Abstract
Angiotensin-I converting enzyme (ACE) is a zinc dipeptidylcarboxypeptidase with two active domains and plays a key role in the regulation of blood pressure and electrolyte homeostasis, making it the principal target in the treatment of cardiovascular disease. More recently, the tetrapetide N-acetyl-Ser–Asp–Lys–Pro (Ac-SDKP) has emerged as a potent antifibrotic agent and negative regulator of haematopoietic stem cell differentiation which is processed exclusively by ACE. Here we provide a detailed biochemical and structural basis for the domain preference of Ac-SDKP. The high resolution crystal structures of N-domain ACE in complex with the dipeptide products of Ac-SDKP cleavage were obtained and offered a template to model the mechanism of substrate recognition of the enzyme. A comprehensive kinetic study of Ac-SDKP and domain co-operation was performed and indicated domain interactions affecting processing of the tetrapeptide substrate. Our results further illustrate the molecular basis for N-domain selectivity and should help design novel ACE inhibitors and Ac-SDKP analogues that could be used in the treatment of fibrosis disorders.
Similar content being viewed by others
Introduction
Angiotensin-1-converting enzyme (ACE, EC 3.4.15.1) is a zinc peptidase and a key enzyme in cardio-vascular physiology1,2. ACE is a central component of the Renin-Angiotensin-Aldosterone System (RAAS), wherein it converts angiotensin I into the vasoactive peptide hormone angiotensin II (AngII)3,4. Interestingly, ACE consists of two catalytically active domains5,6 (referred to as the N- and C-domains based on location on the polypeptide chain) that, while very similar in sequence and structural topology, display differences in substrate processing abilities6,7,8. ACE is also able to cleave the vasodilatory peptide bradykinin and therefore enhance the blood pressure response9. In addition to the cleavage of vasopeptides, ACE is able to cleave a variety of peptides that are outside of its blood pressure role. While these are not the most well-known substrates for ACE, many of these have important physiological roles and therefore warrant careful study.
N-acetyl-Ser-Asp-Lys-Pro (Ac-SDKP) is specifically cleared by ACE10. This process has been reported to be carried out predominantly by the N-domain7. Ac-SDKP was originally discovered as a potent inhibitor of bone marrow derived stem cell differentiation through inhibition of the G1-S phase transition11,12. This suggested a possible role of Ac-SDKP as a co-treatment to reduce the treatment associated severity of some chemotherapies due to this peptide being able to keep stem cells in the quiescent state13. Later findings indicated that Ac-SDKP also possesses potent anti-fibrotic and anti-inflammatory effects in numerous tissues including heart, liver, kidney and lung14,15,16,17,18,19,20,21,22. Ac-SDKP is able to blunt the Smad signalling effects of transforming growth factor-β, a prominent fibrosis marker23,24. Thus, Ac-SDKP appears to have a prominent physiological role.
The N- and C-domains of ACE have been shown to display negative co-operativity in substrate hydrolysis25,26,27. While this is observed with many synthetic and naturally occurring peptides, not all substrates displayed such an effect27. To date, no studies have been performed on the physiologically relevant substrate Ac-SDKP.
The X-ray crystal structures of both individual homologous ACE domains have been previously determined28,29 and more recently the structure of N-domain in complex with the phosphinic tripeptide RXP40730 highlighted the structural requirements for domain specific inhibition. Furthermore, the structure of the C-domain in complex with AngII31 gave the first insight into the mechanism of peptide recognition by ACE and revealed the regulatory role of AngII on the enzyme’s activity. Studying the mechanisms of peptide recognition for each domain of ACE should therefore help our understanding of substrate selectivity and lead to the design of better inhibitors.
The purpose of this study was to understand the kinetic nature of Ac-SDKP hydrolysis and the structural basis for its interaction with the N-domain of ACE. The high resolution (1.8 Å) X-ray crystal structures of the N-domain in complex with the two products of Ac-SDKP cleavage, the dipeptides Ac-SD and KP, were determined. The extent of domain interaction during substrate hydrolysis and the domain selectivity of these processes were further assessed using a fluorescamine assay. Together the data reported here allowed the construction of a comprehensive model of N-domain selective substrate recognition and peptidase activity.
Results
Structure of the Ac-SDKP:N-domain complexes
Co-crystallisation experiments were carried out to assess the interaction between the N-domain and tetrapeptide Ac-SDKP. Surprisingly, two crystal forms were obtained (Table 1). One form presented the same cell dimensions as the previously described minimally glycosylated N-domain30 with 2 chains per asymmetric unit in P1. The structure at 1.8 Å showed electron difference density within the catalytic channel (Fig. 1A), which was interpreted as the Ac-SD product, resulting from the enzymatic reaction. The structure from the second crystal form was solved at a similar resolution, in space group P1 with larger cell dimensions, fitting 4 molecules per asymmetric unit. No significant conformational changes were observed for the N-domain, with only the expected N-terminal hinge region showing signs of disorder in one of the four molecules. However, this crystal form did present an alternative and unambiguous difference electron density at the active site, with the map clearly showing the dipeptidyl carboxypeptidase product of Ac-SDKP hydrolysis (KP, Fig. 1A). The two different fragments occupy a similar position within the S1′-S2′ sub-pockets while only interacting with the zinc ion through water-mediated interactions.
