Abstract
Combating pathological conditions related to hyperactivity of enzymes remains a formidable challenge for health. Small molecules therapy constitutes one of the means to circumvent the medical disorders resulting from enzyme hyperactivity. In this regard, we have synthesized structurally diverse amino acid hybrid Schiff bases (5a–5l and 10a–10k) and evaluated them for carbonic anhydrase II, α-glucosidase, and urease inhibitory potential. These new chemical scaffolds showed variable efficacies against the selected enzymes. The results indicated that compounds 5b (11.8 ± 1.33 µM), 10i (83.3 ± 1.13 µM), and 10f (88.2 ± 2.27 µM) are the most active scaffolds against carbonic anhydrase II, α-glucosidase, and urease, respectively. A structure–activity relationship revealed the most structural features contributing to the overall activities. Molecular docking suggested that these compounds possess excellent binding interactions with the active site residues of the targets by interacting through hydrogen bonding, π–π, and π–cation interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amino acids play an important role for the establishment of innovative drugs using nonproteogenic and unnatural amino acids [1,2,3,4,5,6,7,8]. They are “building blocks of life” and have important biochemical and medicinal applications [9]. Proteogenic amino acid showed physical properties are governed by the structure and nature of their side chains: polar (hydrophilic) vs. nonpolar (hydrophobic); neutral, acidic, or basic; aromatic vs. nonaromatic; essential vs. nonessential; and achiral vs. chiral. For the construction of all peptides and proteins in living system, 20 amino acids are used [10].
L-β-phenylalanine and L-leucine are the amino acids, which have capability to make different Schiff bases. Schiff bases are mostly known as imines or azomethines and act as ligands in various metal complexes. Hugo Schiff, a German chemist, was the first to synthesize Schiff bases in 1864 by condensing primary amines with carbonyl compounds [11]. These biologically dynamic Schiff’s bases and their derivatives are recognized due to their biological activities, mainly as antimicrobial agents [12,13,14,15], antivirus agents [16], antioxidants [17], radical inhibitors [18], antitumor agents [19, 20], carbonic anhydrase (CA) inhibitors [21], xanthine oxidase inhibitors [22], antibacterial [21, 23, 24], plant-growth regulators [25], free radical scavengers [26], trypsin inhibitors [27], inhibitors of cartilage matrix degeneration [28], 5-HT6 antagonists [29], and anti-inflammatory agents [29, 30].
CAs (EC 4.2.1.1) are zinc enzymes strongly involved in regulating cell homeostasis, intracellular pH, fluid secretion, ion transport, and biosynthetic reactions by catalyzing the reversible hydration of carbon dioxide (CO2) to bicarbonate ion (HCO3−) and proton (H+) [31,32,33,34,35,36,37]. These enzymes are common in almost all organisms, from simple to complex [38, 39]. Notably, in tumors, the extracellular pH is more acidic than intracellular pH [40]. In order to maintain the pH, tumor cells generate ion transportations via CA enzymes and ionic channels in between extra and intracellular compartments [40,41,42]. In brain malignant, tumor cells express CA II [43,44,45], similar to that of renal cancer cell line and pancreatic cell carcinomas [43, 45]. With that, CA II has also been involved in glaucoma, epilepsy, leukemia, and cystic fibrosis [46, 47]. Due to the significant role of CAs in life threating diseases, it is important to inhibit the activity of this enzyme to obtain a therapy.
On the other hand, Schiff bases, an important class of compounds, having azomethine (C=N) functional group are known for urease inhibition and are extensively used as mixture with crop fertilizers to inhibit the urea decomposition by soil bacteria [48, 49]. The therapeutic demand of urease inhibition is very important as it has a potential role in curing diseases such as peptic ulcer, urolithiasis, pyelonephritis, and hepatic coma caused by ureolytic bacteria e.g., Helicobacter pylori, Yersinia enterocolitica, and Proteus mirabilis. Urease facilitates the colonial growth of H. pylori by elevating the stomach local pH thereby allowing the bacteria H. pylori to cause gastric infection such as peptic ulcers, gastric lesion, and intestinal cancer [50, 51].
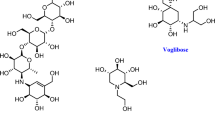
α-Glucosidase enzyme catalyzes the hydrolysis of α-glucosidal bond of complex carbohydrates and releases the monosaccharide (α-D-glucose), which is absorbable in small intestine [52, 53]. Hyperactivity of α-glucosidase enzyme causes elevation in blood glucose in diabetic patients known as hyperglycemia [17]. Inhibition of α-glucosidase enzyme by Schiff bases is one way to treat type II diabetes mellitus by suspending the absorption of glucose in intestine [54]. Acarbose, miglitol, and voglibose are the drugs which are being clinically used for the treatment of type II diabetes mellitus.
The current study aims to explore CA II, α-glucosidase, and urease the inhibition for this purpose, compounds (5a–5l and 10a–10k) were synthesized from two amino acids (L-β-phenylalanine and L-leucine). The newly synthesized heteroaromatic Schiff bases were evaluated against CA II, α-glucosidase, and urease inhibition to check their potencies. In addition, the in vitro enzyme inhibitory results were further confirmed with the aid of molecular docking studies.
Results and discussion
Chemistry
Different amino acid Schiff bases (5a–5l and 10a–10k) were synthesized through esterification of L-β-phenylalanine (1a) and L-leucine (6a) in methanol (MeOH) by adding thionyl chloride, followed by protection of the amino group as tert-butyloxycarbonyl (Boc). The resulting hydrazides (4a and 9a) were then reacted with different benzaldehyde derivatives to furnish different L-α-phenylalanine hydrazones and L-leucine hydrazones derivatives (Schemes 1 and 2; Table 1). The structures of the resulting compounds were confirmed using different spectroscopic techniques.
Enzyme inhibition
The newly synthesized amino acid assembled hybrid structures (5a–5l and 10a–10k) were evaluated against CA II, α-glucosidase, and urease inhibitory activity. The in vitro studies results are summarized in Table 1. The results demonstrate a clear variation in the inhibitory activities ranging from 11.8 to 122.2 µM, 83.3 to 367.8 µM, and 88.2 to 427.3 µM against CA II, α-glucosidase, and urease enzymes, respectively. The structural–activity relationship of the compounds was investigated on the basis of attachment of the functional (electron withdrawing and donating) groups at different positions of phenyl ring of R group.
Structure–activity relationship of CA II enzyme
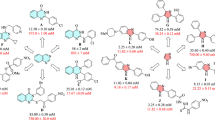
In case of phenylalanine hydrazine derivatives, eight compounds were found active against CA II. Among the active compounds, 5b was the most potent with IC50 value of 11.8 µM, followed by 5d (IC50 = 13.4 µM) and 5c (IC50 = 15.8 µM) even superior than standard acetazolamide (IC50 = 18.2 µM), whilst compounds 5a (IC50 = 30.1 µM), 5e (IC50 = 41.0 µM), and 5i (IC50 = 37.2 µM) showed promising inhibition comparable to standard acetazolamide. Compounds 5g and 5h showed weak activities, while rest of compounds found to be inactive (Table 1). The higher activity of compound 5b could be attributed to the presence of hydroxyl group at ortho position of phenyl ring. When comparing 5b with 5c, the decrease in inhibition is due to the position of methoxy group at para position of the phenyl ring. A variation in activities between 5c and 5d is observed. The higher inhibition of 5d can be accounted for the arrangements of functional groups attached to phenyl ring. The higher activity of 5d than 5i (IC50 = 37.2 µM) and 5e (IC50 = 41.0 µM) is likely due to the replacement of hydroxyl group with methoxy group at meta and para positions of the phenyl ring. Similarly, a higher inhibition of compound 5a (IC50 = 30.1 µM) is seen when compared with compounds 5j‒5l; a fact that can be attributed to the replacement of pyridine ring with phenyl ring.
Similarly, among the leucine hydrazones, compound 10a has displayed an excellent inhibitory activity having an IC50 value of 12.5 µM), followed by 10b (IC50 = 20.5 µM) and 10g (IC50 = 49.8 µM), while 10h (IC50 = 82.1 µM) and 10d (IC50 = 106.5 µM) exhibited weak activities when compared with standard acetazolamide (IC50 = 18.2 µM) (Table 1). Compound 10a (IC50 = 12.5 µM) exerted a stronger inhibition than other compounds in the series, which is possibly due to the presence of pyridine ring. When comparing compounds 10b and 10g, the higher activity of 10b is likely due to the presence of hydroxyl group at ortho position of the phenyl ring. A comparison between 10d with 10g revealed a decrease in inhibition of 10d, which is possibly by the replacement of hydroxyl group with methoxy group attached at the para position of the phenyl ring. Finally, compound 10h possesses better inhibition when compared with 10d due to the presence of methoxy group at the meta position of the phenyl ring.
