Abstract
MADS-box transcription factors play important role in plant growth and development, especially floral organ identities. In our study, a MADS-box gene SlGLO1- tomato floral homeotic protein FBP1-like gene was isolated. Its tissue-specific expression profile analysis showed that SlGLO1 was highly expressed in petals and stamens. RNAi (RNA interference) repression of SlGLO1 resulted in floral organ abnormal phenotypes, including green petals with shorter size and aberrant carpelloid stamens. SlGLO1-silenced lines are male sterile. Total chlorophyll content was increased and chlorophyll biosynthetic genes were significantly up-regulated in SlGLO1-silenced petals and stamens. Furthermore, B-class genes expression analysis indicated that the repressed function of SlGLO1 led to the enhanced expression of TAP3 and the down-regulation of TPI in the petals and stamens, while the expression of TM6 was reduced in petals and increased in stamens and carpels of SlGLO1-RNAi plants. Additionally, pollen grains of transgenic lines were aberrant and failed to germinate and tomato pollen-specific genes were down-regulated by more than 90% in SlGLO1-silenced lines. These results suggest that SlGLO1 plays important role in regulating plant floral organ and pollen development in tomato.
Similar content being viewed by others
Introduction
ABC model of flower development was broadly accepted based on extensive genetic and molecular studies in several model plants1. A-class genes in the first floral whorl specify sepal identity. A- and B-class genes together are required for petal development, B- and C-class genes are both needed for stamen development and C-class genes in the fourth whorl specify carpel identity. Additional regulatory functions have been found, such as D-class genes that are necessary for ovule identity2 and E-class genes that are required for proper floral organ identity in the different whorls3. In Arabidopsis, there are two A-class genes APETALA1 (AP1) and APETALA2 (AP2), two B-class genes APETALA3 (AP3) and PISTILLATA) (PI) and one C-class gene AGAMOUS (AG). The E-class genes contain SEPALLATA1 (SEP1), 2, 3 and 43. All of these genes are MADS box genes except for AP2 and its homologues4. In tomato, MACROCALYX (MC) represents the A-class genes are involved in the development of sepals in the first whorl and in inflorescence determinacy5. Recent studies have suggested that the overexpression of SlFYFL represents longer sepals6. B-class MADS box genes specify petal and stamen identities in several core eudicot species, such as, Solanum lycopersicum GLOBOSA (SlGLO7) (syn. SlGLO1, LePI, TPIB8,9), Tomato PISTILLATA (TPI10) (syn. SlGLO27), Tomato MADS box gene 6 (TM6) (syn. TDR611,12) and Tomato APETALA3 (TAP3) gene (syn. STAMENLESS, SlDEF, LeAP313,10,14). Both TPI- and SlGLO1-silenced plants exhibit aberrant carpelloid stamens while petals are unaffected9. A mutation in TAP3 and the silencing of TM6 both results in the conversion of stamens into carpels and a more or less severe conversion of petals into sepals10. Tomato C-class gene TOMATO AGAMOUS 1 (TAG1) which specified stamen and carpel identities has been identified15. Tomato MADS box gene 5 (TM516) and Tomato AGAMOUS-LIKE gene 2 (TAGL2) (syn. TM2911,17), the two E-class genes, have been described on the basis of their expression patterns and down-regulated phenotypes in tomato.
In tomato, several mutants also exhibit partial or complete homeotic transformations in the second and third floral organ whorls. The sl-218, sl19 and green pistillate (gpi) (syn. pi-2, pistillate 220) are the most investigated mutants for which petal and stamen identity were affected. The petals appear as normal in the sl-2 mutant, while the stamens are twisted and distorted, bearing naked ovules. The sl mutant exhibits sepaloid petals and stamens being replaced by carpels in the third whorl19. The gpi mutant shows a strong and full homeotic transformation of petals into sepals and of stamens into carpels20. The study of Quinet et al.13 indicated that petal and stamen identity in tomato depends on gene-hormone interactions, as mediated by the TAP3 gene through identifying the mutations responsible for the sl phenotypes and investigating how TAP3 interacts with floral meristem identity genes and hormones in tomato.
Although function of many MADS box genes appear to be conserved among flowering plants, some homologous genes in different species have recruited novel functions through evolutionary mechanisms21. One key driving force in evolution is gene duplication22. Duplication of genes often provides a basis for diversification in either molecules or morphology. The diversification usually accompanies the definite fate of duplicated genes that either acquire a new role or undergo subfunctionalization. The Solanaceae possess multiple copies of DEF lineage and GLO lineage genes, allowing for diversification of function. The two duplicates for each lineage of B-class MADS-box genes were found to redundantly partition the role of the B function with a variable subfunctionalization process in Petunia, Nicotiana and Solanum9,10,23,24,25. Recently, specific PFGLO2 silencing in Physalis floridana show no homeotic variation but rather affect pollen maturation, providing insights into functional divergence of the GLOBOSA duplicates within the Solanaceae26. The B-class MADS box genes GLO and DEF together control the organogenesis of petals and stamens. Quantitative levels of B-class genes expression required to obtain petal tissue related to single versus duplicated genes. In plants where the B function genes are duplicated, there is higher redundancy but still quantitative changes in gene expression beyond a certain level also cause observable phenotypes. Changes in gene expression show expression thresholds required for organ formation27,28. In model plant Antirrhinum, thresholds of 11–15% of wild-type levels of expression of DEF or GLO are associated with development of recognizable petal tissue28, supporting the importance of quantitative gene expression levels for floral patterning and organ development.
Previous study by Geuten and Irish showed tomato SlGLO1-RNAi was weakly affected in stamen development in tomato cv. Micro-Tom9. Because Micro-Tom plants has three mutations at least in self-pruning (sp), dwarf (d) and miniature (putative mnt) genes29,30 based on its pedigree and the characterization of one of them, putative mnt, has still been not elucidated, the biochemical and molecular function of SlGLO1 still need further elucidation in tomato cv. Ailsa Craig (a common material for studying tomato). In our study, based on different genetic background we used tomato cv. Ailsa Craig, as the research material which is normal height compared with Micro-Tom. To comprehensively examine the diversification of functions of B-class genes in the Solanaceae, We have isolated SlGLO1 gene (a tomato floral homeotic protein FBP1-like gene) from wild-type tomato (Solanum lycopersicon Mill. cv. Ailsa Craig) flowers and examined its tissue-specific expression. An RNA interference (RNAi) expression vector targeting SlGLO1 was constructed and transformed into tomato plants. Silencing of SlGLO1 in tomato leads to floral organ abnormal phenotypes and defective pollen development, Furthermore, we analyzed the silencing lines at both physiology and molecular levels. This article enhanced our knowledge about the roles of SlGLO1 playing in diverse developmental processes.