Mechanism of Ac-SDKP fragments binding to N-domain ACE.
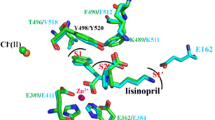
(A) Portions of the electron density map at the site of the bound peptides with left, Ac-SD and right, KP. The maps were generated using REFMAC561 and correspond to the difference weighted 2mFo-DFc density map, contoured at 1.0σ level, in which the peptide atoms were omitted. N-dom in cyan (interacting residues in stick representation), bound dipeptides in grey stick, catalytic zinc ion and water molecules as grey and red spheres respectively. (B) Schematic of peptide binding to N-ACE. Interactions were calculated with LigPlot+64. (C) Superposition of the dipeptide Ac-SD and KP structures. Ac-SD and KP are presented in black and grey sticks, respectively.
The two peptides have their C-terminal end strongly anchored by hydrogen bonds with residues Lys489, Tyr498 and Gln259 forming the S2′ site, amino acids previously shown to be key anchoring residues (Fig. 1B). The aromatic Phe435 and Phe505 also stabilise this end of the peptide by hydrophobic interaction with the Pro or Asp side chains of the substrate. The acidic group of the substrate Asp showed signs of flexibility (Fig. 1A), but may make further water-mediated contacts with the backbone of surrounding enzyme residues Glu431, Thr358 and the Ser260 hydroxyl group. The main difference between the two peptidic substrate fragments resides at their first residue due to the presence of the N-acetyl group on the serine. With the Lys-Pro fragment, the lysine backbone follow a similar bonding pattern to what was observed in the C-domain:Ang II structure, with its oxygen being stabilised by multiple hydrogen bonds with enzyme residues His331, His491 and Tyr501 (Fig. 1B) and its amino group interacting with the catalytic site. The long side chain can make further polar connection directly with Thr358 and water bridges with Asp354, Gln355 and Ser260. On the other hand, the N-acetyl-serine (AcS), of the Ac-Ser-Asp fragment, presents a different orientation. Its Hydroxyl side chain makes direct electrostatic contacts with His331, His491 and Tyr501, as well as a water-mediated interaction with Zn2+. The AcS amino group interacts with a single water molecule itself connected to Thr358 (main chain) and Asp393. The acetyl moiety may be stabilised in the small hydrophobic area composed of Ala332 and Thr358.
Kinetics of Ac-SDKP hydrolysis by ACE
Kinetic analysis was performed to assess the selectivity and possible co-operativity of the N- and C-domains in Ac-SDKP processing. Individual N-domain and C-domain constructs (Fig. 2A) displayed comparable Km values of 199.6 μM and 138.2 μM, respectively (Table 2). The individual N-domain had a turnover rate that was approximately 3-fold higher than that of the C-domain, confirming the N-domain preference for Ac-SDKP, albeit lower than original reports.
Analysis of ACE catalytic efficiency on Ac-SDKP.
(A) Details of ACE constructs. The wild-type ACE (sACE) contains a signal peptide (light grey), the two homologous ectodomains (N-domain in cyan and C-domain in blue), the transmembrane domain (dark grey) is also indicated. Inactive domains are crossed over. (B) Catalytic efficiency of ACE on Ac-SDKP.
Wild-type sACE, with both domains active, displayed a Km value of 239.5 μM while retaining a kcat value (per mole active site) similar to the individual N-domain. Thus, the combined domain turnover rate of sACE (25.2 s−1) was slightly higher than the sum of the kcat values for the individual domains.
In order to further delineate domain selectivity and assess the activity of domain active sites in full length sACE, C-sACE and N-sACE enzymes (these constructs are full length sACE molecules where one of the two active sites has been inactivated by mutation) were employed (Fig. 2A). Interestingly, the Km values of the two domain (X-sACE) enzymes were approximately 3-fold higher than their single domain counterparts and twice that of wild-type sACE, again showing an effect of domain interaction on substrate on-off rates. The kcat values were approximately 2-fold higher in C-sACE and N-sACE constructs than their respective single domains, which may be indicative of an inter-domain influence on substrate release. Overall, this resulted in a similar catalytic efficiency between the individual N-domain and the same domain in full length form (N-sACE), whereas C-sACE was approximately half that of its single domain counterpart (that is, the individual C-domain, Fig. 2B).
So as to better appreciate the role of the C-domain in the full length molecule, the CC-sACE molecule, comprising two active C-domains connected with the interdomain linker (Fig. 2A), was characterised in terms of Ac-SDKP kinetics. This enzyme showed an increased Km compared to wild-type sACE but in a similar range to that of the other full length constructs (Table 2). Interestingly, the turnover rate per unit active site of CC-sACE (19.4 s−1) showed a two-fold increase compared to C-sACE (8.2 s−1). The resulting improved catalytic efficiency therefore suggests a role of the N-domain in lowering C-domain activity in the full length somatic enzyme towards the substrate.