Structure–activity relationship of α-glucosidase enzyme
In the preliminary screening of phenylalanine hydrazones against α-glucosidase, six compounds were found to be more active than standard acarbose. The compound 5k has demonstrated the highest activity with an IC50 value of 87.1 µM, followed by 5c, 5l, and 5g with IC50 values of 166.7, 167.8 and 176.7 µM, respectively (Table 1). However, compounds 5e (IC50 = 270.7 µM) and 5i (IC50 = 317.4 µM) exhibited weak inhibition when compared with standard acarbose (IC50 = 942 µM). Compound 5k was found to be the most active compound of the series, about 11 times better than the standard. This enhanced activity is likely due to the presence of nitro group as electron-withdrawing substituent at ortho position of the phenyl ring. Compound 5l having nitro group at para position has resulted in a significant reduction in α-glucosidase inhibition (IC50 = 167.8 µM). This suggests that the inhibitory activity of α-glucosidase enzyme could be increased with an electron-withdrawing substituent at ortho position. A slight change in activity was observed between compounds 5c and 5g. On the other hand, replacing nitro group at para position of Schiff base moiety (compound 5l) with –OMe group (compound 5e) (IC50 = 270.7 µM), has significantly lowered the α-glucosidase inhibitory activity. Likewise, when comparing 5e with 5i, a higher inhibition of 5e is likely due to the presence of two methoxyl groups at meta and para positions of phenyl ring. In general, the electron-withdrawing groups at ortho position of phenyl group are playing a crucial role in the α-glucosidase inhibition.
Whereas, among the leucine hydrazone derivatives, 10i exerted higher inhibitory activity with IC50 value of 83.3 µM, then 10f, 10b, 10d, and 10e with IC50 values of 351.0, 354.4, 367.8, 397.7 µM, respectively (Table 1). Compound 10i has displayed the most potent activity against α-glucosidase enzyme when compared with other derivatives of the series (10a–10k). The higher inhibition of 10i is possibly due to the presence of chloro group as an electron-withdrawing group at para position of the phenyl group. When 10f is compared with 10e, the higher inhibition of 10f is due to the replacement of dimethoxy with dihydroxy groups at the ortho and para positions of the phenyl group. Likewise, when 10f is compared with 10b, a slight difference was observed indicating that the presence of –OH group at ortho position is crucial in the series. Similarly, when compound 10d is compared with 10e, a decrease in inhibition of 10e (IC50 = 397.7 µM) is observed probably due to the presence of an extra –OCH3 group at meta position of the phenyl ring.
Structure–activity relationship of urease enzyme
The in vitro testing against urease suggested that compound 5g (in case of phenylalanine hydrazine derivatives) was found the most active having IC50 value of 128.6 µM, followed by 5b (IC50 = 173.7 µM), while compounds 5a (IC50 = 304.5 µM) and 5h (IC50 = 427.3 µM) exhibited weak inhibition when compared with the standard thiourea (IC50 = 20.8 µM) (Table 1). The higher activity of compound 5g is due to the presence of two hydroxyl groups at ortho and para positions of the phenyl ring. The lower inhibition of 5a is possibly due to the replacement of substituted phenyl with pyridine ring. When comparing 5g with 5h (IC50 = 427.3 µM), the decrease in inhibition is due to the absence of –OH group at ortho position which further confirm that –OH group substituted at ortho position of the phenyl ring is playing crucial role against urease enzyme in the series. Compounds having –OMe, –NO2, and halo substituted phenyls did not show urease inhibition, which indicates the importance of the hydroxyl group in this series.
In case of leucine hydrazones, compound 10f (IC50 = 88.2 µM) was found to be the most active, whilst compounds 10h (IC50 = 271.7 µM), 10j (IC50 = 308.3 µM), and 10a (IC50 = 320.6 µM) exerted weak inhibition when compared to the standard (Table 1). The higher activity of 10f is due to the presence of two hydroxyl groups at ortho and para positions of the phenyl group, which further justify that the –OH group substituted at ortho and para positions as stated above.
Redocking results
Redocking experiment was performed to evaluate the performance of docking program. The substrate molecule of α-glucosidase was taken from model, while cocrystalized inhibitors of urease and CA II were extracted from their PDB files and redocked in the active site of their respective protein files. After docking, root mean square deviation (RMSD) was automatically calculated by software between the docked and undocked conformations of compounds. The RMSD for of α-glucosidase substrate was 0.49 Å, while RMSD for CA II and urease inhibitors were 0.65 and 0.224 Å, respectively. The redocking results assured that the docking program can produce reliable results. The redocked conformations of compounds are presented in Fig. S1 (Supporting Information).
Molecular docking and interaction analysis of active hits in α-glucosidase, urease, and CA II
Molecular docking of active inhibitors was carried out to predict their mode of binding and interactions with their specific targets including CA II, α-glucosidase, and urease enzymes. Compound 10i was found to be potent against α-glucosidase with IC50 = 83.3 µM. The docked view of 10i showed that the amino group and the carbonyl oxygen of the compound mediate strong hydrogen bonds with the side chains of Glu276 and His348, respectively. Moreover, the compound has also displayed highest docking score (−11.41). The binding interactions of 10i are displayed in Fig. 1. Compound 5K also demonstrated biological activities in range of IC50 = 87.1 µM. The docked view of 5K showed that the carbonyl oxygen and the nitro group of the compound formed hydrogen bonds with a surrounded water molecule and the side chain of His348. Therefore, we can predict that His348 plays important role in the binding and stabilization of active inhibitors in the active site of α-glucosidase [55, 56]. While compounds 5c, 5l, and 5g exhibited moderate inhibitory activities in range of IC50 = 166–176 µM against α-glucosidase. The amino group of 5c mediated bidentate interactions with the side chain of Glu276. Compound 5l exhibited hydrogen bonding with the side chain of Arg312 and a water molecule, while the hydroxyl group of compound 5g formed hydrogen bond with the side chain of Glu304, and the amino group of compound interacted with a water molecule. In addition, 5g also mediated hydrophobic interaction with the phenyl ring of Phe157. Compounds 5b, 5e, 5i, 10f, 10b, 10d, and 10e were among the least active hits. The docked view of these compounds showed that the amino group of 5b and 5e formed hydrogen bond with the side chain of Glu276, while 5i, 10f, 10b, 10d, and 10e interacted with only water molecules. The binding interactions and docking scores of the compounds are tabulated in Table 2. The 2D interactions of the active hits are shown in Fig. S73.
The 3D structure of α-glucosidase is shown in complex with the docked conformation of the most active compound (10i). The protein–ligand interactions are highlighted in the box. The ligand (10i) is shown in green stick model, active site amino acids are depicted in cyan stick model. Hydrogen bonds are shown in green lines
When tested in urease inhibitory assay, compound 10f exhibited highest inhibitory potential with IC50 = 88.2 ± 2.27 µM, while compounds 5b, 10h, 5a, 10j, 10a, and 5h depicted moderate-to-least biological activities. The docked view of 10f (Fig. 2) demonstrated that both the carbonyl groups of the compound interacted with the Ni atoms present in the active site. While amino groups of the compound formed hydrogen bonding with the main chain carbonyl oxygen of Ala636 and a water molecule [56]. The hydroxyl group of 5b donated a hydrogen bond to the side chains of Asp633, while His519 provide π–π interaction to the compound. Similarly, the side chain of His492 and a water molecule interacted with the nitrogen and carbonyl oxygen of 10h. Moreover, Arg609 and His593 provide hydrogen bond to the nitrogen and carbonyl oxygen of 5a and 10j, respectively. While the side chain of His409 and His407 provide π–π interaction to the ring of 10a and 5h, respectively. The docking results of active inhibitors are tabulated in Table 3. The 2D interactions of the active hits at the active site of urease are presented in Fig. S74.
The 3D structure of urease is presented in complex with the docked conformation of the most active compound (10f). The protein–ligand interactions are highlighted in the box. 10f is shown in olive green stick model, active site amino acids are depicted in magenta stick model. Hydrogen bonds are shown in green lines. Metal–ligand interactions are shown in black dotted lines
Among the tested compounds, 5b, 10a, 5d, and 5c exhibited highest inhibitory potential on CA II inhibitory assay. The binding modes of those inhibitors in the active site of CA II showed that the carbonyl moieties of 5b, 10a, and 5d mediated hydrogen bonding with a water molecule, while amino nitrogen of 5b, 5d, and 5c interacted with the side chain of Thr198, His93, and Gln91, respectively, [57]. However, the ring nitrogen of 10a and 5a mediated hydrogen boding with the side chain of His95. Similarly, the amino and carbonyl groups of 10b interacted with the side chain of Thr198 and a water molecule. Furthermore, compounds 5i and 5e only formed water mediated bridging within the active site, however, the amino and –OH group of 10g interacted with a water molecule and Phe94, respectively. The docked view of 10h depicted that the carbonyl group of compound formed multiple hydrogen bonds with the Thr197 and Thr198. Similarly, the carbonyl group of 10g interacted with Thr197 and a water molecule, while His93 stabilized the compounds 10g, 10d, and 5h through hydrophobic interaction. In addition, 10d and 5h also interacted with water molecule in the vicinity. Moreover, 5h also formed a hydrogen bond with the Phe94. The binding interactions and docking results of CA II inhibitors are tabulated in Table 4. The binding mode of the most active hit (5b) is presented in Fig. 3. The 2D interactions of the active hits within the active site of CA II are demonstrated in Fig. S75.
