mostly nonmotile, although the members possessing one polar flagellum (P. homiensis DD-R11T and P. versutus ATCC 25364T) and peritrichous flagellation (P. carotinofaciens E‑396T) have been described. Reproduction occurs by binary fission; the colonies of some species are orange due to carotenoid production. The organisms are catalase- and oxidase-positive, the predominant ubiquinone is Q10, although Q8 is predominant in P. yeei CDC G1212T. Some species are halophilic or halotolerant. Paracocci are aerobic chemoorganotrophs, but some species can utilize nitrate, reducing it to nitrite or molecular nitrogen. The type species Paracoccus denitrificans can grow chemolithoautotrophically, using reduced sulfur compounds as substrates or oxidizing hydrogen. Some Paracoccus species are facultative methylotrophs and utilize methanol or methylamine as a carbon and energy source but use different C1-metabolic pathways. In the case of autotrophic growth, CO2 fixation occurs via the Calvin cycle, and such Paracoccus species possess ribulose bisphosphate carboxylase (RuBisCO); other species are devoid of RuBisCO and use the serine pathway of C1 assimilation (P. aminophilus). Methylamine oxidation is catalyzed by methylamine dehydrogenase or occurs via the N-methylglutamate pathway. Thus, different Paracoccus species vary substantially in their physiological and biochemical properties.
The work was aimed at physiological and biochemical characterization and identification of a novel isolate of facultative methylotrophic bacteria, strain F5Т, attributed to the genus Paracoccus.
MATERIALS AND METHODS
Research subjects. Strain F5Т (VKM B-3226Т = CCUG 71989T) was isolated from coast silt of a small freshwater lake in the vicinity of the town of Pushchino (Moscow oblast, Russia). The silt (3 g) was placed into a 750-mL Erlenmeyer flask with 200 mL of K medium with methylamine hydrochloride 0.3% (wt/vol) and incubated at 28°С, 180 rpm, for 3 days. Enrichment and pure cultures were obtained as described (Doronina et al., 2013). Culture purity was tested by light (Nikon Eclipse Ci, Japan) and electron (JEOL JEM-100B, JEOL, Japan) microscopy, as well as by the uniformity of colonies on agar media with methylamine and glucose/peptone. The studies of morphology, chemotaxonomic, physiological, biochemical, and cultural properties, DNA isolation, 16S rRNA gene sequencing, phylogenetic analysis, determination of DNA G + C content and DNA–DNA hybridization were performed as described previously (Doronina et al., 2013). The K medium contained the following (g/L): KH2PO4, 2.0; (NH4)2SO4, 2.0; NaCl, 0.5; MgSO4 · 7H2O, 0.125; FeSO4 · 7H2O, 0.002; pH 7.4. Cultivation was also carried out on R2A, TSA, and LB media. Paracoccus aminovorans JCM7685T = VKM B-2140Т was used as a reference culture.
Cell extracts and enzyme activities. The cells were harvested in the exponential phase by centrifugation at 6000 g, washed twice with 50 mM Tris-HCl buffer (pH 7.4), and resuspended in the same buffer. The cells were homogenized on a Sonicator S-4000 ultrasonic disintegrator (6 × 1 min, with 1-min breaks) under cooling in ice. Intact cells were separated by centrifugation (14 000 rpm for 30 min, 4°С). Supernatants were used for enzymological analysis.
Amine oxidase was assayed by hydrogen peroxide production (Haywood and Large, 1981). The reaction mixture (2 mL) contained: Tris-HCl buffer, pH 7.5, 150 µmol; 10 U horseradish peroxidase, 1 µmol o‑Dianisidine, 16 µmol methylamine, and the extract. The reaction was started by adding methylamine; the changes in extinction were recorded at 460 nm.