Modelling of the Ac-SDKP substrate interaction with ACE
The crystal structure of the N-domain with the KP fragment gave useful insight into the peptide binding mechanism and allowed us to propose a model for the full substrate interaction (Fig. 3). The lysyl-proline-bound structure was used as the basis of the superposition with the C-domain:AngII (inhibitory peptide resulting from angiotensin I cleavage) structure. The C-terminal end of the Ac-SDKP peptide presents a conserved peptide binding mechanism between the two domains and was thus the best anchor on which to build the complete model (Fig. 4). The C-domain:AngII structure also shows many of the same residues between the two domain interacting along the peptide backbone, so that the rest of the Ac-SDKP peptide was docked based on this mechanism. The Ac-SDKP models for both domains were then refined using the Rosetta FlexPepDock server (which uses a Monte-Carlo with minimization approach)32. The tetrapeptide fits well within the narrow catalytic channel by interacting with ACE domain active sites through its backbone from P2′ to P1 and additional contacts with the P2 Ser and the corresponding N-domain subsite. The acetyl group can be easily accommodated within the wide S2 pocket (Figs 3 and 4). In order to assess the preference of Ac-SDKP for the N-domain, the models were compared to the crystal structures of the N-domain with RXP 407 (an N-domain specific inhibitor33) and C-domain with AngII (which shows C-domain selectivity34) (Fig. 4). Previous structural analysis30 has highlighted the high degree of conservation between the two domains within the S’ binding sites (Table 3). This is important since it highlights the conservation of essential residues for a general role in substrate binding. The specificity for Ac-SDKP appears to reside mostly at the S2 subsite. This subsite, while cavernous, provides contacts in the N-domain that could explain partial N-domain preference of this substrate. There, the hydroxyl group of Ac-Ser is within hydrogen bonding distance of conserved residue His388. The non-conserved Tyr369 can also provide further electrostatic potential compared to the C-domain Phe391 and has been identified previously as a key residue for ligand selectivity35,36. Furthermore, the polar (N-domain) Thr496 may provide additional interaction with the substrate Asp side chains compared to the C-domain enzyme residue Val518. In the P1′ pocket, our structure with Lys-Pro shows that the dipeptide’s ε-amino group can interact with Thr358 which is also not conserved in C-domain ACE (Val380). It should be added that water molecules are expected to play a significant role in peptide binding, as illustrated by several water-mediated bonds in the crystal structures presented here (Fig. 1), but were not included in the models for practical reasons.
Modelling of Ac-SDKP binding to ACE.
(A) Ac-SDKP was manually docked within the catalytic channel of N-dom based on the KP complex structure. The model was refined using the Rosetta FlexPepDock server32. N-dom is shown in cyan and Ac-SDKP in grey stick. (B) Close up view of Ac-SDKP within the catalytic channel, surface charged was calculated with the PyMol/APBS tool65. (C) Comparison of Ac-SDKP models bound to N- and C-domain ACE. N-dom in cyan, C-dom in blue; Ac-SDKP in grey and purple in for the N- and C-domain models, respectively. Residues involved in substrate interaction are labelled with their protein respective colour.
Comparison of domain specific substrate and inhibitors in ACE.
(A) Model of Ac-SDKP binding to N-domain ACE (as above). The catalytic channel is represented with a transparent surface calculated with the program HOLLOW66 (B) Model of Ac-SDKP binding to C-domain ACE, with C-domain ACE in blue and Ac-SDKP in purple. (C) Crystal structure of the N-domain specific RXP 407 inhibitor bound to N-domain ACE (PDB 3NXQ)30, with RXP 407 in pink. (D) Crystal structure of C-domain ACE in complex with AngII (PDB 4APH)31; with AngII in yellow. Non-conserved residues of the catalytic channel are indicated by a framed label. The residues involved in N-domain specificity30 are labelled in red.
ACE belongs to the gluzincin family of metalloproteases and catalysis is expected to follow a general base-type mechanism. The structures determined here allow us to more accurately predict the substrate hydrolysis mechanism employed by ACE. The cleavage of Ac-SDKP is predicted to happen after displacement of the zinc-bound water molecule which then attacks the scissile carbonyl bond to form an oxyanion (Fig. 5). The nucleophilic attack is enhanced with the water being coordinated by the active site Glu362. The resulting intermediate is likely stabilised by Tyr501. As mentioned previously, chloride has an important effect on ACE activity which is also substrate-dependent in the C-domain37. The function of chloride in N-ACE is less understood, but it is unlikely to affect substrate specificity. However, a role for the direct involvement of the conserved chloride-coordinating Arg500 should not be excluded as it is close to the active site and seen making a water-mediated interaction with Tyr501.
Proposed model for the catalytic mechanism of Ac-SDKP hydrolysis by N-domain ACE.
(A) Model of Ac-SDKP (grey) bound to N-domain ACE (cyan) prepared as above. N-domain ACE residues involved in the zinc-mediated activity and stabilisation of the potential transition state intermediates are shown as stick. The water involved in the nucleophilic reaction is shown in red (sphere). (B) Potential role of the chloride ion in activity. Chloride ion is shown in green, with potential hydrogen bond and water mediated-interactions with the catalytic site highlighted as green dashes.