Results
SlGLO1 isolation and expression pattern analysis
In order to explore the potential function of MADS-box genes playing in reproductive organ development in tomato, we isolated a tomato floral homeotic protein FBP1-like gene (namely SlGLO1) from wild-type tomato flowers based on a cDNA clone (GenBank accession no. XM_004245154). Nucleotide sequence analysis showed that SlGLO1 was consistent with the cDNA clone and contained an open reading frame (ORF) of 633 nucleotides and encoded 210 amino acid residues with an estimated molecular mass of 24.74 kDa. The search of conserved structure domains revealed that SlGLO1 protein possesses typical MADS domain and its theoretical isoelectric point is 8.4. The sequence alignment conducted by DNAMAN 5.2.2 program showed that tomato SlGLO1 shared 90.5% identity with the petunia PhFBP1 (Q03488.1) protein at the amino acid level. Phylogenetic and amino acid homology analysis showed that SlGLO1 was highly homologous to PhFBP1 (syn. PhGLO1) and NbGLO1 (Fig. 1A) and belongs to a very conservative MADS-box transcription factor family (Fig. 1B).
Sequence and expression analysis of SlGLO1 in WT.
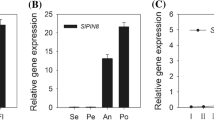
(A) Phylogenetic analysis of SlGLO1 and other MADS-Box proteins was performed by the neighbor-joining method, bootstrap analysis of 1,000 replicates. SlGLO1 is marked with asterisk. Accession numbers for other proteins are listed as follows: PhGLO1 (Q03488.1), NbGLO1 (HQ005417), AmGLO (Q03378.1), PhGLO2 (CAA49568.1), NbGLO2 (HQ005418), TPI (DQ674531), TM6 (X60759), NbTM6 (AY577817), PhTM6 (AAS46017.1), AmDEF (CAA44629.1), NbDEF (DQ437635), PhDEF (Q07472), TAP3 (DQ674532). (B) Multiple sequence alignment of SlGLO1 and other MADS-Box proteins. SlGLO1 is marked with asterisk. Identical amino acids are shaded in black and similar amino acids are shaded in gray. (C) The relative expression patterns of SlGLO1 in WT. RT, root; ST, stem; YL, young leaf; ML, muture leaf; SL, senescent leaf; F, flower; SE, sepal of flower in anthesis; IMG, immature green fruit; MG, mature green fruit; B, breaker fruit; B4, 4 days after breaker fruit; B7, 7 days after breaker fruit. The relative expression of SlGLO1 in the four-whorl floral organs (D) of WT. Se, sepal; Pe, petal; St, stamen; Ca, carpel. Each value represents the mean ± SE of three replicates.
To extend our understanding about the role of SlGLO1 playing in tomato growth and development, we examined its expression patterns in a wide range of tomato organs, including vegetative tissues such as young, mature and senescent leaves, stems and roots and reproductive tissues such as sepals, petals, stamens, carpels and fruits at different stages of development. Quantitative PCR technology was performed to analyze the expression of SlGLO1. The results showed that the expression level of SlGLO1 was higher in flower than other tissues (Fig. 1C). Expression analysis of the four-whorl flower organs in tomato showed that the SlGLO1 mRNA was highly accumulated in petals and stamens (Fig. 1D), which was coincidence with the study of Geuten and Irish where they show SlGLO1 is expressed in incipient petal and stamen primordia in early developmental stages by in situ hybridization9. These results indicate that SlGLO1 may have a specialized function in flower development process.
Silencing of SlGLO1 in tomato causes green petals with shorter size
To explore the physiological role of SlGLO1 in greater depth, we obtained 10 independent SlGLO1-silenced lines by RNA interference (RNAi). To confirm the repression efficiency of SlGLO1 in the transgenic lines, total RNA was extracted from flowers in WT and transgenic lines. Quantitative PCR (qPCR) results showed that SlGLO1 transcripts were significantly reduced in 10 independent transgenic lines compared with the wild type. Among which, the accumulation of SlGLO1 transcripts in lines 3 and 10 was remarkably reduced to roughly 2–10% of control levels, while only about 75–80% of transcript accumulation was repressed in the other lines (Fig. 2). Thus lines 3 and 10 with the strongest down regulation were selected for further investigation.
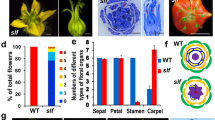
Wide-type tomato flowers usually exhibit green sepals, yellow petals and staminal cones (Fig. 3A). In the second whorl, wild-type petals contain sparse trichomes on the adaxial surface (Fig. 3B). The adaxial epidermal cells of these WT second whorl organs are small and arranged compactly (Fig. 3C). In contrast with the wild type, petals of SlGLO1-silenced lines showed green phenotype (Fig. 3D). Scanning electron microscopy suggested that SlGLO1-RNAi petals developed more trichomes (Fig. 3E) and sparse cells on the adaxial compared with WT (Fig. 3F). In addition, petals of the transgenic plants were also affected in terms of length (Fig. 3G). The length of transgenic petals showed shorter size compared with WT (Fig. 3H). These results suggest that SlGLO1-silenced lines develop flowers showing a classic loss-of-function phenotype of B-class genes consisting of a partial transformation of the petals into sepalloid structures.
Phenotypes of petals in SlGLO1-RNAi lines.
Wild type, (A–C); SlGLO1-RNAi, (D–F). Bars = 300 μm in (B,E), 60 μm in (C,F). (G) is digital photograph and (B,C,E,F), are scanning electron micrographs. (A) Open wild-type flower. (B,C) Wild-type adaxial petal surface. (D) Flower from SlGLO1-RNAi line. (E,F) SlGLO1-RNAi line adaxial petal surface. (G) Silencing of SlGLO1 results in smaller size petal. (H) Petal length of WT and transgenic lines. Error bars represent the standard error of the mean (n = 10). Asterisks indicate a significant difference (P < 0.05) between WT and transgenic lines.