Methylamine dehydrogenase was assayed by DCPIP reduction in the presence of PMS (Meiberg and Harder, 1978). The reaction mixture (2 mL) contained (µmol): K-phosphate buffer, pH 7.5, 150; DCPIP, 0.8; PMS, 4; KCN, 2, methylamine, 7; and the extract. The reaction was started by adding methylamine, and the rate of the changes in extinction was recorded at 600 nm.
N-methylglutamate dehydrogenase was assayed by formaldehyde production measured with the Nash reagent (Nash, 1953). The reaction mixture (2 mL) contained (µmol): K-phosphate buffer, pH 7.5, 100; NAD+, 0.25; and the extract. The reaction was started by adding sodium N-methyl-L-glutamate (5 µmol).
Formaldehyde and formate dehydrogenases were assayed by DCPIP reduction (Johnson and Quayle, 1964). The reaction mixture (2 mL) contained (µmol): K-phosphate buffer, pH 7.5, 50; DCPIP, 0.075; PMS, 0.5; formaldehyde, 10, or formate, 50; and the extract. The reaction was started by adding the substrate.
HAD+-dependent formaldehyde dehydrogenase was assayed by NAD+ reduction at 340 nm (Johnson and Quayle, 1964). The reaction mixture (2 mL) contained (µmol): K-phosphate buffer, pH 7.0, 50; NAD+, 0.25; reduced glutathione, GSH (or without it), 10; formaldehyde, 2; and the extract. The reaction was started by adding formaldehyde.
NAD+-dependent formate dehydrogenase was assayed by NAD+ reduction at 340 nm (Johnson and Quayle, 1964). The reaction mixture (2 mL) contained (µmol): Tris-HCl buffer, pH 7.5, 50; HAD, 0.25; formate, 50; and the extract. The reaction was started by adding formate.
Hydroxypyruvate reductase was assayed by NAD(P)H oxidation (Blackmore and Quayle, 1970). The reaction mixture (2 mL) contained (µmol): Tris-HCl buffer, pH 7.4, 100; NAD(P)H, 0.5; and the extract. The reaction was started by adding 5 µmol of sodium hydroxypyruvate. In the presence of NADH reductase, the result was corrected for NADH oxidation by the extract without hydroxypyruvate.
L-serine glyoxylate aminotransferase was assayed spectrophotometrically by recording the glyoxylate-dependent formation of hydroxypyruvate from L-serine (Blackmore and Quayle, 1970). The reaction mixture (2 mL) contained (µmol): Tris-HCl buffer, pH 7.5, 100; pyridoxal phosphate, 0.02; NADH, 0.5; glyoxylate, 10; and the extract. The reaction was started by adding 10 µmol of L-serine. The appropriate correction was made for the result in the presence of glyoxylate reductase activity in the extracts.
Ribulose bisphosphate carboxylase was assayed by the radioisotope method by monitoring the dynamics of NaH14CO3 inclusion into the acid-resistant product. The reaction mixture (1 mL) contained (µmol): Tris-HCl buffer, pH 7.6, 100; MgCl2, 2.5; reduced glutathione (GSH), 10; ribulose-1,5-bisphosphatte (sodium salt), 0.25; and the extract; the reaction was started by adding 18 µmol NaH14CO3 (20 µCu). The reaction was carried out at 30°С; 0.02-mL samples were taken during 20 min with 2-min intervals, applied to Whatman GF/F glass microfiber paper squares, fixed with 0.05 mL of 6 N HCl, and dried; then the radioactivity was measured.
Hexulose phosphate synthase was assayed spectrophotometrically (Ferenci et al., 1974) by NADP+ reduction in the glucose-6-phosphate dehydrogenase- and glucose phosphate isomerase-mediated reaction. The reaction mixture (2 mL) contained (µmol): K‑phosphate buffer, pH 7.0; MgCl2, 8; NADP+, 0.5; rabbit muscle phosphoglucose isomerase, 1.68 µmolar units; glucose-6-phosphate dehydrogenase (type 7, Sigma), 0.15 µmolar units; ribose-5-phosphate; and the extract. The reaction was started by adding 5 µmol of formaldehyde.
Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase were assayed by NADP+ or NAD+ reduction (Korndberg and Horecker, 1955). The reaction mixture (2 mL) contained (µmol): K‑phosphate buffer, pH 7.5, 150; MgCl2, 10; NAD(P)+, 1.0; glucose-6-phosphate sodium salt or 6‑phosphogluconate sodium salt, 10; and the extract. The reaction was started by adding the appropriate substrate.
Fructose-1,6-bisphosphate aldolase was assayed with the coupling enzyme glycerophosphate dehydrogenase (van Dijken and Quayle, 1977). The reaction mixture (2 mL) contained (µmol): Tris-HCl buffer, pH 7.5, 100; CoCl2, 2; NADH, 0.5; glycerophosphate dehydrogenase, 0.36 U; and the extract. The reaction was started by adding 2 µmol of fructose-1,6-bisphosphate.
2-Keto-3-deoxy-6-phospohgluconate aldolase was assayed with the coupling enzyme lactate dehydrogenase (Wood, 1971). The reaction mixture (2 mL) contained (µmol): imidazole buffer, pH 8.0, 50; 6-phosphogluconate (sodium salt), 10; NADH, 0.5; MgCl2, 5; dithiotreitol, 2; lactate dehydrogenase from pig muscle (Reanal), 5 U; and the extract. The reaction was started by adding 6-phosphogluconate. The control was the reaction mixture without 6-phosphogluconate.
Isocitrate lyase was assayed spectrophotometrically by glyoxylate phenylhydrazone formation at 324 nm (Dixon and Kornberg, 1959). The reaction mixture (2 mL) contained (µmol): K-phosphate buffer, pH 6.8, 150; MgCl2, 10; phenylhydrazine, 6.5; cysteine-HCl, 4; and the extract. The reaction was started by adding 5 µmol of potassium isocitrate.
Glutamate dehydrogenase was assayed in the reaction mixture (2 mL) containing (µmol): Tris-HCl buffer, pH 7.5, 100; NH4Cl, 80; NAD(P)H, 0.5; and the extract. The reaction was started by adding 10 µmol of α-ketoglutarate.
Glutamate synthase was assayed by NAD(P)H oxidation (Meers and Tempest, 1970). The reaction mixture (2 mL) contained (µmol): Tris-HCl buffer, pH 7.6, 100; α-ketoglutarate, 10; NADH or NADPH, 0.25; and the extract. The reaction was started by adding 25 µmol of glutamine.
Glutamine synthetase was assayed by the modified colorimetric method (Elliott, 1955) in the γ-glutamine transferase reaction. The reaction mixture (7.52 mL) contained (µmol): imidazole buffer, pH 7.15; MgCl2, 0.27; sodium arsenate, 12.5; ADP, 0.18; hydroxylamine, 20; and the extract. The reaction was started by adding 20 µmol of L-glutamine; the mixture was incubated for 10 min at 30°С, followed by the addition of 0.6 mL of solution containing (g/L): 55 g of FeCl3, 20 g of trichloroacetic acid, and 21 mL of HCl. The mixture without L-glutamine was used as a control. The formed precipitate was removed by centrifugation and the absorption was measured at 540 nm. The value of 0.63 absorbance unit corresponded to 1 µmol/mL of γ-glutamyl hydroxamate (the reaction product).
Optical measurements were performed on a Shimadzu UV-1700 spectrophotometer (Japan) in a thermostated cuvette at 30°С. Enzyme activities were expressed as a number of nanomoles of transformed substrate or formed product in 1 min per 1 mg of protein. Specific activity was assayed with the amount of protein providing linear dependence of the rate on the enzyme concentration. The calculations were made with the following molar extinction coefficients, µmol–1 cm–1: NAD(P)H (at 340 nm), 6.22; DCPIP (at 600 nm), 21.9; o-Dianisidine, (at 460 nm) 28.8. Radiometric measurements were made with a LS6500 Multi-Purpose Scintillating Counter (Beckman Coulter, United States) in the mixture containing 4 g of 2,5-diphenyloxazole (PPO) and 0.05 g of 1,3-bis(5-phenyloxazole-2yl)benzene (POPOP) in 1 L of toluene.