Discussion
Ac-SDKP is an important substrate of ACE that, through specific inhibition, could lead to beneficial therapeutic effects in the treatment of fibrosis disorders. Therefore, a thorough enzymatic and structural analysis would be useful in assisting our understanding of the requirements for hydrolysis of tetrapeptide substrates. Previous work involving Ac-SDKP kinetics involved the use of paper chromatography or HPLC, neither of which allow for medium to high throughput kinetic characterisation7,38,39. A modified fluorescamine assay was previously applied to ACE substrates of N-acetyl-AngI and substance P40,41, as well as to other enzyme substrates42,43. Recently, our group employed this derivatising agent to assess simple enzyme activity of a newly identified polymorphism44. Here we employed fluorescamine to assess the detailed kinetics of Ac-SDKP processing by ACE.
The N-domain selectivity of Ac-SDKP processing was found to be markedly reduced compared to some previous reports. Literature reports differ on the degree of catalytic efficiencies and selectivities of Ac-SDKP hydrolysis7,38. The Km values determined in this study are generally higher than the original publication presenting Ac-SDKP hydrolysis as a highly N-domain specific process7 also determined Km values that differ depending on the enzymatic construction and methodology employed38. The selectivity of Ac-SDKP reported here for the individual domains (N-domain and C-domain) is only 2-fold. Deddish et al. also found a decrease in selectivity, with Ac-SDKP having approximately 8-fold N-selectivity38. In this study, the C-domain enzymes had a considerably higher turnover rate than previously described, thus decreasing the overall selectivity. Increased C-domain activity could be due to the assay conditions used here that differs to other studies, where we sought to best mimic the NaCl physiological context (generally 100 mM NaCl). In the original publication of the N-selectivity of Ac-SDKP processing, despite showing that maximal activity of both domains occurs at 100 mM NaCl, a concentration of 50 mM NaCl was used in the assay buffers7. Indeed, lower chloride concentrations seemed to result in increased preference for N-domain activity38. Thus, a contributor to higher C-domain activity could possibly be due to the use of an optimal chloride concentration for the C-domain6,37. Interestingly, the in vivo N-domain inactivation results in a 4–7 fold increase in plasma Ac-SDKP concentrations22,45,46. This indicates that the selectivity observed in the current report is consistent with more physiologically relevant observations. Importantly, while it seems that the selectivity of Ac-SDKP for the N-domain is less than originally reported, the in vivo studies state that the modest increase in serum peptide levels through the use of N- domain selective inhibitors can still possibly provide the desired therapeutic effects.
The structural basis of Ac-SDKP’s preferential hydrolysis by the N-domain was investigated by analysing the molecular interactions of the N-domain with the peptide. The two peptides occupied the same area of the S′ catalytic sub-pocket and displayed noticeable differences in their mode of binding within the S1′ site while showing common features in carboxy-terminal ends binding within with the S2′ sites. This common anchoring in the S2′ site is homologous to that of peptide binding in C-domain ACE31 and of the peptide-based N-domain inhibitors RXP 40733 and 33RE35. Surprisingly, the substrate binding pockets of N-domain ACE do not present any changes at all between the two structures and can accommodate the two very different peptides without any conformational rearrangement (Fig. 1). Interestingly, phosphinic inhibitors were recently showed to fit to the conserved substrate binding pocket of the two domains of ACE and its Drosophila homologue (AnCE) with the enzymes showing little plasticity47. This unspecific mechanism of peptide recognition may explain the wide range of substrates cleaved by this enzyme.
Using the structural information above, we were able to generate a model for Ac-SDKP binding into the enzyme active site and revealed the importance of the S2 site in providing possible unique interactions for preferential processing. Further, it suggests a minimal set of amino acids that are responsible for enzyme selectivity that, if properly exploited, could result in domain selective inhibitors and/or drugs. Some of these residues have been implicated in selective inhibitor binding35,36 and thus this study also serves the prioritisation of optimal interactions with this site. The N-acetyl group is easily accommodated in the cavernous S2 site with the primary contacts in this region being carried out by the P2 Ser of the substrate. Our structural model provides visual insight for observations of other kinetic studies, whereby selectivity of Ac-SDKP analogues could be influenced by changing the unprimed substrate amino acids while having benzoyl derivatives (considerably larger than an acetyl group) on the substrate N-terminus8,48. Taken together, this suggests a limited role of the acetyl group in contributing to preferential processing.
The two active sites within human and bovine somatic ACE exhibit negative co-operativity with certain, but not all, substrates26,27,49,50. The current study shows that the catalytic efficiencies of sACE in Ac-SDKP hydrolysis appears overall to be additive between the two domains. Similar to other kinetic observations with physiological substrates, such as Ang(1–9) and BK, the degree of co-operativity between domains in Ac-SDKP processing is considerably less than many synthetic peptides observed previously27,51. The sACE CC-domain enzyme possessed a catalytic ability per active site similar to the individual domains, indicating an additive effect between the two C-domains. The analysis of the CC-sACE enzyme also indicated that the N-domain has some inhibitory effect on the C-domain in the somatic isoform, an effect noticed in both kinetics of fluorogenic substrates and in the proteolytic shedding of ACE50. It seems plausible that such an effect is relevant in the physiological setting as well, possibly either by affecting substrate access, product release, or by reducing the overall dynamics of the domain required for efficient processing.