Silencing of SlGLO1 produces green and aberrant stamens
Wide-type tomato stamen usually exhibit yellow staminal cone in which long anthers are joined by lateral hairs (Fig. 4A,B). In the third whorl, the proximal regions of the anthers contain more elongated epidermal cells on the adaxial surface (Fig. 4C), whereas the epidermal cells are irregular in the distal regions (Fig. 4D). By contrast, SlGLO1-silenced stamen showed green and twisty staminal cone (Fig. 4E), suggesting that stamens are partially transformed into carpel. Besides, transgenic stamen did not close, as a result of the absence of interweaving lateral hairs in the proximal region (Fig. 4F). Occasionally, the third whorl organs, instead of forming a cone as in the wild type, were splayed out and fully transformed carpelloid stamens (Fig. 4G). Scanning electron microscopy revealed that the adaxial epidermal cells were rounded and convex in the proximal region of transgenic anthers (Fig. 4H) and concave in the distal region (Fig. 4I). These results suggest that SlGLO1-silenced lines develop flowers showing a typical loss-of-function phenotype of B-class genes consisting of a partial or complete transformation of the stamens into carpel-like organs. In addition, no effects were observed in sepals, pistils and vegetative organs.
Phenotypes of stamens in SlGLO1-RNAi lines.
Wild type, (A–D); SlGLO1-RNAi, (E–I). Bars = 60 μm in (C,D,H,I). (A,B,E–G) are digital photographs and (C,D,H,I), are scanning electron micrographs. (A) Wild-type stamen. (B) Split wild-type stamen. (C) Wild-type adaxial stamen proximal end. (D) Wild-type adaxial stamen distal end. (E) Stamen from SlGLO1-RNAi line. (F) Stamens of SlGLO1-RNAi lines are unclosed. (G) Silencing of SlGLO1 results in carpelloid stamens. (H) SlGLO1-RNAi line adaxial stamen proximal end. (I) SlGLO1-RNAi line adaxial stamen distal end.
Chlorophyll content is increased and chlorophyll biosynthetic genes are significantly up-regulated in petals and stamens of SlGLO1-silenced lines
To determine whether the green petals phenotype represented a change in total chlorophyll content between control and SlGLO1 RNAi lines, total chlorophyll was extracted from petals of fully opened flowers. The total chlorophyll in petal of SlGLO1-RNAi lines increased approximately 3- to 4-fold compared with WT (Fig. 5A), accounting for the green color of petals in transgenic lines. Furthermore, we examined the expression of positive regulators of chlorophyll biosynthetic pathway in petals of wild-type and transgenic plants, such as DEFECTIVE CHLOROPLASTS AND LEAVES (SlDCL) (GenBank: U55219), which is required for both chloroplast development and palisade cell morphogenesis in leaf mesophyll31, Golden2-like1 (SlGLK1) (GenBank: JQ316460) and Golden2-like2 (SlGLK2) (GenBank: JQ316459)32, which are expressed in cotyledons, sepals and leaves. The results showed that these three chlorophyll biosynthetic genes were up-regulated significantly in petals of transgenic plants (Fig. 5B–D). Among which, SlGLK1 and SlGLK2 genes were up-regulated by approximate 6 folds.
Total chlorophyll content of petals and chlorophyll biosynthetic genes expression of wild-type and silenced-SlGLO1 lines.
(A) Total chlorophyll content of petals from wild-type and silenced-SlGLO1 lines. (B–D) respectively represents expression analysis of SlDCL, SlGLK1, SlGLK2 in petals of wild-type and transgenic lines. Each value represents the mean ± SE of three replicates. Asterisks indicate a significant difference (P < 0.05) between WT and transgenic lines.
Similarly, we assessed the total chlorophyll content in wild-type and SlGLO1-silenced stamens and found that total chlorophyll was also significantly increased in stamens of silenced lines (Fig. 6A). We further examined the expression of chlorophyll biosynthetic genes (SlDCL, SlGLK1 and SlGLK2) in wild-type and SlGLO1-silenced stamens. The data showed that expression level of these three genes were all increased by approximately 3 to 8-fold in stamens of transgenic plants (Fig. 6B–D). These results suggest that the increased expression of chlorophyll biosynthetic genes may affect the chlorophyll biosynthesis, thus form the green petals and stamens in transgenic lines.
Total chlorophyll content of stamens and chlorophyll biosynthetic genes expression of wild-type and silenced-SlGLO1 lines.
(A) Total chlorophyll content of stamens from wild-type and silenced-SlGLO1 lines. (B–D) respectively represents expression analysis of SlDCL, SlGLK1, SlGLK2 in stamens of wild-type and transgenic lines. Each value represents the mean ± SE of three replicates. Asterisks indicate a significant difference (P < 0.05) between WT and transgenic lines.
Transcriptional analysis of tomato floral organ identity genes in the transgenic lines
Given that the SlGLO1 gene was highly expressed at the second-whorl petal and third-whorl stamen and green petal and aberrant carpelloid stamen phenotype of SlGLO1 RNAi plant, we examined the expression of three known tomato B-class genes (TAP3, TPI and TM6) involved in the floral organ development in WT and two SlGLO1 RNAi lines. For TAP3 and TPI genes, their expression was nearly absent from first-whorl sepals but was strong in second- and third-whorl organs in wild-type floral organs. By contrast, TM6 expression was slightly weaker in mature tomato sepals and stamens but was strongly expressed in the petals and carpels (Fig. 7A–C). In SlGLO1 RNAi flowers, expression of TPI was reduced in the second-whorl organs, while TAP3 expression was slight high in second- and third-whorl organs, expression of TM6 was reduced in second-whorl organs and increased in the third- and fourth-whorl organs, suggesting that SlGLO1 may regulates directly or indirectly all three other B-class genes (Fig. 7A–C).
Expression Patterns of other floral organ identity genes in wild-type and RNAi Lines.