The 3D structure of CA II is presented in complex with the docked conformation of the most active compound (5b). The protein–ligand interactions are highlighted in the box. The inhibitor is shown in olive green stick model, active site amino acids are depicted in coral stick model. Hydrogen bonds are shown in black lines
Conclusion
In summary, a series of new amino acid hybrid Schiff bases (5a–5l and 10a–10k) were synthesized and their CA II, α-glucosidase, and urease inhibitor activities were evaluated in vitro. Among the phenylalanine hydrazine derivatives, compounds 5b (11.8 µM) proved to be the most potent against CA II enzyme, while 10i (83.3 µM) and 10f (88.2 µM) were found to be the most active scaffolds against α-glucosidase and urease, respectively. The structure–activity relationship was discussed. Some compounds proved to be more potent than the control groups. Furthermore, the molecular docking studies showed that all the active compounds accommodate well in the active site of these enzymes by interacting with key amino acids. These results should be evaluated in vivo to further assess their potential as enzymes inhibitory drugs.
Experimental
General
All reagents were obtained from Sigma-Aldrich chemical company, Germany. Solvents used for chemical reactions were purified and dried by standard procedures. Melting point was determined using Stuart SMP10 digital melting point apparatus). The masses of the compounds were confirmed from high-resolution electrospray ionization mass spectrometry (Agilent 6530 LC Q-TOF, country of origin USA/EU, made in Singapore). The NMR spectra (1H-NMR (600 MHz) and 13C-NMR (150 MHz)) were recorded on nuclear magnetic resonance (BRUKER, Zürich, Switzerland) spectrometer using the solvent peaks as internal reference (CDCl3, δH: 7.26; δC: 77.2–76.8; DMSO-d6, δH: 2.49; δC: 40.0–39.1). The following abbreviations: s: singlet, d: doublet, dd: doublet of doublet, t: triplet, m: multiplet, J: coupling constant were used to explain NMR signals. Chemical shifts were expressed in parts per million (δ) values and coupling constants (J) are given in Hertz (Hz). ATR-Tensor 37 spectrophotometer (Bruker, Ettlingen, Baden-Württemberg, Germany) was used the determination of infrared (IR) spectra. All reactions were monitored by thin-layer chromatography (TLC). TLC plates were visualized under the UV light at 254 and 366 nm. Solvents for chromatography were of commercial grade and distilled prior to use.
General procedure for the synthesis of amino acids Schiff bases derivatives
Amino acid Schiff bases were synthesized in four steps following published protocol [58]. L-β-phenylalanine (1.65 g, 10 mmol)) and L-leucine (1.3 g, 10 mmol) were used as a starting materials for the synthesis of amino acid Schiff bases (5a–5l and 10a–10k). Esterification of carboxylic group was done by the addition of thionyl chloride (2.1 mL, 15 mmol) slowly to a solution of L-β-phenylalanine (1.65 g, 10 mmol) and methanol (20 mL) at 0 °C. The reaction solution was stirred (at room temperature) for overnight. Reaction was thoroughly observed by TLC for assuring that all amount of amino acid changed to ester or not. After reaction completion, the organic solvent was evaporated under vacuum and the resultant product was filtered. Same procedure was used for the esterification of L-leucine (1.3 g, 10 mmol) using SOCl2 (2.1 mL, 15 mmol) and methanol (15 mL).
Second step was to protect the amino group through the reaction of tert-Boc reagent. In this step, amino acid methyl ester (1.75 g, 9.8 mmol) was added to a mixture of anhydrous sodium carbonate (1.2 g, 12 mmol) and water (40 mL) in the round bottom flask (100 mL). Then, tert-Boc (2.1 g, 10 mmol) was dissolved in ethyl acetate (20 mL) and then added to that solution and the reaction was allowed to stir for 24 h at room temperature. The progress of the reaction was checked by TLC. When the reaction was completed, adjusts PH at 5.5 with oxalic acid and then extracted the desire product through EtOAc. The solvent was dried over anhydrous MgSO4 and evaporated under reduced pressure to obtain Boc-phenylalanine methyl ester (91%). Same procedure was used for the synthesis of Boc-leucine methyl ester (yield, 92%) using anhydrous Na2CO3 (1.2 g, 12 mmol), water (30 mL), and Boc (2.1 g, 10 mmol).
Third step involved the formation of Boc-phenylalanine hydrazide. Hydrazine monohydrate (2 mL, 32 mmol) was added to a solution of Boc-phenylalanine methyl ester (2.5 g, 9.1 mmol) and methanol (20 mL) and stirred under room temperature for 24 h. The reaction mixture was thoroughly monitored by TLC. After reaction completion, the solvent was evaporated under reduced pressure and the product was washed with methanol (89%). Similarly, the same procedure was used for the synthesis of Boc-leucine hydrazide using hydrazine monohydrate (2 mL, 32 mmol) and methanol (25 mL).
Tert-butyl(S, E)-(3-oxo-1-phenyl-3-(2-(pyridine-2-ylmethylene)hydrazineyl)propyl)carbamate (5a)
2-Pyridinecarboxaldehyde (32.1 mg, 0.3 mmol) was added to a stirred solution of Boc-phenylalanine hydrazide (55.4 mg, 0.2 mmol) in EtOH (5 mL) at room temperature for 24 h. The progress of the reaction was monitored with TLC. After completion reaction, the product (5a) was filtered, and washed with n-hexane for removing excess amount of aldehydes (Yield 88%). The same procedure was used for the synthesis of other compounds (5b–5l) using different types of benzaldehydes moities (0.3 mmol). Similarly, same benzaldehydes were used for the synthesis of compounds (10a–10k) using Boc-leucine hydrazide (50 mg, 0.2 mmol) in 5 mL EtOH at room temperature for 24 h. All compounds were obtained in good yields ranging from 78 to 94%. The structures of all compounds were established by HRMS, IR, and 1H- and 13C-NMR.
Light yellow powder; Yield: 88%; m.p. 130–135 °C; FT-IR (solid, cm−1): 3321, 3250, 1666, 1514, 1461, 1377, 1335, 1290, 1233, 1160; 1H-NMR (600 MHz, DMSO-d6): δ 8.19 (1H, s, NH), 8.59 (1H, CH=N), 7.99, 8.01 (2H, m), 7.99, 7.87 (2H, m), 7.58–7.13 (5H, m), 5.09 (1H, br.s, NH-Boc), 4.20 (1H, br.s, CH–NH), 2.77, 2.97 (2H, m), 1.30 (9H, s); 13C-NMR (150 MHz, DMSO-d6): δ 28.1, 36.6, 52.7, 78.2, 119.5, 124.2, 126.3, 128.1, 129.1, 136.8, 137.7, 143.8, 147.2, 149.5, 153.0, 155.4, 168.7, 173.4; HRMS (ESI+) m/z: 369.1846 [M+H]+.
Tert-butyl(S,E)-(3-(2-(2-hydroxybenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5b)
White powder; Yield: 86%; m.p. 115–120 °C; FT-IR (solid, cm−1): 3333, 2975, 1663, 1614, 1520, 1367, 1264, 1162, 1092; 1H-NMR (CDCl3, 600 MHz): δ 10.99 (1H, s, NH), 8.03 (1H, CH=N), 7.29‒7.16 (5H, m), 7.05 (2H, d, J = 7.2 Hz), 9.68 (2H, d = 7.8 Hz), 6.80 (1H, t, J = 7.2 Hz), 5.68 (1H, br.s, NH-Boc), 4.51 (1H, br.s, CH–NH), 3.22‒2.98 (2H, m), 1.37 (9H, s); 13C-NMR (150 MHz, CDCl3): δ 28.3, 38.5, 55.2, 80.7, 117.0, 117.6, 119.7, 127.0, 128.5, 129.3, 131.3, 132.1, 136.3, 149.4, 151.2, 156.2, 157.8, 158.5, 167.9, 172.9; HRMS (ESI+) m/z: 384.1908 [M+H]+.
Tert-butyl(S,E)-(3-(2-(2-hydroxy-4-methoxybenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5c)
Yellow amorphous powder; Yield: 80%; m.p. 166–170 °C; FT-IR (solid, cm−1): 3341, 3246, 1659, 1607, 1572, 1513, 1447, 1373, 1343, 1306, 1277, 1163; 1H-NMR (CDCl3, 600 MHz): δ 10.39 (1H, s, NH), 7.94 (1H, CH=N), 7.23–7.15 (5H, m), 6.92 (1H, d, J = 9.0 Hz), 6.40 (1H, s), 6.33 (1H, dd = 8.4, 1.6 Hz), 5.85 (1H, br.s, NH-Boc), 4.53 (1H, d, J = 6.6 Hz, CH–NH), 3.74 (3H, s, OCH3), 3.13‒2.98 (2H, m), 1.37 (9H, s); 13C-NMR (150 MHz, CDCl3): δ 28.3, 38.6, 55.3, 52.3, 80.6, 101.3, 106.9, 126.9, 127.1, 128.5, 129.5, 132.1, 149.4, 151.1, 156.2, 159.8, 160.5, 162.8, 163.0, 167.8, 172.6; HRMS (ESI+) m/z: 414.2033 [M+H]+.