In summary, the detailed molecular interactions between Ac-SDKP and ACE were characterised. We have found that the tetrapeptide is indeed preferentially processed by the N-domain although this preference is less than some previous reports. The observations of the dipeptide fragments resulting from Ac-SDKP cleavage provided the structural basis to offer a complete model of domain preferential substrate hydrolysis, from peptide recognition by specific domain amino acids to a detailed catalytic mechanism. Further, we emphasize important sets of amino acids: conserved anchoring residues that appear to play a role in ACE substrate binding generally and N-domain unique residues that could provide contacts for preferential positioning. Domain interactions were also shown to influence ACE activity with an observed inhibitory effect of the N-domain over C-domain cleavage of Ac-SDKP. Overall, these findings enhance our understanding of peptide hydrolysis generally and should help the development of N-domain selective inhibitors that could be useful in the treatment of tissue injury and fibrosis.
Methods
Enzymes
For single, soluble enzymatic domains: a modified tACE construct, tACEΔ36NJ, that lacks the transmembrane region and unique 36 amino acid N-terminus (and therefore identical to the sACE C-domain; hereon referred to as C-domain) had been generated previously52. A soluble form of the N-domain, consisting of amino acids 1 to 629 of somatic ACE (hereon referred to as N-domain), in vector pECE was a kind gift from Dr. Sergei Danilov UIC, Chicago) and was cloned into sequencing vector pBlueScript SK+ (Invitrogen) as previously described29,53.
The full length domain knock outs of sACE were a kind gift from Dr. Vincent Dive6. A full length molecule with only an active N-domain active site (N-sACE) has zinc binding residues His361 and His365 converted to Lys. Similarly a full length molecule with only an active C-domain active site (C-sACE) possesses conversions of the corresponding His residues. The CC-sACE molecule consists of two C-domains joined by the sACE inter-domain linker region and was constructed as described previously50. All full length constructs have the complete signal, transmembrane and stalk region corresponding to wild type sACE.
Expression and purification of enzymes
All enzymes were expressed in Chinese Hamster Ovary (CHO) cells using standard tissue culture approaches as formerly described54. All enzymes were purified using lisinopril-Sepharose affinity chromatography as previously described55 with the following considerations: single domains were isolated from the harvest medium while full length enzymes were purified from whole cell triton lysates. N-domain constructs required the addition of 800 mM NaCl to medium/lysates for effective purification56. ACE activity was detected using the substrate Cbz-Phe-His-Leu (Z-FHL)57 and pooled enzyme dialysed twice with 2 litres of 5 mM Hepes (pH 7.5). All enzymes were concentrated and stored at 4 °C in 50 mM Hepes (pH 7.5). Enzyme integrity and purity were assessed by SDS-PAGE and subsequent Coomassie staining.
In order to determine loss enzymatic activity due to storage, specific activities were calculated immediately after purification assuming that the enzyme which is eluted off the lisinopril-Sepharose column must be active in order to bind the ligand. Specific activities were re-determined prior to kinetic analysis and active protein concentrations adjusted accordingly.
Determination of kinetic parameters
A plate adapted fluorescamine assay was employed as published44. Thirty microlitres of AcSDKP substrate in Hepes buffer (50 mM Hepes (pH 7.5), 100 mM NaCl, 10 μM ZnSO4 buffer), ranging in concentration from 0 to 1000 μM, were warmed to 37 °C in a 96-well plate. The assay was commenced by the addition of 10 μl pre-warmed enzyme (0.2 pmols in Hepes buffer) and incubated for 15 minutes for N-domain and sACE enzymes and 30 minutes for the C-domain enzymes. The assay was stopped by the addition of 50 μl 1 M HCl. The solution was neutralised by the addition of 50 μl 1 M NaOH and the pH increased to 8.3 by the addition of 100 μl 500 mM K2HPO4/KH2PO4 buffer pH 8.3. Ten microlitres of fluorescamine (2 mg/ml in acetone, Sigma-Aldrich Co.) was added and the resulting mixture incubated for 3 minutes at room temperature. Fluorescence intensities were measured at λex = 390 nm and λem = 475 nm using a Cary Eclipse spectrofluorimeter (Varian Inc.). Changes in fluorescence compared to unhydrolysed substrate were converted to reaction velocities by the use of a standard curve obtained by complete AcSDKP hydrolysis and kinetic constants calculated from nonlinear regression analysis (v 4.01, GraphPad Prism®). For full length ACE constructs, the presence of 2 moles active sites for every 1 mole of enzyme was taken into account when calculating turnover rates27.
X-ray crystallography
The crystals of N-ACE in complex with the peptides were obtained by co-crystallization with a 2.5 mM concentration of Ac-SDKP (Biorbyt, orb70378). Crystals were grown with 1 μl of the N-ACE:Ac-SDKP sample (protein at 8 mg/ml in 50 mm HEPES, pH 7.5, 0.1 mm PMSF) mixed with an equal volume of reservoir solution consisting of 30% PEG550 MME/PEG20000, 100 mM Tris/Bicine, pH 8.5 and 0.06 M divalent cations (Molecular Dimensions) and suspended above the well as a hanging drop.