Se, sepal; Pe, petal; St, stamen; Ca, carpel. (A,B) respectively represents expression analysis of TAP3, TPI, TM6 (B-class genes) in wild-type and transgenic lines. (D–G) respectively represents expression analysis of MC (A-class gene), TAG1(C-class gene), TM5 and TAGL2 (E-class genes) in wild-type and transgenic lines. Each value represents the mean ± SE of three replicates. Asterisks indicate a significant difference (P < 0.05) between WT and transgenic lines.
To further characterize the potential function of SlGLO1 in flora organ development at molecular level, a set of tomato floral homeotic genes were examined in wide type and transgenic tomato mature floral organs. MC, one A-class gene which is involved in sepal development5, was significantly up-regulated in the second-whorl petals in SlGLO1 RNAi lines (Fig. 7D). Silencing of SlGLO1 enhanced remarkably transcription level of TAG1 (Fig. 7E), one C-class gene which has been identified for its role in the specification of stamen and carpel identities16. In addition, TM516 and TAGL2 (syn. TM2911,17), two E-class genes, were also up-regulated with various degrees in SlGLO1 RNAi lines (Fig. 7F,G).
SlGLO1-silenced tomato plants are male sterile
The development and function of the pollen is crucial important in the reproductive process of most plant species. In our study, SlGLO1-silenced tomato lines cannot produce fruit set (Fig. 8A). To examine whether or not the pollen collected from transgenic lines germinated, a pollen germination experiment was performed between WT and transgenic lines. The results showed that most of pollen grains in these transgenic lines were aberrant and uninflated and failed to germinate (Fig. 8B), suggesting pollen development is markedly affected by SlGLO1 silencing. Transgenic flowers which displayed transformation of stamens to carpelloid tissue were male sterile, but seeds developed normally when such flowers were manually crossed with wild-type pollen (Fig. 8C). This observation indicates that the ovules of such transgenic plants are functional.
Fruit phenotype, pollen germination and cross between WT and SlGLO1-RNAi lines.
(A) The SlGLO1-silenced lines are male sterile. (B) Pollen germination of WT and SlGLO1-RNAi lines. Pollen grains are germinated following 5 h culture at 25 °C in vitro containing liquid culture medium with 120 g/L sucrose, 120 mg/L boric acid, 4 mg/L gibberellin and 0.5 mg/L thiamine. Bars = 20 μm. (C) Fruit of the transgenic line produces normally seeds through manual crossing assay.
Further, we assessed the expression of tomato pollen-specific genes involved in the pollen development in WT and SlGLO1-silenced lines. SlCRK1 is one of the cysteine-rich receptor-like kinases, is important in pathogen defense and programmed cell death33; Pectin methylesterase inhibitor (SlPMEI) is key regulators of pectin methylesterase (PME)34; Pollen-specific receptor kinases gene LePRK3 is likely to be involved in perceiving extracellular cues during pollen tube growth35; Exogenous rapid alkalinization factor (SlPRALF) acts as a negative regulator of pollen tube elongation within a specific developmental window36 and LAT52 may play a role during germination or early tube growth37. The quantitative PCR results showed that these genes were all down-regulated by more than 90% in SlGLO1-silenced lines (Fig. 9), which suggest that silencing of SlGLO1 represses the expression of these genes, subsequently leads to pollen sterility.
Expression analysis of tomato pollen-specific genes in the pollen of wild-type and transgenic plants.
(A–E) respectively represents expression of pollen-specific genes SlCRK1, SlPMEI, LePRK3, SlPRALF and LAT52 in pollen of wild-type and transgenic lines. Each value represents the mean ± SE of three replicates. Asterisks indicate a significant difference (P < 0.05) between WT and transgenic lines.
Analysis of putative cis-acting elements within the SlGLO1 promoter
To date, many cis-elements have been reported for their critical roles in determining the tissue-specific expression profiles of plant genes38,39. In this study, a 1534-bp region upstream of the SlGLO1 start codon was analyzed to identify cis-acting elements, using the information in two public databases ( http://www.dna.affrc.go.jp/PLACE and http://intra.psb.ugent.be:8080/Plant CARE). Statistical analysis revealed that there were nine pollen-specific activation-related elements POLLEN1LELAT52 (Lat52, AGAAA)40 and eight late pollen gene g10-related elements (G10, GTGA)41 enriched in the promoter of SlGLO1 gene, which may be crucial for pollen development (Fig. S1).
Discussion
To date, it has been demonstrated that five classes of MADS-box genes (A, B, C, D and E) determine the identities of floral organ1,42. In the ABCDE model, petal and stamen structures are specified by the genes of B-class. The B-class floral homeotic genes has been reported to be implicated in flora organs development processes in many model plants, such as, Arabidopsis43,44, Antirrhinum45,46, Petunia26,47, Nicotiana9 and tomato8,9,10. For instance, the flowers of transgenic petunia plants in which PhFBP1 (syn. PhGLO1) expression is inhibited by a co-suppression approach, exhibit homeotic conversions of petals towards sepals and stamens towards carpels47. In petunia, loss of function of GREENPETALS (GP, syn. PhDEF) affects petal identity, whereas TM6 is required for stamen identity26,48. In Nicotiana, DEF-silenced plants display a marked transformation of petals into sepals and stamens into carpeloid structures49. Most Nicotiana flowers of NbGLO1-VIGS plants develop sepaloid tissues in petals and carpelloid tissues in stamens9. Here, we characterized the function of tomato SlGLO1 by RNAi-mediated gene silencing. Similar to the reports in Nicotiana and Petunia9,47, the silencing of SlGLO1 in tomato causes floral organ abnormal phenotypes, including green petals and stamens, shorter petals size, aberrant carpelloid stamens and defective pollen grains (Figs 3, 4 and 8), suggesting SlGLO1 plays an important role in diverse developmental processes.