Protein assay was performed by the Lowry method (Lowry et al., 1951) using Sigma reagents (United States).
RESULTS AND DISCUSSION
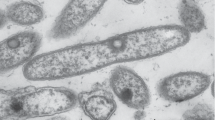
Isolate morphology. The cells of strain F5Т were gram-negative, nonmotile, coccoid, 0.4–0.7 × 0.5–0.9 µm (Figs. 1a, 1b), with a capsule. Did not form spores and pigment, reproduced by binary fission. The colonies on R2A agar medium were circular, white, glossy, transparent, 2–3 mm in diameter (day 3, 29°С), with wavy edges and viscous consistency.
Cultural, physiological, biochemical, and chemotaxonomic properties. Strain F5Т grew in the liquid K medium with methylamine without cell aggregation, did not form pigment. It was aerobic, catalase- and oxidase-positive, needed no vitamins or other growth factors. Grew in the temperature range from 17 to 37°С (optimal 28–30°С) and pH 5.5–8.0 (optimal pH 7.0–7.5). Used the following broad range of substrates as the carbon and energy sources: methylamine, trimethylamine, ethanol, glucose, galactose, fructose, maltose, mannose, trehalose, ribose, sucrose, succinate, pyruvate, α-ketoglutarate, malate, acetate, betaine, alanine, serine, valine, inositol, mannitol, acetamide, and glucuronic acid. No growth occurred on methanol, dimethylamine, formate, dimethyl sulfoxide, dimethylformamide, dichloromethane, arabinose, lactose, xylose, glutamate, aspartate, and in the H2/CO2/O2 gas mixture. Ammonium and nitrates were used as nitrogen sources. The isolate was capable of nitrate reduction. Starch was hydrolyzed. Gelatinase was not present. The optimal NaCl concentration in the medium was 0.05%; growth was observed up to 2.5% NaCl and was completely absent at 3.5% NaCl.
The strain was sensitive to the following antibiotics (µg/disc): ampicillin (10), gentamycin (10), lincomycin (2), nalidixic acid (30), novobiocin (30) and streptomycin (10). Resistant to kanamycin (30), neomycin (30) and erythromycin (15). The fatty acid composition of the cells included C18:1ω7c (72.3%), C16:0 (11.6%), C18:0 (3.2%), and C19:0cyc (1.9%) fatty acids. The predominant phospholipids were phosphatidylcholine, phosphatidylglycerol, diphosphatidylglycerol and phosphatidylethanolamine (Fig. 2). The predominant ubiquinone was Q10.
Two-dimensional chromatography of the polar lipids of strain Paracoccus simplex F5T. Direction 1, chloroform–methanol–water (65 : 25 : 4); direction 2, chloroform–methanol–acetic acid–water (85 : 12 : 15 : 4). The assay of amino-containing lipids with 0.2% ninhydrin in acetone (a); detection of phosphorus-containing lipids with molybdenum blue (b). Designations: phosphatidylcholine (PC), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), diphosphatidylethanolamine (PDE), diphosphatidylglycerol (DPG), unidentified phospholipids (PL1–4), unidentified amino lipids (AL).
Metabolic characteristics. The results of enzymological analysis of the cells grown on methylamine are presented in Table 1. It was established that strain F5Т had a high methylamine dehydrogenase activity demonstrating the activity with phenazine methosulfate (PMS). The product of this reaction (formaldehyde) was oxidized to formate by NAD+-glutathione-dependent dehydrogenase and then to CO2 by NAD+-dependent formate dehydrogenase. Amine oxidase and N-methylglutamate dehydrogenase were absent.