X-ray diffraction data were collected on station IO3 at the Diamond Light Source (Oxon, UK) equipped with a PILATUS-6M detector (Dectris, Switzerland). Crystals were kept at constant cryo-temperature (100 K) during data collection. Raw data images were processed and scaled with MOSFLM58 and SCALA using the CCP4 suite 6.559. Molecular replacement with the coordinates of N-domain (PDB code 3NXQ)30 was used to determine initial phases for structure solution in PHASER60. The working models were refined using REFMAC561 and manually adjusted with COOT62. Water molecules were added at positions where Fo − Fc electron density peaks exceeded 3σ and potential hydrogen bonds could be made. Validation was performed with MOLPROBITY63. Crystallographic data statistics are summarized in Table 1. All figures were drawn with PyMOL (Schrödinger, LLC, New York). Hydrogen bonds were verified with the program LigPlot+64.
Additional Information
How to cite this article: Masuyer, G. et al. Structural basis of Ac-SDKP hydrolysis by Angiotensin-I converting enzyme. Sci. Rep. 5, 13742; doi: 10.1038/srep13742 (2015).
References
Acharya, K. R., Sturrock, E. D., Riordan, J. F. & Ehlers, M. R. ACE revisited: a new target for structure-based drug design. Nat. Rev. Drug Discov. 2, 891–902 (2003).
Anthony, C. S., Masuyer, G., Sturrock, E. D. & Acharya, K.R. Structure based drug design of angiotensin-I converting enzyme inhibitors. Curr. Med. Chem. 19, 845–855 (2012).
Skeggs, L. T., Kahn, J. R. & Shumway, N. P. The preparation and function of the hypertensin-converting enzyme. J. Exp. Med. 103, 295–299 (1956).
Zaman, M. A., Oparil, S. & Calhoun, D. A. Drugs targeting the renin-angiotensin-aldosterone system. Nat. Rev. Drug Discov. 1, 621–636 (2002).
Soubrier, F. et al. Two putative active centers in human angiotensin I-converting enzyme revealed molecular cloning. Proc. Natl. Acad. Sci. U S A 85, 9386–9390 (1988).
Wei, L., Alhenc-Gelas, F., Corvol, P. & Clauser, E. The two homologous domains of human angiotensin I-converting enzyme are both catalytically active. J. Biol. Chem. 266, 9002–9008 (1991).
Rousseau, A., Michaud, A., Chauvet, M.-T., Lenfant, M. & Corvol, P. The hemoregulatory peptide N-Acetyl-Ser-Asp-Lys-Pro Is a natural and specific substrate of the N-terminal active site of human angiotensin-converting enzyme. J. Biol. Chem. 270, 3656–3661 (1995).
Araujo, M. C. et al. Peptidase specificity characterization of C- and N-terminal catalytic sites of angiotensin I-converting enzyme. Biochemistry 39, 8519–8525 (2000).
Yang, H. Y., Erdös, E. G. & Levin, Y. A dipeptidyl carboxypeptidase that converts angiotensin I and inactivates bradykinin. Biochim. Biophys. Acta. 214, 374–376 (1970).
Rieger, K. et al. Involvement of human plasma angiotensin I-converting enzyme in the degradation of the haemoregulatory peptide N-acetyl-seryl-aspartyl-lysyl-proline. Biochem. J. 296, 373–378 (1993).
Lenfant, M. et al. Inhibitor of hematopoietic pluripotent stem cell proliferation: purification and determination of its structure. Proc. Natl. Acad. Sci. U S A 86, 779–782 (1989).
Lombard, M. N., Sotty, D., Wdzieczak-Bakala, J. & Lenfant, M. In vivo effect of the tetrapeptide, N-acetyl-Ser-Asp-Lys-Pro, on the G1-S transition of rat hepatocytes. Cell Tissue Kinet. 23, 99–103 (1990).
Bogden, A. E. et al. Amelioration of chemotherapy-induced toxicity by cotreatment with AcSDKP, a tetrapeptide inhibitor of hematopoietic stem cell proliferation. Ann. N Y Acad. Sci. 628, 126–139 (1991).
Peng, H., Carretero, O. A., Brigstock, D. R., Oja-Tebbe, N. & Rhaleb, N. E. Ac-SDKP reverses cardiac fibrosis in rats with renovascular hypertension. Hypertension 42, 1164–1170 (2003).
Yang, F. et al. Ac-SDKP reverses inflammation and fibrosis in rats with heart failure after myocardial infarction. Hypertension. 43, 229–236 (2004).
Rasoul, S. et al. Antifibrotic effect of Ac-SDKP and angiotensin-converting enzyme inhibition in hypertension. J. Hypertens. 22, 593–603 (2004).
Peng, H. et al. Angiotensin-converting enzyme inhibitors: a new mechanism of action. Circulation 112, 2436–2445 (2005).
Peng, H., Carretero, O. A., Liao, T. D., Peterson, E. L. & Rhaleb, N. E. Role of N-acetyl-seryl-aspartyl-lysyl-proline in the antifibrotic and anti-inflammatory effects of the angiotensin-converting enzyme inhibitor captopril in hypertension. Hypertension 49, 695–703 (2007).
Lin, C. X. et al. Prevention of aortic fibrosis by N-acetyl-seryl-aspartyl-lysyl-proline in angiotensin II-induced hypertension. Am. J. Physiol. Heart Circ. Physiol. 295, H1253–H1261 (2008).
Liao, T. D. et al. N-acetyl-seryl-aspartyl-lysyl-proline attenuates renal injury and dysfunction in hypertensive rats with reduced renal mass: council for high blood pressure research. Hypertension 55, 459–467 (2010).