Tert-butyl(S,E)-(3-(2-(4-hydroxy-3-methoxybenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5d)
Colorless solid; Yield: 82%; m.p. 118–123 °C; FT-IR (solid, cm−1): 3339, 2972, 1663, 1595, 1515, 1456, 1376, 1274, 1164, 1124, 1030; 1H-NMR (CDCl3, 600 MHz): δ 9.53 (1H, s, NH), 7.94 (1H, CH=N), 7.23‒7.15 (5H, m), 7.02 (1H, d = 1.6 Hz), 6.90 (1H, dd, J = 7.8, 1.6 Hz), 6.84 (1H, d, J = 7.8 Hz), 5.43 (1H, br.s, NH-Boc), 4.44 (1H, t, J = 7.8 Hz, CH–NH), 3.94 (3H, s, OCH3), 3.25‒2.96 (2H, m), 1.38 (9H, s); 13C-NMR (150 MHz, CDCl3): δ 28.3, 38.5, 52.5, 56.1, 76.7, 79.6, 107.6, 114.0, 123.0, 125.8, 126.7, 128.2, 129.3, 145.1, 147.0, 149.2, 155.2, 167.8, 173.3; HRMS (ESI+) m/z: 414.2021 [M+H]+.
Tert-butyl (S,E)-(3-(2-(4-methoxybenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5e)
Yellow amorphous powder; Yield: 83%; m.p. 173–178 °C; FT-IR (solid, cm−1): 3330, 3250, 1659, 1601, 1516, 1452, 1374, 1309, 1252, 1164, 1024; 1H-NMR (600 MHz, CDCl3): δ 9.08 (1H, s, NH), 7.62 (1H, CH=N), 7.62‒7.56 (2H, dd, J = 8.4 Hz), 7.58‒7.13 (5H, m), 6.91–6.85 (2H, dd, J = 8.4 Hz), 5.29 (1H, br.s, NH-Boc), 4.40 (1H, dd, J = 7.8, 7.2 Hz, CH–NH), 3.84 (3H, s, OCH3), 5.29 (1H, br.s, CH–NH), 3.21‒3.01 (2H, m), 1.39 (9H, s); 13C-NMR (150 MHz, CDCl3): δ 28.3, 38.6, 52.3, 55.3, 79.6, 114.0, 125.9, 126.0, 126.7, 128.7, 129.4, 129.5, 136.5, 144.4, 148.5, 155.1, 161.4, 167.5, 173.1; HRMS (ESI+) (m/z): 398.2077 [M+H]+.
Tert-butyl(S,E)-(3-(2-(2,5-dimethoxybenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5f)
Light yellow powder; Yield: 89%; m.p. 160–165 °C; FT-IR (solid, cm−1): 3342, 3248, 1659, 1512, 1453, 1415, 1368, 1309, 1257, 1219, 1164, 1090; 1H-NMR (600 MHz, DMSO-d6): δ 8.50 (1H, s, NH), 8.30 (1H, CH=N), 7.35 (1H, d, J = 2.4 Hz), 7.31‒7.23 (5H, m), 7.10 (1H, d, J = 8.4 Hz), 6.99 (1H, dd, J = 8.4, 2.4 Hz), 5.03 (1, d, J = 6.0 Hz, NH-Boc), 4.16 (1H, br.s, CH–NH), 3.79 (3H, s, OCH3), 3.73 (3H, s, OCH3), 3.00–2.992 (1H, m), 2.82–2.73 (1H, m), 1.32 (9H, s); 13C-NMR (150 MHz, DMSO-d6): δ 28.2, 36.2, 56.3, 55.5, 55.4, 78.1, 108.6, 113.6, 117.7, 126.3, 128.1, 129.0, 129.3, 138.9, 142.2, 152.2, 152.3, 153.3, 168.3, 173.1; HRMS (ESI+) m/z: 428.2195 [M+H]+.
Tert-butyl(S,E)-(3-(2-(2,4-dihydroxybenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5g)
Dark brown solid powder; Yield: 84%; m.p. 110–120 °C; FT-IR (solid, cm−1): 3214, 2976, 1666, 1621, 1505, 1450, 1366, 1316, 1230, 1159; 1H-NMR (600 MHz, DMSO-d6): δ 8.27 (1H, s, NH), 9.95 (1H, CH=N), 7.27‒7.26 (5H, br.s), 7.18 (1H, s), 6.33 (2H, d, J = 9.0 Hz), 4.19 (1H, dd, J = 9.0, 4.8 Hz, CH–NH), 2.96–2.72 (2H, m), 1.30 (9H, s); 13C-NMR (150 MHz, DMSO-d6): δ 28.2, 37.4, 39.9, 40.0, 52.7, 55.0, 78.2, 102.5, 107.7, 110.4, 126.3, 128.1, 131.1, 137.9, 148.4, 155.3, 158.1, 159.4, 160.6, 167.7, 172.4; HRMS (ESI+) m/z: 400.1862 [M+H]+.
Tert-butyl(S, E)-(3-(2-(4-hydroxybenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5h)
Colorless solid; Yield: 85%; m.p. 190–200 °C; FT-IR (solid, cm−1): 3324, 1689, 1649, 1594, 1504, 1432, 1369, 1332, 1258, 1206, 1163; 1H-NMR (600 MHz, DMSO-d6): δ 8.07 (1H, s, NH), 7.87 (1H, CH=N), 7.50 (2H, dd, J = 7.8, 2.4 Hz), 7.29‒7.22 (5H, br.s), 6.82 (2H, dd, J = 8.4, 2.4 Hz), 5.04 (1H, br.s, NH-Boc), 4.19 (1H, dd, J = 9.0, 4.8 Hz, CH–NH), 2.96‒2.73 (2H, m), 1.30 (9H, s); 13C-NMR (150 MHz, DMSO-d6): δ 28.2, 36.5, 37.4, 39.8, 52.9, 55.1, 78.1, 115.8, 124.9, 126.3, 128.1, 128.8, 129.1, 129.3, 138.0, 143.8, 147.2, 155.4, 159.7, 168.0, 172.8; HRMS (ESI+) m/z: 384.1894 [M+H]+.
Tert-butyl(S,E)-(3-(2-(3,4-dimethoxybenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5i)
Colorless solid; Yield: 91%; m.p. 168–173 °C; FT-IR (solid, cm−1): 3324, 1690, 1649, 1595, 1511, 1433, 1368, 1331, 1257, 1204, 1163; 1H-NMR (600 MHz, CDCl3): δ 9.14 (1H, s, NH), 7.61 (1H, CH=N), 7.40 (1H, s), 7.28‒7.21 (5H, br.s), 7.07 (1H, dd, J = 8.4, 2.4 Hz), 6.84 (1H, d, J = 8.4 Hz), 5.28 (1H, br.s, NH-Boc), 4.41 (1H, dd, J = 7.8, 7.2 Hz, CH–NH), 3.95 (6H, s, 2 × OCH3), 3.25–2.95 (2H, m), 1.39 (9H, s); 13C-NMR (150 MHz, DMSO): δ 28.3, 38.5, 52.5, 55.9, 56.1, 78.1, 108.2, 110.4, 122.3, 126.2, 126.7, 128.7, 129.3, 136.6, 144.7, 149.4, 151.3, 155.2, 167.5, 173.2; ESI-HRMS (m/z): 428.2181 [M+H]+.
Tert-butyl(S,E)-(3-(2-(4-chlorobenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5j)
Colorless solid; Yield: 89%; m.p. 186–192 °C; FT-IR (solid, cm−1): 3338, 1669, 1596, 1527, 1432, 1369, 1314, 1264, 1169, 1054, 1018; 1H-NMR (DMSO-d6, 600 MHz): δ 8.13 (1H, s, NH), 7.87 (1H, CH=N), 7.61 (2H, dd, J = 9.0, 2.4 Hz), 7.43 (2H, dd, J = 9.0, 2.4 Hz), 7.20–7.13 (5H, m), 4.98 (1H, br.s, NH-Boc), 4.13 (1H, br.s, CH–NH), 2.85‒2.66 (2H, m), 1.20 (9H, s); 13C-NMR (150 MHz, DMSO): δ 28.2, 36.6, 52.7, 78.1, 126.3, 128.1, 128.7, 129.1, 130.8, 132.9, 133.2, 134.3, 137.9, 142.1, 145.6 155.4, 168.6, 173.3; HRMS (ESI+) m/z: 424.1403 (Cl35), 426.1408 (Cl37) [M+Na]+.