Hydroxypyruvate reductase and serine-glyoxylate aminotransferase activities were detected. 3-Hexulose phosphate synthase and ribulose-1,5-bisphosphate carboxylase activities were not detected. Isocitrate lyase activity was very low. This enzyme was probably not associated with C1 metabolism. Consequently, strain F5Т implementd the isocitrate lyase-negative (icl–) variant of the serine pathway. Isocitrate dehydrogenase was NADP+-dependent. Primary involvement of ammonium nitrogen was implemented via the glutamate pathway and by glutamate dehydrogenase. The enzyme activities of carbohydrate metabolism were very low.
Phylogenetic analysis. The 16S rRNA gene sequencing of the studied strain F5Т (1405 bp) showed the high level of similarity with representatives of the genus Paracoccus: 98.0% with P. aminovorans JCM 7685Т (D32240), 97.9% with P. huijuniae FLN-7Т (EU725799), and 97.5% with P. limosus NB88Т (HQ336256) (Fig. 3). The DNA G + C content in strain F5Т was 65.8 mol %. The level of DNA–DNA homology between strain F5Т and P. aminovorans JCM 7685Т was only 21 ± 3%. Strain F5Т was also phylogenetically characterized using mauA, the gene encoding the small subunit of methylamine dehydrogenase. The phylogenetic position of strain F5Т based on the comparison of MauA amino acid sequences is shown on Fig. 4. Strain F5Т had the maximum similarity (96–100%) in the MauA protein with representatives of the genus Paracoccus and some bacteria of other genera: Methylobacterium (90–92%), Methylopila (89–91%), and Methylophaga (77–78%).
The phylogenetic position of strain F5Т based on the results of comparative analysis of the 16S rRNA gene sequences. The scale corresponds to 10 nucleotide substitutions per every 100 nucleotides (evolutionary distance). The neighbor-joining method was used. The root was determined by including the Rhodobacter capsulatus ATCC 11166T (D16428) sequence as an outgroup.
The phylogenetic position of the strain F5Т based on comparison of amino acid sequences of the MauA protein. The scale corresponds to 10 amino acid substitutions per every 100 amino acids (evolutionary distance). The root was determined by including the sequence of Methylacidiphilum infernorum V4 (YP_001940651) as an outgroup. The numbers show the statistical confidence of branching order determined by the “bootstrap” analysis of 100 alternative trees.
The results of polyphasic analysis indicated that strain F5Т was a novel species of the genus Paracoccus, with the proposed name of Paracoccus simplex sp. nov.
The differentiating characteristics of P. simplex sp. nov. and of the phylogenetically closest members of the genus Paracoccus are given in Table 2.
P. simplex sp. nov., in contrast to P. aminovorans JCM 7685Т, has a tryptophan deaminase, utilizes the broader range of polycarbon substrates, and does not grow on dimethylamine and dimethylformamide; in contrast to P. huijuniae FLN-7Т, it cannot grow at 42°С and utilize methanol. The studied strain F5Т differs from P. limosus NB88Т in gluconate and maltose utilization. Moreover, unlike the latter two species, it cannot grow at 4% NaCl in the medium.