RNAi repression of SlGLO1 resulted in green petals phenotype (Fig. 3D). Similarly, repression of PhGLO1 (syn. PhFBP1) in Petunia results in green tips at the end of the midveins of the petals47. Similar alteration of petals coloration phenotype is also observed in Petunia phglo1 mutant: the midveins of the petals become broader and greener, especially toward the edge of the corolla at the abaxial side of the petals24. Furthermore, Comparing the total chlorophyll contents of transgenic petals with wild type mature flowers, total chlorophyll contents in transgenic lines were all significantly increased, while that in wild-type remain at low level (Fig. 5A). Expression levels of chlorophyll biosynthetic genes SlDCL, SlGLK1 and SlGLK2, were increased in petals of transgenic plants compared with wild-type (Fig. 5B–D). These results suggest that the silencing of SlGLO1 leads to the activation of the chlorophyll biosynthesis and then results in green petals. Thence, we speculate that SlGLO1 may be involved in the activation of chlorophyll synthesis. Besides, the SlGLO1-silenced lines represented shorter petals size (Fig. 3G,H). A decrease in cell proliferation (Fig. 3F) in the SlGLO1-RNAi plants may partially explain the phenotype of the smaller petals size. Similar phenotype is observed in Petunia PhGLO1 (syn. PhFBP1) transgenic plants47, tomato TM6 RNAi Line10 and Nicotiana NbGLO1-VIGS plants9. However, previous studies showed that in tomato cv. Micro-Tom (a dwarf cultivar of tomato), silencing of SlGLO1 has no obvious effect on petal development9. The tomato cultivar Micro-Tom was produced for ornamental purposes by crossing Florida Basket and Ohio 4013-3 cultivars and displays a very dwarf phenotype with small and red ripened fruits. Based on its pedigree, the phenotype of Micro-Tom plants is due to at least three mutations: self-pruning (sp), dwarf (d) and miniature (putative mnt)29,30. The characterization of one of them, putative mnt, has still been not clarified. Recent study indicated that Micro-Tom seedlings in the dark present a weak photomorphogenic phenotype (disappearance of hook and cotyledon opening)50. We speculate that mutant tomato Micro-Tom results in a weak photomorphogenesis and thus some genes participating in light signal transduction pathways may be mutated. In our study, we used tomato cv. Ailsa Craig (a near-isogenic tomato line), as the research material which is normal height compared with Micro-Tom. This may explain the difference in phenotype observed as compared to the study previously mentioned.
In our study, SlGLO1 also impacts stamens development. The SlGLO1-silenced lines produced green stamens (Fig. 4E). Total chlorophyll content in transgenic lines was significantly increased, while that in wild-type accumulated little (Fig. 6A). Expression levels of chlorophyll biosynthetic regulatory genes including SlDCL, SlGLK1 and SlGLK2, were increased in stamens of transgenic plants compared with wild-type (Fig. 6B–D). Besides, transgenic stamen showed an unclosed phenotype (Fig. 4F). However, these phenotypes were not found in previous studies in Micro-Tom by Geuten and Irish9. Aberrant carpelloid stamens were also observed in transgenic lines (Fig. 4E,G). These results suggest that SlGLO1-silenced lines develop flowers showing a classic B-class gene loss-of-function phenotype consisting of a partial or complete transformation of the stamens into carpel-like organs.
Whether silencing of SlGLO1 affects other floral organ identity genes? We investigated the expression of these genes in WT and transgenic flowers. We performed a quantitative analysis of the transcripts of TM6 (syn. TDR611,12), TPI10 (syn. SlGLO27) and TAP3 gene (syn. SlDEF, LeAP310,13) in the WT and SlGLO1 RNAi lines (Fig. 7A–C). The results found that repressed SlGLO1 increased expression of TAP3 and reduced the expression of TPI in the second- and third-whorl organs of SlGLO1-RNAi plants, which was consistent with the study of Geuten and Irish9. In the study of Geuten and Irish, knockdown of SlGLO1 results in reduced second-whorl TPI/SlGLO2 expression and knockdown of TPI/SlGLO2 causes a reduction in SlGLO1 expression, suggesting cross-activation of TPI/SlGLO2 and SlGLO1 in the second-whorl organs. Besides, silencing of SlGLO1 down-regulated the expression of TM6 in the second-whorl organs of SlGLO1-RNAi plants. Therefore, these results suggest that SlGLO1 may regulate all three other B-class genes directly or indirectly. According to our results, silencing of SlGLO1 modified the expression profiles of TAP3, TPI and TM6 in flowers, indicating that the transgenic flower phenotype may be due to a coordinated misregulation of TAP3, TPI and TM6. Furthermore, to further characterize the potential molecular regulation mechanism of SlGLO1 in flora organ development, the other floral organ identity were tested in mature floral organs of wide type and transgenic tomato (Fig. 7D–G). Our results showed that silencing of SlGLO1 affected significantly the expression of A-class (MC), C-class (TAG1) and E-class (TM5 and TAGL2) genes. Silencing of SlGLO1 increased the transcript levels of MC, TAG1, TM5 and TAGL2 significantly, suggesting that SlGLO1 may be involved in the repression of these genes. Together, these results indicate that the SlGLO1 in tomato affects floral organ development possibly via regulating the expression of the other floral organ identity genes.
B-class MADS-box genes are also involved in the pollen development. For instance, specific PFGLO2 or/and PFTM6 silencing show a reduction in mature pollen in Physalis floridana25. In our study, we performed a pollen germination experiment between WT and transgenic lines. The result showed that most of pollen grains in these transgenic lines were aberrant and failed to germinate (Fig. 8B), suggesting silencing of SlGLO1 affects markedly pollen grains development and forms defective pollen grains. In addition, SlGLO1-silenced stamens were twisty and seem to become shorter, relatively. These alterations in stamens and pollen grains were susceptible for pollination. Therefore, the SlGLO1-silenced lines were male sterile. However, it is demonstrated that the ovules of transgenic lines were functional through manual crossing assay (Fig. 8C). Previous studies about two pollen-specific genes including SlCRK1 and SlPMEI showed that their promoters region possess lots of pollen-specific cis-acting elements, such as, the pollen-specific activation-related elements POLLEN1LELAT52 (Lat52, AGAAA) and late pollen gene g10-related elements (G10, GTGA)33,34. Kim et al.33,34 also confirmed promoters of SlCRK1 and SlPMEI have strong pollen-specific activity in the homozygous transgenic plants of tomato. Similarly, we analyzed putative cis-acting elements within the SlGLO1 promoter. The result showed that there were nine pollen-specific activation-related elements POLLEN1LELAT52 (Lat52, AGAAA) and eight late pollen gene g10-related elements (G10, GTGA) in the promoter of SlGLO1 gene (Fig. S1). This result demonstrates further that SlGLO1 may be involved in the pollen development. Furthermore, expression analysis of tomato pollen-specific genes in transgenic lines suggest that SlGLO1 may regulates directly or indirectly all these pollen-specific genes through combining with these genes promoters or interacting with other transcription factors.