Tert-butyl(S,E)-(3-(2-(2-nitrobenzylidene)hydrazineyl)-3-oxo-1-phenylpropyl)carbamate (5k)
Dark yellow amorphous powder; Yield: 87%; m.p. 195–200 °C; FT-IR (solid, cm−1): 2973, 1668, 1521, 1339, 1260, 1220, 1169, 1056; 1H-NMR (600 MHz, DMSO-d6): δ 8.60 (1H, s, NH), 7.37 (1H, CH=N), 8.07 (1H, t, J = 9.0, 8.4 Hz), 8.20 (1H, d, J = 7.8 Hz), 7.82 (1H, dd, J = 8.4, 7.8 Hz), 7.67 (1H, t, J = 7.8, 7.2 Hz), 7.30–7.17 (5H, m), 5.06 (1H, br.s, NH-Boc), 4.23 (1H, br.s, CH–NH), 2.98 (1H, m), 2.82 (1H, m), 1.30 (9H, s); 13C-NMR (150 MHz, DMSO): δ 28.2, 36.5, 52.7, 78.1, 124.7, 126.3, 127.8, 128.5, 129.2, 130.5, 133.7, 138.2, 142.3, 148.1, 155.4, 168.9, 173.5; HRMS (ESI+) m/z: 435.1363 [M+Na]+.
Tert-butyl(S,E)-(4-methyl-1-(2-(2-nitrobenzylidene)hydrazineyl)-1-oxopentan-2-yl)carbamate (5l)
Yellow solid powder; Yield: 94%; m.p. 201–208 °C; FT-IR (solid, cm−1): 2973, 1668, 1521, 1339, 1260, 1220, 1169, 1056; 1H-NMR (600 MHz, DMSO-d6): δ 8.37 (1H, s, NH), 7.37 (1H, CH=N), 8.07 (1H, t, J = 9.0, 8.4 Hz), 8.31 (2H, t, J = 9.0 Hz), 7.96 (2H, dd, J = 8.4 Hz), 7.32‒7.26 (4H, m), 7.21 (1H, m), 5.13 (1H, br.s, NH-Boc), 4.26 (1H, br.s, CH–NH), 2.99 (1H, m), 2.87 (1H, m), 1.32 (9H, s); 13C-NMR (150 MHz, DMSO): δ 28.2, 36.7, 37.2, 39.3, 40.0, 52.7, 55.4, 78.1, 124.0, 126.3, 127.7, 128.1, 129.2, 137.9, 140.6, 141.0, 144.5, 147.7, 155.4, 169.2, 173.7; HRMS (ESI+) m/z: 435.1363 [M+Na]+.
Tert-butyl (S,E)-(4-methyl-1-oxo-1-(2-(pyridine-2-ylmethylene)hydrazineyl)pentan-2-yl)carbamate (10a)
White powder; Yield: 82%; m.p. 67–72 °C; FT-IR (solid, cm−1): 2962, 1671, 1509, 1463, 1366, 1241, 1162, 1043; 1H-NMR (CDCl3, 600 MHz): δ 10.12 (1H, s, NH), 8.56 (1H, dd, J = 10.8, 4.2 Hz), 7.99 (1H, s, CH=N), 7.96 (1H, d, J = 7.8 Hz), 7.68 (1H, t, J = 7.2 Hz), 7.25 (1H, br.s), 5.39 (1H, br.s, NH-Boc), 4.29 (1H, br.s, CH–NH), 1.80 (1H, br.s), 1.53 (1H, br.s), 1.65 (1H, m), 1.40 (9H, s), 1.04 (6H, d, J = 6.6 Hz); 13C-NMR (150 MHz, CDCl3): δ 21.6, 23.3, 28.3, 42.1, 49.7, 80.0, 120.0, 124.2, 136.5, 145.2, 149.1, 152.8, 155.7, 175.7; HRMS (ESI+) m/z: 334.20 [M+H]+.
Tert-butyl(S,E)-(1-(2-(2-hydroxybenzylidene)hydrazineyl)-4-methyl-1-oxopentan-2-yl)carbamate (10b)
Light Yellow solid powder; Yield: 86%; m.p. 80–90 °C; FT-IR (solid, cm−1): 2962, 1666, 1615, 1525, 1489, 1362, 1270, 1239, 1204, 1160; 1H-NMR (CDCl3, 600 MHz): 10.77 (1H, s, NH), 8.15 (1H, s, CH=N), 7.20 (1H, t, J = 7.2 Hz), 7.05 (1H, dd, J = 8.4, 4.2 Hz), 6.85 (1H, d, J = 7.8 Hz), 6.75 (1H, t, J = 8.4, 6.0 Hz), 5.73 (1H, br.s, NH-Boc), 4.30 (1H, br.s, CH–NH), 1.74 (1H, m), 1.72 (1H, br.s), 1.65 (1H, m), 1.41 (9H, s), 0.95 (6H, d, J = 6.6 Hz); 13C-NMR (150 MHz, CDCl3): δ 22.8, 23.3, 28.3, 40.9, 52.2, 80.7, 117.0, 119.2, 130.9, 131.7, 149.4, 151.1, 156.6, 158.5, 169.2; HRMS (ESI+) m/z: 350.1920 [M+H]+.
Tert-buty(S,E)-(1-(2-(4-hydroxy-3-methoxybenzylidene)hydrazineyl)-4-methyl-1-oxopentan-2-yl)carbamate (10c)
Colorless solid; Yield: 84%; m.p. 120–125 °C; FT-IR (solid, cm−1): 2960, 1663, 1594, 1511, 1461, 1428, 1370, 1276, 1162, 1119, 1035; 1H-NMR (600 MHz, CDCl3): δ 9.24 (1H, s, NH), 7.67 (1H, s), 7.34 (1H, s, CH=N), 6.97 (1H, d, J = 7.8 Hz), 6.89 (1H, d, J = 7.8 Hz), 6.85 (1H, d, J = 7.8 Hz), 5.91 (1H, br.s, NH-Boc), 5.22 (1H, br.s, CH–NH), 3.88 (3H, s, OCH3), 1.85 (1H, m), 1.80 (1H, m), 1.68 (1H, m), 1.42 (9H, s), 0.94 (6H, d, J = 6.6 Hz); 13C-NMR (150 MHz, CDCl3): δ 22.7, 24.9, 28.3, 42.4, 49.8, 55.9, 56.1, 79.5, 107.2, 114.0, 123.2, 126.0, 144.7, 149.1, 155.8, 175.0; HRMS (ESI+) m/z: 380.2064 [M+H]+.
Tert-butyl(S,E)-(1-(2-(4-methoxybenzylidene)hydrazineyl)-4-methyl-1-oxopentan-2-yl)carbamate (10d)
Light brown amorphous powder; Yield: 88%; m.p. 92–100 °C; FT-IR (solid, cm−1): 2961, 1665, 1604, 1562, 1508, 1460, 1365, 1304, 1246, 1163, 1028; 1H-NMR (CDCl3, 600 MHz): δ 9.44 (1H, s, NH), 7.82 (1H, s, CH=N), 7.58 (2H, d, J = 8.4 Hz), 6.88 (2H, d, J = 8.4 Hz), 5.22 (1H, br.s, NH-Boc), 4.22 (1H, br.s, CH–NH), 3.82 (3H, s, OCH3), 1.82 (1H, m), 1.70 (1H, m), 1.62 (1H, m), 1.42 (9H, s), 0.94 (6H, d, J = 6.6 Hz); 13C-NMR (150 MHz, CDCl3): δ 21.7, 22.7, 28.3, 42.2, 49.2, 55.3, 79.2, 114.1, 14.2, 114.3, 126.2, 128.8, 129.4, 144.4, 161.4, 175.2; HRMS (ESI+) m/z: 364.2227 [M+H]+.
Tert-butyl(S,E)-(1-(2-(2,5-dimethoxybenzylidene)hydrazineyl)-4-methyl-1-oxopentan-2-yl)carbamate (10e)
Colorless solid; Yield: 84%; m.p. 88–95 °C; FT-IR (solid, cm−1): 2959, 1669, 1495, 1462, 1425, 1362, 1253, 1218, 1164, 1111, 1042; 1H-NMR (DMSO-d6, 600 MHz): δ 11.23 (1H, s, NH), 8.28 (1H, s, CH=N), 7.30 (1H, d, J = 1.8 Hz), 7.06 (2H, d, J = 8.4 Hz), 4.92 (1H, br.s, NH-Boc), 3.99 (1H, br.s, CH–NH), 3.99 (3H, s, OCH3), 3.79 (3H, s, OCH3), 1.71 (1H, m), 1.61 (1H, m), 1.50 (1H, m), 1.44 (9H, s), 0.96 (6H, d, J = 6.6 Hz); 13C-NMR (150 MHz, DMSO): δ 21.2, 22.9, 28.2, 39.5, 40.1, 49.4, 52.0, 55.3, 78.0, 108.5, 113.4, 117.5, 122.8, 138.7, 142.1, 152.1, 155.6, 174.3; HRMS (ESI+) m/z: 394.2340 [M+H]+.