Description of Paracoccus simplex sp. nov. Paracoccus simplex sp. nov. (sim’plex. L. adj. simplex, simple). The cells are gram-negative, non-motile, non-spore-forming rods 0.4–0.7 × 0.5–0.9 μm, capsule-forming, reproducing by binary fission. The colonies on the R2A medium are round, white, glossy, transparent, 2–3 mm in diameter with a wavy edge and a viscous consistency. Grows at a temperature of 17 to 37°C with an optimum at 28–30°C and at pH 5.5 to 8.0, optimally at pH 7.0–7.5. Does not grow at 3.5% NaCl in the medium. Grows on R2A, TSA, and LB agar. Obligate aerobic bacterium. Does not grow under anaerobic conditions and does not use nitrate as an electron acceptor. Oxidase and catalase positive, urease-negative. Nitrates are reduced to nitrites. Negative for β‑galactosidase, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, but positive for tryptophandeamine, produces indole and acetoin. Grows on methylamine, trimethylamine, ethanol, glucose, galactose, fructose, maltose, mannose, trehalose, ribose, sucrose, succinate, pyruvate, α-ketoglutarate, malate, acetate, betaine, alanine, serine, valine, inositol, mannitol, acetamide, glucuronic acid. Does not grow on methanol, dimethylamine, formate, dimethyl sulfoxide, dimethylformamide, dichloromethane, arabinose, lactose, xylose, glutamate, aspartate, and also in the H2/CO2/O2 atmosphere. No vitamins or other growth factors are required. Implements the isocitrate lyase-negative (icl–) variant of the serine pathway. The dominant respiratory quinone is Q10. In the fatty acid composition of cells, more than 70% is C18:1ω7c. There are also C16:0 (11.6%), C18:0 (3.2%) and C19cyc (1.9%) acids. Phospholipid composition of the cells includes phosphatidylcholine, phosphatidylglycerol, diphosphatidylglycerol, and phosphatidylethanolamine. The DNA G + C content of the type strain F5T is 65.8 mol %. The 16S rRNA and mauA genes sequences of the strain F5T were deposited in the GenBank/EMBL/DDBL under accession numbers MG938051 and MH059568, respectively. The type strain, F5T (= VKM B-3226T = CCUG 71989T), was isolated from a coastal freshwater lake in the vicinity of Pushchino, Moscow Region (Russia).
REFERENCES
Blackmore, M.A. and Quayle, J.R., Microbial growth on oxalate by a route not involving glyoxylate carboligase, Biochem. J., 1970, vol. 118, pp. 53–59.
Davis, D.H., Doudoroff, M., Stanier, R.Y., and Mandel, M., Proposal to reject the genus Hydrogenomonas: taxonomic implications, Int. J. Syst. Evol. Microbiol., 1969, vol. 19, pp. 375–390.
Dixon, G.H. and Kornberg, H.L., Assay methods for key enzymes of the glyoxylate cycle, Proc. Biochem. Soc., 1959, p. 3.
Doronina, N.V., Kaparullina, E.N., and Trotsenko, Y.A. Methylopila musalis sp. nov., an aerobic facultatively methylotrophic bacterium isolated from banana fruit, Int. J. Syst. Evol. Microbiol., 2013, vol. 63, pp. 1847–1852.
Elliott, W.H., Glutamine synthesis: glutamic acid + ATP + ammonia → glutamine + ADP + phosphate. Methods in Enzymology, Colowick, S.P. and Kaplan, N.O., Eds., New York: Academic, 1955, vol. 2, pp. 337−342.
Ferenci, T., Strom, T., and Quayle, J.R., Purification and properties of 3-hexulose phosphate synthase and phospho-3-hexuloisomerase from Methylococcus capsulatus, Biochem. J., 1974, vol. 144, pp. 477–486.
Haywood, G.W. and Large, P.J., Microbial oxidation of amines, Biochem. J., 1981, vol. 199, pp. 187–201.
Johnson, P.A. and Quayle, J.R., Microbial growth on C1-compounds. Oxidation of methanol, formaldehyde and formate by methanol-grown Pseudomonas AM1, Biochem. J., 1964, vol. 93, pp. 281–290.
Katayama, Y., Hiraish, A., and Kuraishi, H., Paracoccus thiocyanatus sp. nov., a new species of thiocyanate-utilizing facultative chemolithotroph, and transfer of Thiobacillus versutus to the genus Paracoccus as Paracoccus versutus comb. nov. with emendation of the genus, Microbiology (UK), 1995, vol. 141, pp. 1469–1477.