In conclusion, this study concerned the morphological, physiological and molecular features of SlGLO1-RNAi transgenic tomato. Though a large amount of study focused on B-class genes, the specific biological functions of most plant B-class genes still remain to be elucidated. Clearly, understanding the role of SlGLO1 gene will not only extend knowledge of the biological function of B-class genes, but also provide new insight into exploring further the significance of SlGLO1 in regulating plant floral organ development.
Materials and Methods
Plant materials and growth conditions
In our study, we used tomato (Solanum lycopersicon Mill. cv. Ailsa Craig), a near-isogenic tomato line, as the wild type. The plants were grown in a greenhouse and managed routinely. Transgenic cultures grew under standard greenhouse conditions (16-h-day/8-h-night cycle, 25 °C/18 °C day/night temperature, 80% humidity and 250 μmol m−2 s−1 light intensity). Tomato plants of the first generation (T0) came from tissue culture were used in our study. For organ-specific expression profiling of SlGLO1, the tissues of roots, stems, leaves, sepals, flowers and fruits of different period from tomato plant were collected. The ripening stages of tomato fruits were divided following the method described by Xie et al.6. The four-whorl mature floral organs (sepal, petal, stamen and carpel) in the wild type and silenced lines were also collected. All plant samples were immediately frozen with liquid nitrogen, mixed and stored at –80 °C until further use.
Isolation and sequence analysis of SlGLO1
The primers: Forward, 5′ AAATAGGGGAAGTGTTTGAGC 3′ and Reverse, 5′ GAACCAAACCACATCACAAGA 3′ were used for cloning the coding region of SlGLO1. The PCR procedure was performed at 94 °C for 5 min followed by 35 cycles of 30 s at 94 °C, 30 s at 56 °C and 30 s at 72 °C and a final extension at 72 °C for 10 min. The amplified products were subcloned into pMD18-T vector (Takara, Dalian, China) and sequenced. The translation of SlGLO1 gene was used by the ExPASy translate tool, while grand average of hydropathicity (GRAVY) and theoretical molecular weight (Mw) were calculated using the ExPASy ProtParam tool ( http://www.expasy.org/). Sequence alignment was conducted using the DNAMAN 5.2.2 program. Conserved structure domains were annotated according to ScanProsite (scanprosite/). An unrooted neighbor-joining tree was constructed using the MEGA 5.05 software. To analyze putative cis-elements in the promoter region of SlGLO1 gene, promoter sequence (1500 bp regions upstream the 5′ end of the predicted ORF) of SlGLO1 gene was extracted from SGN database and searched against the promoter database PLACE ( http://www.Dna.Affrc.Go.jp/PLACE/signalscan.html)51 and plant CARE ( http://bioinformatics.Psb.ugent.be/webtools/plantcare/html/).
Construction of the SlGLO1 RNAi vector and plant transformation
In order to repress the expression of the SlGLO1 gene, an RNAi vector was constructed. A 395-bp specific DNA fragment of SlGLO1 was amplified from wild-type flower cDNA with the following primers: Forward, 5′-GGTACCAAGCTTAAATAGGGGAAGTGTTTGAGC-3′ and Reverse, 5′-CTCGAGTCTAGAGAACCAAACCACATCACAAGA-3′, which had been tailed with KpnI/HindIII and XhoI/XbaI restriction sites at the 5′ end, respectively. Then, the amplified products were digested with HindIII/XbaI and KpnI/XhoI and inserted into the pHANNIBAL plasmid at the HindIII/XbaI restriction site in the sense orientation and at the KpnI/XhoI restriction site in the antisense orientation. Finally, the double-stranded RNA expression unit, containing the cauliflower mosaic virus (CaMV) 35 S promoter, SlGLO1 fragment in the antisense orientation, PDK intron, SlGLO1 fragment in the sense orientation and OCS terminator, was purified and linked into the plant binary vector pBIN19 with SacI and XbaI restriction sites.
The generated binary plasmids were transformed into WT tomato cotyledon explants via Agrobacterium strain LBA44046952. Positive transgenic lines were screened for kanamycin (50 mg L−1) resistance. The transgenic plants were detected with primers NPTII-F (5′-GACAATCGGCTGCTCTGA-3′) and NPTII-R (5′-AACTCCAGCATGAGATCC-3′). The positive transgenic plants were selected and used for subsequent experiments.
Quantitative real-time PCR analysis
Total RNA was isolated using Trizol (Invitrogen, Shanghai, China) according to the manufacturer’s instructions and was treated with the Dnase-I (Promega, Beijing, China) for 30 min at 25 °C. The first strand cDNA synthesis was performed using M-MLV reverse transcriptase (Promega, Beijing, China) with oligo (dT)18 primer. Quantitative RT-PCR was carried out using the CFX96TM Real-Time System (Bio-Rad, USA). All reactions were performed using the SYBR Premix Ex Taq II kit (Takara) in a 10 μL total sample volume (5.0 μL of 2 × SYBR Premix Ex Taq, 1.0 μL of primers, 1.0 μL of cDNA and 3 μL of distilled, deionized water). Quantitative RT-PCR reactions were performed using a two-step method: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. To remove the effect of genomic DNA and the template from the environment, no-template control and no-reverse transcription control experiments were performed. Tomato CAC gene was selected as internal standard for organ-specific expression analysis53. The primers SlGLO1-Q-F and SlGLO1-Q-R (Table S1) were used to determine the expression levels of SlGLO1 in the wild type and transgenic lines. The analysis of relative gene expression levels were detected using the 2−ΔΔСT method54. Additionally, all samples were taken in three biological replicates and standard curves were run simultaneously. Primers used for quantitative RT-PCR are showed in Supplementary Table S1.