Tert-butyl(S,E)-(1-(2-(2,4-dihydroxybenzylidene)hydrazineyl)-4-methyl-1-oxopentan-2-yl)carbamate (10f)
Dark brown solid powder; Yield: 82%; m.p. 101–110 °C; FT-IR (solid, cm−1): 2966, 1667, 1622, 1511, 1457, 1366, 1321, 1235, 1162; 1H-NMR (DMSO-d6, 600 MHz): δ 11.22 (1H, s, NH), 8.28 (1H, s, CH=N), 7.24 (1H, d, J = 8.4 Hz), 7.05 (1H, d, J = 8.4 Hz), 6.30 (1H, s), 4.83 (1H, br.s, NH-Boc), 4.00 (1H, br.s, CH–NH), 1.69 (1H, m), 1.61 (1H, m), 1.52 (1H, m), 1.36 (9H, s), 0.91 (6H, d, J = 6.6 Hz); 13C-NMR (150 MHz, DMSO): δ 21.2, 24.3, 28.2, 39.5, 51.8, 78.0, 102.5, 107.7, 110.3, 127.9, 131.2, 142.2, 148.4, 155.4, 158.2, 160.6, 168.6; HRMS (ESI+) m/z: 366.1768 [M+H]+.
Tert-butyl(S,E)-(1-(2-(4-hydroxybenzylidene)hydrazineyl)-4-methyl-1-oxopentan-2-yl)carbamate (10g)
Light brown amorphous powder; Yield: 89%; m.p. 105–112 °C; FT-IR (solid, cm−1): 2965, 1661, 1604, 1511, 1444, 1366, 1239, 1161, 1055, 1022; 1H-NMR (DMSO-d6, 600 MHz): δ 11.26 (1H, s, NH), 7.85 (1H, s, CH=N), 7.49 (2H, d, J = 8.4 Hz), 6.84 (2H, d, J = 8.4 Hz), 4.92 (1H, br.s, NH-Boc), 4.00 (1H, t, J = 8.4, 5.4 Hz, CH–NH), 1.71 (1H, m), 1.61 (1H, m), 1.51 (1H, m), 1.44 (9H, s), 0.95 ((3H, d, J = 6.6 Hz), 0.88 (3H, d, J = 6.6 Hz); 13C-NMR (150 MHz, DMSO): δ 21.7, 22.9, 28.3, 39.4, 40.1, 49.3, 51.9, 78.0, 115.7, 125.1, 128.3, 143.5, 147.2, 155.4, 159.5, 159.5, 174.0; HRMS (ESI+) m/z: 350.2070 [M+H]+.
Tert-butyl(S,E)-(1-(2-(3,4-dimethoxybenzylidene)hydrazineyl)-4-methyl-1-oxopentan-2-yl)carbamate (10h)
Colorless solid; Yield: 79%; m.p. 99–105 °C; FT-IR (solid, cm−1): 2961, 1667, 1602, 1572, 1510, 1460, 1418, 1389, 1364, 1265, 1237, 1162, 1052; 1H-NMR (DMSO-d6, 600 MHz): δ 11.12 (1H, s, NH), 7.88 (1H, s, CH=N), 7.28 (1H, s), 7.00 (1H, d, J = 8.4 Hz), 6.87 (1H, d, J = 8.4 Hz), 4.92 (1H, br.s, NH-Boc), 4.01 (1H, dd, J = 8.4, 6.0 Hz, CH–NH), 4.01 (3H, s, OCH3), 3.78 (3H, s, OCH3), 1.72 (1H, m), 1.63 (1H, m), 1.52 (1H, m), 1.36 (9H, s), 0.99 (3H, d, J = 6.6 Hz), 0.88 (3H, d, J = 6.6 Hz); 13C-NMR (150 MHz, DMSO): δ 21.7, 22.9, 28.3, 39.4, 40.1, 49.4, 52.0, 55.2, 78.0, 107.6, 111.5, 121.5, 127.0, 143.3, 147.0, 149.0, 155.4, 174.1; HRMS (ESI+) m/z: 394.1983 [M+H]+.
Tert-butyl(S,E)-(1-(2-(4-chlorobenzylidene)hydrazineyl)-4-methyl-1-oxopentan-2-yl)carbamate (10i)
Colorless solid; Yield: 85%; m.p. 151–155 °C; FT-IR (solid, cm−1): 2958, 1667, 1523, 1489, 1458, 1392, 1366, 1319, 1279, 1236, 1091, 1054, 1012; 1H-NMR (600 MHz, DMSO-d6): δ 11.30 (1H, s, NH), 7.96 (1H, s, CH=N), 7.70 (2H, dd, J = 8.4 Hz), 7.50 (2H, dd, J = 8.4 Hz), 6.87 (1H, d, J = 8.4 Hz), 4.93 (1H, br.s, NH-Boc), 4.02 (1H, dd, J = 8.4, 5.4 Hz, CH–NH), 1.70 (1H, m), 1.61 (1H, m), 1.50 (1H, m), 1.36 (9H, s), 0.94 (3H, d, J = 6.6 Hz), 0.88 (3H, d, J = 6.6 Hz); 13C-NMR (150 MHz, DMSO-d6): δ 21.3, 22.9, 28.3, 39.5, 40.1, 49.3, 52.0, 79.2, 128.2, 128.9, 133.2, 134.3, 142.0, 145.5, 155.5, 174.4; HRMS (ESI+) m/z: 390.1563 (Cl35), 392.1540 (Cl37) [M+Na]+.
Tert-butyl(S, E)-(4-methyl-1-(2-(2-nitrobenzylidene)hydrazineyl)-1-oxopentan-2-yl)carbamate (10j)
Light yellow powder; Yield: 80%; m.p. 164–168 °C; FT-IR (solid, cm−1): 2973, 1668, 1521, 1339, 1260, 1220, 1169, 1056; 1H-NMR (600 MHz, DMSO-d6): δ 8.38 (1H, s, NH), 8.67 (1H, s, CH=N), 8.06 (2H, t, J = 8.4 Hz), 7.80 (1H, t, J = 7.8, 7.2 Hz), 7.66 (1H, dd, J = 7.2 Hz), 4.91 (1H, br.s, NH-Boc), 4.03 (1H, dd, J = 8.4, 6.0 Hz, CH–NH), 1.71 (1H, m), 1.63 (1H, m), 1.54 (1H, m), 1.37 (9H, s), 0.92 (3H, d, J = 6.6 Hz), 0.87 (3H, d, J = 6.6 Hz); 13C-NMR (150 MHz, DMSO-d6): δ 21.2, 24.3, 28.2, 39.7, 49.4, 52.1, 78.1, 124.7, 127.4, 128.6, 130.5, 133.6, 138.6, 142.1, 148.2, 155.5, 174.7; HRMS (ESI+) m/z: 401.1812 [M+Na]+.
Tert-butyl(S,E)-(4-methyl-1-(2-(4-nitrobenzylidene)hydrazineyl)-1-oxopentan-2-yl)carbamate (10k)
Light yellow powder; Yield: 82%; m.p. 153–158 °C; FT-IR (solid, cm−1): 2973, 1668, 1521, 1339, 1260, 1220, 1169, 1056; 1H-NMR (DMSO-d6, 600 MHz): δ 8.10 (1H, s, NH), 8.36 (1H, s, CH=N), 8.32 (2H, d, J = 9.0 Hz), 7.96 (1H, d, J = 8.4 Hz), 7.92 (1H, d, J = 8.4 Hz), 4.97 (1H, br.s, NH-Boc), 4.07 (1H, dd, J = 8.4, 6.0 Hz, CH–NH), 1.72 (1H, m), 1.63 (1H, m), 1.52 (1H, m), 1.38 (9H, s), 0.97 (3H, d, J = 6.6 Hz), 0.90 (3H, d, J = 6.6 Hz); 13C-NMR (150 MHz, DMSO): δ 21.4, 22.9, 28.2, 39.7, 40.0, 49.3, 52.1, 78.1, 79.2, 124.0, 127.5, 140.8, 144.4, 147.7, 155.5, 174.8; HRMS (ESI+) m/z: 401.1478 [M+Na]+.
CA II inhibition assay
The total reaction mixture comprised of 20 µL (0.5 mmol/well) of test compounds (10% DMSO in total), then 140 µL of HEPES–Tris buffer (20 mmol, pH = 7.4), 20 µL of purified bovine erythrocyte CA II (1 mg/mL, 0.1 units/well) prepared in buffer, and finally 20 µL substrate 4-nitrophenyl acetate (4-NPA, 0.7 mmol) were added to attain final volume of 200 µL/well [59]. Enzyme (EC 4.2.1.1, Sigma-Aldrich, St. Louis, MO, USA) along with tested compounds were incubated for 15 min in a 96-well plate. Then, the reaction was started by adding 20 µL of substrate (4-nitrophenyl acetate) and continuously monitoring the rate (velocities) of product formation for 30 min with the intervals of 1 min, at 25 °C by using a microplate reader (Bio-Rad, Molecular Devices, CA, USA).