Kelly, D., Rainey, F., and Wood, A., The Genus Paracoccus, in The Prokaryotes, vol. 5, 3rd edn, Proteobacteria— alpha and beta subclasses, Rosenberg, E., Stackebrandt, E., Thompson, F., Lory, S., and DeLong, E., Eds., Berlin: Springer, 2006a, pp. 232–249.
Kelly, D.P., Euzéby, J.P., Goodhew, C.F., and Wood, A.P., Redefining Paracoccus denitrificans and Paracoccus pantotrophus and the case for a reassessment of the strains held by international culture collections, Int. J. Syst. Evol. Microbiol., 2006b, vol. 56, pp. 2495–2500.
Kornberg, A. and Horecker, B.L., Glucose-6-phosphate dehydrogenase, Methods in Enzymology, 1955, vol. 1, pp. 323−327.
Lee, M.J. and Lee, S.S., Paracoccus limosus sp. nov., isolated from activated sludge in a sewage treatment plant, Int. J. Syst. Evol. Microbiol., 2013, vol. 63, pp. 1311–1316.
Liu, Z.P., Wang, B.J., Liu, X.Y., Dai, X., Liu, Y.H., and Liu, S.J., Paracoccus halophilus sp. nov., isolated from marine sediment of the South China Sea, China, and emended description of the genus Paracoccus Davis 1969, Int. J. Syst. Evol. Microbiol., 2008, vol. 58, pp. 257–261.
Lowry, O.H., Rosebrough, N.J., Farr, A.L., and Randall, R.J., Protein measurement with the Folin phenol reagent, J. Biol. Chem., 1951, vol. 193, pp. 265–275.
Meers, J.L., Tempest, D.W., and Brown, C.M., Glutamine (amide): α-oxoglutarate aminotransferase oxidoreductase (NADP) an enzyme involved in biosynthesis of glutamate by some bacteria, J. Gen. Microbiol., 1970, vol. 64, pp. 187–194.
Meiberg, J.B.M. and Harder, W., Aerobic and anaerobic metabolism of trimethylamine, dimethylamine and methylamine in Hyphomicrobium sp. X, J. Gen. Microbiol., 1978, vol. 106, pp. 265–276.
Nash, T., The colorimetric estimation of formaldehyde by means of the Hantzsch reaction, Biochem. J., 1953, vol. 55, pp. 416–421.
Pujalte, M.J., Lucena, T., Ruvira, M.A., Arahal, D.R., and Macián, M.C., The Paracoccus Group, in The Prokaryotes—Alphaproteobacteria and Betaproteobacteria, 4rd edn., Rosenberg, E., DeLong, E., Lory, S., Stacke-brandt, E., and Thompson, F., Eds., Berlin: Springer, 2014, p. 460.
Sun, L.N., Zhang, J., Kwon, S.W., He, J., Zhou, S.G., and Li, S.P., Paracoccus huijuniae sp. nov., an amide pesticide-degrading bacterium isolated from activated sludge of a wastewater biotreatment system, Int. J. Syst. Evol. Microbiol., 2013, vol. 63, pp. 1132–1137.
Urakami, T., Araki, H., Oyanagi, H., Suzuki, K.I., and Komagata, K., Paracoccus aminophilus sp. nov. and Paracoccus aminovorans sp. nov., which utilize N,N-dimethylformamide, Int. J. Syst. Bacteriol., 1990, vol. 40, pp. 287–291.
Van Dijken, J.P. and Quayle, J.R., Fructose metabolism in four Pseudomonas species, Arch. Microbiol., 1977, vol. 114, pp. 281–286.
Wood, W.A., Assay of enzymes representative of metabolic pathways, Methods Microbiol., 1971, vol. 6A, p. 421.
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by E. Makeeva
Rights and permissions
About this article
Cite this article
Doronina, N.V., Kaparullina, E.N., Chemodurova, A.A. et al. Paracoccus simplex sp. nov., a New Methylamine-Utilizing Facultative Methylotroph. Microbiology 87, 662–671 (2018). https://doi.org/10.1134/S0026261718050077
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261718050077