Extraction and quantitation of petal and stamen chlorophyll
Weighted 0.5 g fresh petals of wild-type and transgenic lines, pounded to powder with liquid nitrogen, extracted with 5 ml 80% acetone for 24 h in dark, centrifuged 5000 rpm for 15 min at 4 °C. The absorbance of the supernatant was measured at 645 and 663 nm in a PerkinElmer Lambda 900 UV/VIS/NIR spectrophotometer using above-mentioned 80% acetone as a blank. Total chlorophyll content were calculated using the formulas: Chl (mg g−1) = 20.29A645 + 8.02A66355. The chlorophyll of each sample was extracted and measured in triplicate. Chlorophyll contents of stamens were measured using the same method.
Scanning electron microscopy
Petals and stamens of wild type and transgenic plants at anthesis were fixed in 70% ethanol/acetic acid/formaldehyde (18:1:1, by volume; FAA) and subsequently dehydrated in an ethanol-water series. After dissection, the materials were dried and sputtered gold for scanning electron microscopy according to a previously published protocol56.
Statistics of petals length
In our study, we found that transgenic petals were shorter than the wild type. We measured the length of petals at the fully open flowers, at least 10 flowers per plant were measured.
Pollen collection, handling and germination
Pollen was collected in early July from mature floral organs of tomato plants cultivated in the glasshouses. Flowers were collected during the early morning and pollen was extracted within 1–2 h of collection as described by Karapanos et al.57. The pollen was then enclosed in vitro containing silica gel and stored for up to one week at 4 °C. Liquid culture medium for tomato pollen germination was composed of 120 g/L sucrose, 120 mg/L boric acid, 4 mg/L gibberellin and 0.5 mg/L thiamine58. After 5 hour incubation at 25 ± 1 °C in dark condition, 2 μl of pollen culture solution were placed on microscope slides and photographs were taken at 4 sites per slide, with the aid of an optical microscope (Olympus BX 40, Olympus Corp., Tokyo, Japan). A pollen grain was considered germinated when the pollen tube was equal or larger than the grain diameter (25–30 μm)59,60. The experiments were repeated three times.
Cross assay
Unopened flower buds (about 1 cm in length) from transgenic lines were sliced open lengthwise and emasculated with forceps. Mature pollen from the wild type tomato was transferred by brushing anthers onto the stigmas of transgenic lines. Pollinated flowers were labeled and bagged with small plastic bags to prevent uncontrolled cross-pollination.
Statistical analysis
The mean values of data were measured from three replicates and ‘Standard Error’ of the means was calculated. Data were analyzed by Origin 8.0 software and t test (SAS 9.2) was used for assessing significant differences among the means.
Additional Information
How to cite this article: Guo, X. et al. The tomato floral homeotic protein FBP1-like gene, SlGLO1, plays key roles in petal and stamen development. Sci. Rep. 6, 20454; doi: 10.1038/srep20454 (2016).
References
Coen, E. S. & Meyerowitz, E. M. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37 (1991).
Colombo, L. et al. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7, 1859–1868 (1995).
Pelaz, S., Tapia-Lopez, R., Alvarez-Buylla, E. R. & Yanofsky, M. F. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11, 182–184 (2001).
Theissen, G. et al. A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149 (2000).
Vrebalov, J. et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346 (2002).
Xie, Q. et al. Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Sci. Rep. 4, 4367 (2014).
Mazzucato, A., Olimpieri, I., Siligato, F., Picarella, M. E. & Soressi, G. P. Characterization of genes controlling stamen identity and development in a parthenocarpic tomato mutant indicates a role for the DEFICIENS ortholog in the control of fruit set. Physiol. Plantarum 132, 526–537 (2008).
Leseberg, C. H. et al. Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59, 2253–2265 (2008).
Geuten, K. & Irish, V. Hidden variability of floral homeotic B genes in Solanaceae provides a molecular basis for the evolution of novel functions. Plant Cell 22, 2562–2578 (2010).
de Martino, G. et al. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18, 1833–1845 (2006).
Busi, M. V. et al. MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 52, 801–815 (2003).
Pnueli, L. et al. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1, 255–266 (1991).
Quinet, M. et al. Transcriptional and hormonal regulation of petal and stamen development by STAMENLESS, the tomato (Solanum lycopersicum L.) orthologue to the B-class APETALA3 gene. J. Exp. Bot. 65, 2243–56 (2014).
Kramer, E. M., Dorit, R. L. & Irish, V. F. Molecular evolution of petal and stamen development, Gene duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149, 765–783 (1998).
Pnueli, L., Hareven, D., Rounsley, S. D. & Yanofsky, M. F. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6, 163–173 (1994a).
Pnueli, L., Hareven, D., Broday, L. & Hurwitz, C. The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6, 175–186 (1994b).
Ampomah-Dwamena, C., Morris, B. A., Sutherland, P., Veit, B. & Yao, J. L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 130, 605–617 (2002).
Sawhney, V. K. The role of temperature and its relationship with gibberellic acid in the development of floral organs of tomato (Lycopersicum esculentum). Can. J. Bot. 61, 1258–1265 (1983).
Gómez, P. et al. Stamenless, a tomato mutant with homeotic conversions in petals and stamens. Planta 209, 172–179 (1999).
Rasmussen, N. & Green, P. B. Organogenesis in flowers of the homeotic green pistillate mutant of tomato (Lycopersicon esculentum). Am. J. Bot. 80, 805–813 (1993).
Smaczniak, C., Immink, R. G. H., Angenent, G. C. & Kaufmann, K. Developmental and evolutionary diversity of plant MADS domain factors: insights from recent studies. Development 139, 3081–3098 (2012).
Rijpkema, A. S., Gerats, T. & Vandenbussche, M. Evolutionary complexity of MADS complexes. Curr. Opin. Plant Biol. 10, 32–38 (2007).
Rijpkema, A. S., Royaert, S., Zethof, J., van der Weerden, G., Gerats, T. & Vandenbussche, M. Analysis of the petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18, 1819–1832 (2006).
Vandenbussche, M., Zethof, J., Royaert, S., Weterings, K. & Gerats, T. The duplicated b-class heterodimer model: Whorl specific effects and complex genetic interactions in Petunia hybrid flower development. Plant Cell 16, 741–754 (2004).
Zhang, S. H., Zhang, J. S., Zhao, J. & He, C. H. Distinct subfunctionalization and neofunctionalization of the B-class MADS-box genes in Physalis floridana. Planta 241, 387–402 (2015).