The percent inhibition was calculated from the formula given below
α-Glucosidase inhibition assay
This study was carried out by following the published methodology [55, 60] with slight modifications. In this assay, test compounds (20 µL/well) of different concentrations, α-glucosidase enzyme solution (20 µL/well) from Saccharomyces cerevisiae, and phosphate-buffered saline buffer (pH 6.8) (135 µL/well), were employed to a 96-well plate followed by incubation at 37 °C for 15 min and blank absorbance without the substrate was recorded. Changes in absorbance were recorded for 30 min at 400 nm after addition of substrate (25 µL).
The percent inhibition was calculated from the formula given below
Urease inhibition protocol
The in vitro assay of urease inhibition was performed in total 200 µL reaction by incubating 25 µL urease enzyme (1 unit/well) from Canavalia ensiformis (Jack bean), 5 µL of test compounds (0.5 mmol), and 55 substrate urea (100 mmol) at 30 °C for 15 min. To each vial, phenolic reagent (A) 45 μL and 70 μL of alkali reagent (B) were added [56]. Phenolic reagent (A) was a combination of 1% w/v phenol and 0.005% w/v sodium nitroprusside. While Alkali reagent (B) was of composition (0.5% w/v NaOH and 0.1% w/v NaOCl). To observe urease inhibition of new derivatives, method published by Weatherburn was used considering release of ammonia upon hydrolysis. Absorbance values were taken after 50 min using microplate reader (xMark™ Microplate Spectrophotometer, BIO-RAD). After 50 min, the increasing absorbance at 630 nm was measured in a microplate reader (xMark™ Microplate Spectrophotometer, BIO-RAD). Each reaction was performed in triplicate using a volume of 200 μL at end. As a standard urease inhibitor, we have used thiourea.
The percent inhibition was calculated from the formula given below
Molecular docking
Molecular docking of active inhibitors was performed on their respective targets by Molecular Operating Environment software (MOE, 2014.09). The X-ray crystallographic structures of CA II (PDB ID: 1BN1) and urease (PDB ID: 4H9M) were retrieved from RCSB Protein Data Bank. The homology model α-glucosidase was developed by using the primary sequence of yeast (S. cerevisiae) from UniProtKB (http://www.uniprot.org/) with accession ID = P53341. Homology search was built by SwissModel server using the crystallographic structure of S. cerevisiae isomaltase (3AJ7) as a template, which depicted 72.4% sequence similarity with our query. Later, model was refined by energy minimization (up to 0.1 RMS gradients). The refined structure was scrutinized by Ramachandaran plot which showed that 86.7, 12.3, 0.6, and 0.4% residues are present in most favored, allowed, generously allowed, and generously disallowed regions (Ala278 and Thr566, which are not a part of active site), respectively. Thus, the model is good to be used in docking studies. The 3D structures of protein targets were prepared for molecular docking by 3D protonate command of MOE, which add hydrogens and atomic charges on proteins using MMFF94x force field (RMSD gradient = 0.1 kcal × mol−1 Å−1). For docking, active site was defined around the 3 Å vicinity of the cocrystalized ligands of CA II and urease and the docked substrate of α-glucosidase model. The molecular docking of newly synthesized amino acid clubbed Schiff bases was carried out by applying the triangle matcher algorithm and London dG scoring function (MOE, 2014.09). The docking protocol was tested by redocking experiments for each target prior to the docking of newly synthesized inhibitors. The best docked solution of each compound against its specific target was selected on the basis of docking score and binding interactions [61].
References
El-Faham A, El Massry AM, Amer A, Gohar YM. A versatile synthetic route to chiral quinoxaline derivatives from amino acids precursors. Lett Pept Sci. 2002;9:49–54.
Khattab SN. Synthesis and biological activity of novel amino acid-(N’-benzoyl) hydrazide and amino acid-(N’-nicotinoyl) hydrazide derivatives. Molecules. 2005;10:1218–28.
Marsham PR, Wardleworth JM, Boyle FT, Hennequin LF, Kimbell R, Brown M, et al. Design and synthesis of potent non-polyglutamatable quinazoline antifolate thymidylate synthase inhibitors. J Med Chem. 1999;42:3809–20.
Okada Y, Tsukatani M, Taguchi H, Yokoi T, Bryant SD, Lazarus LH. Amino acids and peptides. LII. Design and Synthesis of opioid mimetics containing a pyrazinone ring and examinaiton of their opioid receptor binding activity. Chem Pharm Bull. 1998;46:1374–82.
Polyak F, Lubell WD. Rigid dipeptide mimics: synthesis of enantiopure 5- and 7-benzyl and 5, 7-dibenzyl Indolizidinone amino acids via enolization and alkylation of δ-Oxo α, ω-Di-[N-(9-(9-phenylfluorenyl)) amino] azelate esters. J Org Chem. 1998;63:5937–47.
Sun G, Uretsky NJ, Wallace LJ, Shams G, Weinstein DM, Miller DD. Synthesis of chiral 1-(2′-amino-2′-carboxyethyl)-1, 4-dihydro-6, 7-quinoxaline-2, 3-diones: α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionate receptor agonists and antagonists. J Med Chem. 1996;39:4430–8.
Xia Y, Yang ZY, Xia P, Bastow KF, Nakanishi Y, Lee K-H. Antitumor agents. part 202: novel 2′-amino chalcones: design, synthesis and biological evaluation. Bioorg Med Chem Lett. 2000;10:699–701.
Barrett D, Tanaka A, Harada K, Ohki H, Watabe E, Maki K, et al. Synthesis and biological activity of novel macrocyclic antifungals: acylated conjugates of the ornithine moiety of the lipopeptidolactone FR901469. Bioorg Med Chem Lett. 2001;11:479–82.
Hughes AB, editor. Amino acids, peptides and proteins in organic chemistry. Weinheim: Wiley-VCH; 2009.
O’Donnell MJ. Benzophenone Schiff bases of glycine derivatives: versatile starting materials for the synthesis of amino acids and their derivatives. Tetrahedron. 2019;75:3667–96.
Zafar H, Ahmad A, Khan AU, Khan TA. Synthesis, characterization and antimicrobial studies of Schiff base complexes. J Mol Struct. 2015;1097:129–35.
Cheng K, Zheng QZ, Qian Y, Shi L, Zhao J, Zhu HL. Synthesis, antibacterial activities and molecular docking studies of peptide and Schiff bases as targeted antibiotics. Bioorg Med Chem. 2009;17:7861–71.
Jarrahpour A, Khalili D, De Clercq E, Salmi C, Brunel JM. Synthesis, antibacterial, antifungal and antiviral activity evaluation of some new bis-Schiff bases of isatin and their derivatives. Molecules. 2007;12:1720–30.
Panchal PK, Pansuriya PB, Patel M. In-vitro biological evaluation of some ONS and NS donor Schiff’s bases and their metal complexes. J Enzym Inhib Med Chem. 2006;21:453–8.
Avupati VR, Yejella RP, Parala VR, Killari KN, Papasani VM, Cheepurupalli P, et al. Synthesis, characterization and in vitro biological evaluation of some novel 1, 3, 5-triazine–Schiff base conjugates as potential antimycobacterial agents. Bioorg Med Chem Lett. 2013;23:5968–70.
Sashidhara KV, Rosaiah JN, Bhatia G, Saxena J. Novel keto-enamine Schiffs bases from 7-hydroxy-4-methyl-2-oxo-2H-benzo [h] chromene-8, 10-dicarbaldehyde as potential antidyslipidemic and antioxidant agents. Eur J Med Chem. 2008;43:2592–6.
Vančo J, Švajlenová O, Račanská E, Muselík J, Valentová J. Antiradical activity of different copper (II) Schiff base complexes and their effect on alloxan-induced diabetes. J Trace Elem Med Bio. 2004;18:155–61.
Ambike V, Adsule S, Ahmed F, Wang Z, Afrasiabi Z, Sinn E, et al. Copper conjugates of nimesulide Schiff bases targeting VEGF, COX and Bcl-2 in pancreatic cancer cells. J Inorg Biochem. 2007;101:1517–24.
Gajula MR, Reddy YVR. Synthesis, characterization and in vitro biological evaluation of some new 1, 3, 5-triazine-bis-azomethine hybrid molecules as potential antitubercular agents. Eur J Chem. 2014;5:374–9.
Sinha D, Tiwari AK, Singh S, Shukla G, Mishra P, Chandra H, et al. Synthesis, characterization and biological activity of Schiff base analogues of indole-3-carboxaldehyde. Eur J Med Chem. 2008;43:160–5.
You ZL, Shi DH, Xu C, Zhang Q, Zhu HL. Schiff base transition metal complexes as novel inhibitors of xanthine oxidase. Eur J Med Chem. 2008;43:862–71.
Amin R, Krammer B, Abdel-Kader N, Verwanger T, El-Ansary A. Antibacterial effect of some benzopyrone derivatives. Eur J Med Chem. 2010;45:372–8.
Bharti S, Nath G, Tilak R, Singh S. Synthesis, anti-bacterial and anti-fungal activities of some novel Schiff bases containing 2, 4-disubstituted thiazole ring. Eur J Med Chem. 2010;45:651–60.
Ye XX, Chen ZF, Zhang AJ, Zhang LX. Synthesis and biological evaluation of some novel Schiff’s bases from 1, 2, 4-triazole. Molecules. 2007;12:1202–9.