Wang, M., Liu, R., Jia, X., Mu, S. & Xie R. N-acetyl-seryl-aspartyl-lysyl-proline attenuates renal inflammation and tubulointerstitial fibrosis in rats. Int. J. Mol. Med. 26, 795–801 (2010).
Li, P. et al. Angiotensin-converting enzyme N-terminal inactivation alleviates bleomycin-induced lung injury. Am. J. Pathol. 177, 1113–1121 (2010).
Liu, Y. H. et al. N-acetyl-seryl-aspartyl-lysyl-proline prevents cardiac remodeling and dysfunction induced by galectin-3, a mammalian adhesion/growth-regulatory lectin. Am. J. Physiol. Heart Circ. Physiol. 296, H404–H412 (2009).
Peng, H., Carretero, O. A., Peterson, E. L. & Rhaleb, N. E. Ac-SDKP inhibits transforming growth factor-beta1-induced differentiation of human cardiac fibroblasts into myofibroblasts. Am. J. Physiol. Heart. Circ. Physiol. 298, H1357–H1364 (2010).
Binevski, P. V., Sizova, E. A., Pozdnev, V. F. & Kost, O. A. Evidence for the negative cooperativity of the two active sites within bovine somatic angiotensin-converting enzyme. FEBS Lett. 550, 84–88 (2003).
Skirgello, O. E., Binevski, P. V., Pozdnev, V. F. & Kost, O. A. Kinetic probes for inter-domain co-operation in human somatic angiotensin-converting enzyme. Biochem. J. 391, 641–647 (2005).
Rice, G. I., Thomas, D. A., Grant, P. J., Turner, A. J. & Hooper, N. M. Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem. J. 383, 45–51 (2004).
Natesh, R., Schwager, S. L., Sturrock, E. D. & Acharya K. R. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature 421, 551–554 (2003).
Corradi, H. R., Schwager, S. L., Nchinda, A. T., Sturrock, E. D. & Acharya K. R. Crystal structure of the N domain of human somatic angiotensin I-converting enzyme provides a structural basis for domain-specific inhibitor design. J. Mol. Biol. 357, 964–974 (2006).
Anthony, C. S. et al. The N domain of human angiotensin-I-converting enzyme: the role of N-glycosylation and the crystal structure in complex with an N domain-specific phosphinic inhibitor, RXP407. J. Biol. Chem. 285, 35685–35693 (2010).
Masuyer, G., Schwager, S. L., Sturrock, E. D., Isaac, R. E. & Acharya, K. R. Molecular recognition and regulation of human angiotensin-I converting enzyme (ACE) activity by natural inhibitory peptides. Sci. Rep. 2, 717 (2012).
London, N., Raveh, B., Cohen, E., Fathi, G. & Schueler-Furman, O. Rosetta FlexPepDock web server - high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 39, W249–W253 (2011).
Dive, V. et al. RXP 407, a phosphinic peptide, is a potent inhibitor of angiotensin I converting enzyme able to differentiate between its two active sites. Proc. Natl. Acad. Sci. U S A. 96, 4330–4335 (1999).
Fuchs, S. et al. Angiotensin-converting enzyme C-terminal catalytic domain is the main site of angiotensin I cleavage in vivo. Hypertension 51, 267–274 (2008).
Douglas, R. G. et al. Fragment-based design for the development of N-domain-selective angiotensin-1-converting enzyme inhibitors. Clin. Sci. (Lond) 126, 305–313 (2014).
Kröger, W. L., Douglas, R. G., O’Neill, H. G., Dive, V. & Sturrock, E.D. Investigating the domain specificity of phosphinic inhibitors RXPA380 and RXP407 in angiotensin-converting enzyme. Biochemistry 48, 8405–8412 (2009).
Yates, C. J. et al. Molecular and thermodynamic mechanisms of the chloride-dependent human angiotensin-I-converting enzyme (ACE). J. Biol. Chem. 289, 1798–1814 (2014).
Deddish, P. A. et al. Single-domain angiotensin I converting enzyme (kininase II): characterization and properties. J. Pharmacol. Exp. Ther. 279, 1582–1589 (1996).
Michaud, A., Williams, T. A., Chauvet, M. T. & Corvol, P. Substrate dependence of angiotensin I-converting enzyme inhibition: captopril displays a partial selectivity for inhibition of N-acetyl-seryl-aspartyl-lysyl-proline hydrolysis compared with that of angiotensin I. Mol. Pharmacol. 51, 1070–1076 (1997).
Conroy, J. M. & Lai, C. Y. A rapid and sensitive fluorescence assay for angiotensin-converting enzyme. Anal. Biochem. 87, 556–561 (1978).
Yokosawa, H., Endo, S., Ogura, Y. & Ishii, S. A new feature of angiotensin-converting enzyme in the brain: hydrolysis of substance P. Biochem. Biophys. Res. Commun. 116, 735–742 (1983).
Wang, W. et al. A fluorescence-based homogeneous assay for measuring activity of UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. Anal Biochem. 290, 338–346 (2001).