Zhang, J. S. et al. Deciphering the Physalis floridana double-layeredlantern1 mutant provides insights into functional divergence of the GLOBOSA duplicates within the Solanaceae. Plant Physiol. 164, 748–764 (2014a).
Bey, M. et al. Characterization of antirrhinum petal development and identification of target genes of the class B MADS box gene DEFICIENS. Plant Cell 16, 3197–3215 (2004).
Manchado-Rojo, M., Delgado-Benarroch, L., Roca, M.J., Weiss, J. & Egea-Cortines, M. Quantitative levels of Deficiens and Globosa during late petal development show a complex transcriptional network topology of B function. Plant J. 72, 294 (2012).
Pnueli, L. et al. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989 (1998).
Meissner, R. et al. A new model system for tomato genetics. Plant J. 12, 1465–1472 (1997).
Keddie, J. S., Carroll, B., Jones, J. D. & Gruissem, W. The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 15, 4208–4217 (1996).
Powell, A. L. T. et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336, 1711–1715 (2012).
Kim, W. B. et al. Identification of a pollen-specific gene, SlCRK1 (RFK2) in tomato. Genes Genom. 36, 303–311 (2014).
Kim, W. B. et al. SlPMEI, a pollen-specific gene in tomato. Can. J. Plant Sci. 94, 73–83 (2014).
Covey, P. A. et al. A pollen-specific RALF from tomato that regulates pollen tube elongation Plant Physiol. 153, 703–715 (2010).
Kim, H. U. et al. New pollen-specific receptor kinases identified in tomato, maize and Arabidopsis: the tomato kinases show overlapping but distinct localization patterns on pollen tubes. Plant Mol. Biol. 50, 1–16 (2002).
Twell, D., Wing, R., Yamaguchi, J. & McCormick, S. Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 217, 240–245 (1989).
Annadana, S. et al. Cloning of the chrysanthemum UEP1 promoter and comparative expression in florets and leaves of Dendranthema grandiflora. Transgenic Res. 11, 437–445 (2002).
Faktor, O., Kooter, J. M., Dixon, R. A. & Lamb, C. J. Functional dissection of a bean chalcone synthase gene promoter in transgenic tobacco plants reveals sequence motifs essential for floral expression. Plant Mol. Biol. 32, 849–859 (1996).
Yu, D., Chen, C. & Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1540 (2001).
Rogers, H. J. et al. Functional analysis of cis regulatory elements within the promoter of the tobacco late pollen gene g10. Plant Mol. Biol. 45, 577–585 (2001).
Theiben, G. & Saedler, H. Floral quartets. Nature 409, 469–471 (2001).
Jack, T., Brockman, L. & Meyerowitz, E. M. The homeotic gene apetala3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697 (1992).
Goto, K. & Meyerowitz, E. M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560 (1994).
Schwarz-Sommer, Z. et al. Characterization of the Antirrhinum floral homeotic mads-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11, 251–263 (1992).
Tröbner, W. et al. GLOBOSA: A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11, 4693–4704 (1992).
Angenent, G. C., Franken, J., Busscher, M., Colombo, L. & van Tunen, A. J. Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J. 4, 101–112 (1993).
Tsuchimoto, S., Mayama, T., van der Krol, A. & Ohtsubo, E. The whorl-specific action of a petunia class B floral homeotic gene. Genes Cells 5, 89–99 (2000).
Liu, Y. et al. Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol. Biol. 54, 701–711 (2004).
Martí, E., Gisbert, C., Bishop, G. J., Dixon, M. S. & García-Martínez, J. L. Genetic and physiological characterization of tomato cv. Micro-Tom. J. Exp. Bot. 57, 2037–2047 (2006).
Higo, K., Ugawa, Y., Iwamoto, M. & Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300 (1999).
Chen, G. et al. Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 136, 2641–2651 (2004).
Expósito-Rodríguez, M., Borges, A. A., Borges-Pérez, A. & Pérez, J. A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8, 131 (2008).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time Quantitative PCR and the 2 - Δ Δ C T method. Methods 25, 402–408 (2001).
Arnon, D. I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1 (1949).
Irish, V. F. & Sussex, I. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2, 741–753 (1990).
Karapanos, I. C., Fasseas, K., Olympios, C. & Passam, H. C. Factors affecting the efficacy of agar-based substrates for the study of tomato pollen germination. J. Hortic. Sci. Biotech. 81, 631–638 (2006).
Wang F., Li W., Qin G. & Yuan Z. Screening of the optimal liquid culture medium for tomato pollen germination. Northern Hortic. 11, 143–145 (2008).
Dafni, A. & Firmage, D. Pollen viability and longevity: practical, ecological and evolutionary implications. Plant Syst. Evol. 222, 113–132 (2000).
Nepi, M. & Franchi, G. G. Cytochemistry of mature angiosperm pollen. Plant Syst. Evol. 222, 45–62 (2000).
Acknowledgements
This work was supported by National Natural Science Foundation of China (no. 31572129) and the Natural Science Foundation of Chongqing of China (cstc2015jcyjA80026) and Chongqing University Postgraduates’ Innovation Project (2015).
Author information
Authors and Affiliations
Contributions
G.C. and Z.H. designed and managed the research work and improved the manuscript. W.Y., X.Y., Z.Z. and J.Z. performed the experiments. X.G. performed the experiments and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Guo, X., Hu, Z., Yin, W. et al. The tomato floral homeotic protein FBP1-like gene, SlGLO1, plays key roles in petal and stamen development. Sci Rep 6, 20454 (2016). https://doi.org/10.1038/srep20454
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep20454
- Springer Nature Limited
This article is cited by
-
Hindered tomato reproductive development by altered brassinosteroid sensitivity1 mutant
Plant Growth Regulation (2022)
-
SlGT11 controls floral organ patterning and floral determinacy in tomato
BMC Plant Biology (2020)
-
Ectopic expression of miRNA172 in tomato (Solanum lycopersicum) reveals novel function in fruit development through regulation of an AP2 transcription factor
BMC Plant Biology (2020)
-
Overexpression of SlOFP20 affects floral organ and pollen development
Horticulture Research (2019)
-
B-class MADS-box TM6 is a candidate gene for tomato male sterile-1526
Theoretical and Applied Genetics (2019)