Gacche R, Gond D, Dhole N, Dawane B. Coumarin Schiff-bases: as antioxidant and possibly anti-inflammatory agents. J Enzym Inhib Med Chem. 2006;21:157–61.
Toyota E, Sekizaki H, Takahashi Y-U, Itoh K, Tanizawa K. Amidino-containing Schiff base copper (II) and iron (III) chelates as a thrombin inhibitor. Chem Pharm Bull. 2005;53:22–26.
Cardile V, Panico A, Geronikaki A, Gentile B, Ronsisvalle G. In vitro evaluation of thiazolyl and benzothiazolyl Schiff bases on pig cartilage. Il Farmaco. 2002;57:1009–13.
Doddareddy MR, Cho YS, Koh HY, Pae AN. CoMFA and CoMSIA 3D QSAR analysis on N1-arylsulfonylindole compounds as 5-HT6 antagonists. Bioorg Med Chem. 2004;12:3977–85.
Jayashree B, Anuradha D, Venugopala K. Synthesis and characterization of Schiff bases of 2’-amino-4’-(6-chloro-3-coumarinyl)-thiazole as potential NSAIDs. Asian J Chem. 2005;17:2093.
Bhandari SV, Bothara KG, Raut MK, Patil AA, Sarkate AP, Mokale VJ. Design, synthesis and evaluation of antiinflammatory, analgesic and ulcerogenicity studies of novel S-substituted phenacyl-1, 3, 4-oxadiazole-2-thiol and Schiff bases of diclofenac acid as nonulcerogenic derivatives. Bioorg Med Chem. 2008;16:1822–31.
D’Ascenzio M, Guglielmi P, Carradori S, Secci D, Florio R, Mollica A, et al. Open saccharin-based secondary sulfonamides as potent and selective inhibitors of cancer-related carbonic anhydrase IX and XII isoforms. J Enzym Inhib Med Chem. 2017;32:51–59.
Gokcen T, Gulcin I, Ozturk T, Goren AC. A class of sulfonamides as carbonic anhydrase I and II inhibitors. J Enzym Inhib Med Chem. 2016;31:180–8.
Gul HI, Mete E, Taslimi P, Gulcin I, Supuran CT. Synthesis, carbonic anhydrase I and II inhibition studies of the 1, 3, 5-trisubstituted-pyrazolines. J Enzym Inhib Med Chem. 2017;32:189–92.
Ho YT, Purohit A, Vicker N, Newman SP, Robinson JJ, Leese MP, et al. Inhibition of carbonic anhydrase II by steroidal and non-steroidal sulphamates. Biochem Biophys Res Commun. 2003;305:909–14.
Ibrahim DA, Lasheen DS, Zaky MY, Ibrahim AW, Vullo D, Ceruso M, et al. Design and synthesis of benzothiazole-6-sulfonamides acting as highly potent inhibitors of carbonic anhydrase isoforms I, II, IX and XII. Bioorg Med Chem. 2015;23:4989–99.
Akocak S, Lolak N, Nocentini A, Karakoc G, Tufan A, Supuran CT. Synthesis and biological evaluation of novel aromatic and heterocyclic bis-sulfonamide Schiff bases as carbonic anhydrase I, II, VII and IX inhibitors. Bioorg Med Chem. 2017;25:3093–7.
Sağlık BN, Çevik UA, Osmaniye D, Levent S, Çavuşoğlu BK, Demir Y, et al. Synthesis, molecular docking analysis and carbonic anhydrase I-II inhibitory evaluation of new sulfonamide derivatives. Bioorg Chem. 2019;91:103153.
Aggarwal M, Boone CD, Kondeti B, McKenna R. Structural annotation of human carbonic anhydrases. J Enzym Inhib Med Chem. 2013;28:267–77.
Supuran CT. Carbonic anhydrases-an overview. Curr Pharm Des. 2008;14:603–14.
Webb S, Sherratt J, Fish R. Mathematical modelling of tumor acidity: regulation of intracellular pH. J Theor Biol. 1999;196:237–50.
Lee AH, Tannock IF. Heterogeneity of intracellular pH and of mechanisms that regulate intracellular pH in populations of cultured cells. Cancer Res. 1998;58:1901–8.
Montcourrier P, Silver I, Farnoud R, Bird I, Rochefort H. Breast cancer cells have a high capacity to acidify extracellular milieu by a dual mechanism. Clin Exp Metastasis. 1997;15:382–92.
Frazier ML, Lilly BJ, Wu EF, Ota T, Hewett-Emmett D. Carbonic anhydrase II gene expression in cell lines from human pancreatic adenocarcinoma. Pancreas. 1990;5:507–14.
Parkkila AK, Herva R, Parkkila S, Rajaniemi H. Immunohistochemical demonstration of human carbonic anhydrase isoenzyme II in brain tumours. Histochem J. 1995;27:974–82.
Parkkila S, Rajaniemi H, Parkkila AK, Kivelä J, Waheed A, Pastoreková S, et al. Carbonic anhydrase inhibitor suppresses invasion of renal cancer cells in vitro. Proc Natl Acad Sci USA. 2000;97:2220–4.
Achal V, Pan X. Characterization of urease and carbonic anhydrase producing bacteria and their role in calcite precipitation. Curr Microbiol. 2011;62:894–902.
Şentürk M, Gülçin İ, Beydemir Ş, Küfrevioğlu Öİ, Supuran CT. In vitro inhibition of human carbonic anhydrase I and II isozymes with natural phenolic compounds. Chem Biol Drug Des. 2011;77:494–9.
Khan KM, Wadood A, Ali M, Ul-Haq Z, Lodhi MA, Khan M, et al. Identification of potent urease inhibitors via ligand-and structure-based virtual screening and in vitro assays. J Mol Graph Model. 2010;28:792–8.
Qin Y, Cabral JM. Kinetic studies of the urease-catalyzed hydrolysis of urea in a buffer-free system. Appl Biochem Biotech. 1994;49:217–40.
Mobley H, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Mol Biol Rev. 1995;59:451–80.
Saleem M, Hareem S, Khan A, Naheed S, Raza M, Hussain R, et al. Dual inhibitors of urease and carbonic anhydrase-II from Iris species. Pure Appl Chem. 2019;91:1695–707.
Gao C, Li B, Zhang B, Sun Q, Li L, Li X, et al. Synthesis and biological evaluation of benzimidazole acridine derivatives as potential DNA-binding and apoptosis-inducing agents. Bioorg Med Chem. 2015;23:1800–7.
Gao H, Huang YN, Xu PY, Kawabata J. Inhibitory effect on α-glucosidase by the fruits of Terminalia chebula Retz. Food Chem. 2007;105:628–34.
Chiasson J-L. Prevention of type 2 diabetes: fact or fiction? Expert Opin Pharm. 2007;8:3147–58.
Rehman NU, Khan A, Al-Harrasia A, Hussain H, Wadood A, Riaz M, et al. New α-glucosidase inhibitors from the resins of Boswellia species with structure–glucosidase activity and molecular docking studies. Bioorg Chem. 2018;79:27–33.
Rehman NU, Khan A, Al-Harrasi A, Khiat M, Hidayat Hussain H, Wadood A, et al. Natural urease inhibitors from Aloe vera resin and Lycium shawii and their structural-activity relationship and molecular docking study. Bioorg Chem. 2019;88:102955.
Rehman N, Halim SA, Khan M, Hussain H, Yar Khan H, Khan A, et al. Antiproliferative and carbonic anhydrase II inhibitory potential of chemical constituents from Lycium shawii and aloe vera: evidence from in silico target fishing and in vitro testing. Pharmaceuticals. 2020;13:94.
Golmohammadi F, Balalaie S, Hamdan F, Maghari S. Efficient synthesis of novel conjugated 1, 3, 4-oxadiazole–peptides. N J Chem. 2018;42:4344–51.
Shank RP, Doose DR, Streeter AJ, Bialer M. Plasma and whole blood pharmacokinetics of topiramate: the role of carbonic anhydrase. Epilepsy Res. 2005;63:103–12.
Oki T, Matsui T, Osajima Y. Inhibitory effect of α-glucosidase inhibitors varies according to its origin. J Agric Food Chem. 1999;47:550–3.
Rehman NU, Rafiq K, Khan A, Ahsan Halim S, Ali L, Al-Saady N, et al. α-Glucosidase inhibition and molecular docking studies of natural brominated metabolites from marine macro brown alga Dictyopteris hoytii. Mar Drugs. 2019;17:666.
Acknowledgements
The authors are grateful to The Research Council (TRC), Oman, for funding through the project (BFP/RGP/CBS/18/011) and University of Nizwa, Oman, for the generous support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Rafiq, K., Khan, M., Muhammed, N. et al. New amino acid clubbed Schiff bases inhibit carbonic anhydrase II, α-glucosidase, and urease enzymes: in silico and in vitro. Med Chem Res 30, 712–728 (2021). https://doi.org/10.1007/s00044-020-02696-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-020-02696-0