Kang, T. et al. Autolytic processing at Glu586-Ser587 within the cysteine-rich domain of human adamalysin 19/disintegrin-metalloproteinase 19 is necessary for its proteolytic activity. J. Biol. Chem. 277, 48514–48522 (2002).
Danilov, S. M. et al. A novel angiotensin I-converting enzyme mutation (S333W) impairs N-domain enzymatic cleavage of the anti-fibrotic peptide, AcSDKP. PLoS One 9, e88001 (2014).
Junot, C. et al. RXP 407, a selective inhibitor of the N-domain of angiotensin I-converting enzyme, blocks in vivo the degradation of hemoregulatory peptide acetyl-Ser-Asp-Lys-Pro with no effect on angiotensin I hydrolysis. J. Pharmacol. Exp. Ther. 297, 606–611 (2001).
Fuchs, S. et al. Role of the N-terminal catalytic domain of angiotensin-converting enzyme investigated by targeted inactivation in mice. J. Biol. Chem. 279, 15946–15953 (2004).
Masuyer, G. et al. Crystal structures of highly specific phosphinic tripeptide enantiomers in complex with the angiotensin-I converting enzyme. FEBS J. 281, 943–956 (2014).
Michaud, A., Chauvet, M.T. & Corvol, P. N-domain selectivity of Angiotensin I-converting enzyme as assessed by structure–function studies of its highly selective substrate, N-Acetyl-Seryl-Aspartyl-Lysyl-Proline. Biochem. Pharmacol. 57, 611–618 (1999).
Binevski, P. V., Sizova, E. A., Pozdnev, V. F. & Kost, O. A. Evidence for the negative cooperativity of the two active sites within bovine somatic angiotensin-converting enzyme. FEBS Lett. 550, 84–88 (2003).
Woodman, Z. L. et al. The N domain of somatic angiotensin-converting enzyme negatively regulates ectodomain shedding and catalytic activity. Biochem. J. 389, 739–744 (2005).
Jaspard, E., Wei, L. & Alhenc-Gelas, F. Differences in the properties and enzymatic specificities of the two active sites of angiotensin I-converting enzyme (kininase II). Studies with bradykinin and other natural peptides. J. Biol. Chem. 268, 9496–9503 (1993).
Ehlers, M. R. et al. Proteolytic release of membrane-bound angiotensin-converting enzyme: role of the juxtamembrane stalk sequence. Biochemistry 35, 9549–9559 (1996).
Balyasnikova, I. V., Metzger, R., Franke, F. E. & Danilov, S. M. Monoclonal antibodies to denatured human ACE (CD 143), broad species specificity, reactivity on paraffin sections and detection of subtle conformational changes in the C-terminal domain of ACE. Tissue Antigens 61, 49–62 (2003).
Gordon, K. et al. Deglycosylation, processing and crystallization of human testis angiotensin-converting enzyme. Biochem. J. 371, 437–42 (2003).
Ehlers, M. R., Chen, Y. N. & Riordan, J. F. Purification and characterization of recombinant human testis angiotensin-converting enzyme expressed in Chinese hamster ovary cells. Protein Expr. Purif. 2, 1–9 (1991).
Deddish, P. A. et al. Naturally occurring active N-domain of human angiotensin I-converting enzyme. Proc. Natl. Acad. Sci. U S A 91, 7807–7811 (1994).
Schwager, S. L., Carmona, A. K. & Sturrock, E. D. A high-throughput fluorimetric assay for angiotensin I-converting enzyme. Nat. Protoc. 1, 1961–1964 (2006).
Leslie, A. G. W. & Powell, H. R. Processing diffraction data with Mosflm. Evolving Methods for Macromolecular Crystallography 245, 41–51 (2007).
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 (1994).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53, 240–255 (1997).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and Development of Coot. Acta Crystallogr. D66, 486–501 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D66, 12–21 (2010).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U S A 98, 10037–10041 (2001).
Ho, B. K. & Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008).
Acknowledgements
We thank the scientists at station I03 of Diamond Light Source, Didcot, Oxfordshire (UK), for their support during X-ray diffraction data collection. We gratefully acknowledge the expert technical support of Sylva Schwager at the IDM, University of Cape Town (South Africa). K.R.A. and E.D.S. also thank the University of Cape Town (South Africa) and University of Bath (UK) respectively for the Visiting Professorships. This work was supported by the Medical Research Council (U.K.) Project Grant G1001685 (to K.R.A.) and the National Research Foundation (South Africa) CPRR grant 13082029517 (to E.D.S.).
Author information
Authors and Affiliations
Contributions
G.M. performed all the crystallography experiments, analysed the data and wrote the manuscript. R.G.D. carried out all the protein expression and kinetics, analysed the data and wrote the manuscript. E.D.S. conceived the project, supervised the biochemical work, analysed the data and edited the manuscript. K.R.A. conceived the project, supervised the structural study, analysed the data and edited the manuscript. All authors contributed to the conceptualisation of the study and reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Masuyer, G., Douglas, R., Sturrock, E. et al. Structural basis of Ac-SDKP hydrolysis by Angiotensin-I converting enzyme. Sci Rep 5, 13742 (2015). https://doi.org/10.1038/srep13742
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep13742
- Springer Nature Limited
This article is cited by
-
The potential role of renin angiotensin system in acute leukemia: a narrative review
Molecular Biology Reports (2024)