Abstract
There is a strong pressure to reduce nitrogen (N) fertilizer inputs while maintaining or increasing current cereal crop yields. We show that overexpression of TaDWF4-B, the dominant shoot expressed homoeologue of OsDWF4, in wheat can increase plant productivity by up to 105% under a range of N levels on marginal soils, resulting in increased N use efficiency (NUE). We show that a two to four-fold increase in TaDWF4 transcript levels enhances the responsiveness of genes regulated by N. The productivity increases seen were primarily due to the maintenance of photosystem II operating efficiency and carbon assimilation in plants when grown under limiting N conditions and not an overall increase in photosynthesis capacity. The increased biomass production and yield per plant in TaDWF4 OE lines could be linked to modified carbon partitioning and changes in expression pattern of the growth regulator Target Of Rapamycin, offering a route towards breeding for sustained yield and lower N inputs.
Similar content being viewed by others
Introduction
Wheat (Triticum aestivum L.) is a major crop worldwide providing 23% of human dietary protein1. In many parts of the world, on-farm wheat yields have plateaued since the mid-1990s, although the yield capacity of cultivars has continued to increase2. Modern agricultural practices include the development of high-yielding varieties of cereal grains, expansion of irrigation infrastructure, modernization of management techniques, distribution of hybridized seeds, and the addition of synthetic fertilizers and pesticides to increase and protect yields. However, this approach has led to direct losses of reactive N to the aquatic environment which lower water quality through eutrophication, coupled with the loss of N2O and the high CO2 emissions during fertilizer production, so the environmental consequences are substantial3,4,5. As it has been estimated that only 33% of N applied to a field is taken up by the crop, a multi-faceted approach is crucial to reducing the necessary application of fertilizer as well as losses of the applied fertilizer6,7. Improving N use efficiency (NUE) (defined as the ratio of grain produced per unit of N supply) can be achieved by improving yield under a constant N supply. Thus far, strategies to improve the yield capacity of crop cultivars have focused on photosynthesis8,9,10, with gains in biomass and yield reaching 40%11,12. However, this is not sufficient, and reducing our reliance on synthetic fertilizer will also rely on identifying ways to reduce wheat N requirement, i.e., being able to produce wheat with lower N input without a reduction in yield or grain quality2. While identifying genes and pathways regulating specific aspects such as N uptake, assimilation or remobilization can provide some advantages, approaches that target regulatory components of N metabolism are more promising. In particular, understanding and identifying genes that can regulate N responsiveness, i.e., the overall capacity of plants to induce morphological and physiological changes according to the external availability of N, are eagerly sought6,13.
Many domesticated crop species are polyploid and it is thought that the genome duplication events and the resultant chromosome rearrangements have been a major driving force creating biological complexity, novelty, and adaptation to environmental changes14,15,16. Wheat, an allohexaploid with genome AABBDD, is the product of two ancient hybridization events: Triticum uratu (A) and Aegilops spp. (B) hybridized 0.5 million years ago17, to form the tetraploid Triticum turgidum, which then underwent a second hybridization ~10,000 years ago with Aegilops tauschii (D) to form Triticum aestivum18. A more nuanced process of continuous diversification and allopolyploid speciation to produce the network of polyploid Triticum wheats has also been described19 and with the advent of whole-genome resequencing, the B-subgenome has been shown to exhibit most variation with multiple alien introgressions and deletions20. The presence of homeoalleles in hexaploid wheat increases both coding sequence variation per se and regulatory variation with the new promoter and transcription factor combinations and functional diversification of the duplicated genes21,22. This genome asymmetry and variation have consequences for quality traits such as the control of wheat seed storage proteins, biotic and abiotic traits, agronomic traits, and response to pests and diseases23,24,25.
Phytohormones are known to modify agronomically important traits, in particular, brassinosteroids (BR)26,27. BR have been shown to positively affect traits including tiller number, leaf size and angle, photosynthesis, and yield28,29,30,31 plus other desirable traits, including NUE, disease resistance, and end-use quality traits31,32,33,34.
Manipulating levels of DWF4/CYP90B, the rate-limiting step in the BR synthesis pathway, has been shown to increase yield, biomass production, tiller number, and quality traits in diverse plant species30,31,34,35. The mechanism of how altering BR levels impacts yield is still not completely understood, but in many plant species an increased rate of carbon fixation has been observed, suggesting that photosynthesis is key to the increased yields30,34.
An explanation of how the BR pathway controls so many different agronomically important phenotypes was shown through the direct interaction between BZR1 (Brassinazole Resistant 1), the transcription factor activating BR related genes, and the Target Of Rapamycin (TOR), the growth regulator sensing the carbon (C) status of the plant36,37. Overexpression of TOR in plants also mirrors many of the same phenotypes as modification of the BR pathway, suggesting that TOR may be the mode of increased growth and yields shown by many BR overexpressors. The BR and TOR pathways directly interact through the BR transcription factor BZR1 and the TOR kinase. The TOR kinase can maintain the activation potential of BZR1 and its role in growth promotion, by keeping the BZR1 protein stable37. Reducing TOR expression by RNAi silencing led to a decreased ability of BR regulated genes to be upregulated, arrested plant growth and, abolished the ability of increased BZR1 to promote growth when high levels of sugar were present38.
While many studies have shown that modification of the BR pathway can increase yields, plants have typically been assessed under nutritionally replete conditions. In order to assess if the same yield gains could be achieved in wheat grown on either marginal soils, or with lower fertilizer inputs, we generated over-expression lines to test whether modification of DWF4 levels could drive productivity gains and/or increase NUE.
Results
The wheat DWF4 gene family has seven members
To identify putative orthologs of DWF4/CYP90B in wheat, the amino acid sequence for the rice DWF4/CYP90B (locus Os03g0227700) was used in a BLASTP search using wheat genomics resources at Ensembl (http://plants.ensembl.org/Triticum_aestivum/Info/Index). Seven amino acid sequences were identified with high homology to the rice DWF4 amino acid sequence (e value < 1e−50, and >70% homology), these were encoded by four sequences located on chromosome 3 (one on chromosome 3A, one on chromosome 3B, two sequential copies on chromosome 3D) and three sequences located on chromosome 4 (Suppl. Fig. 1a). The amino acid sequences were clustered by chromosome group (Suppl. Fig. 1a). Putative proteins encoded by sequences on chromosome group 4 tended to be more similar to each other (98.62–99.01% identity) compared to those on chromosome group 3 (87.97–95.77% identity). Within chromosome group 3, TaDWF4-3D2 was the most distinct within the family (Suppl. Table 1).
Comparison of the group 4 homoeologue putative protein sequences revealed that 4A contains two large insertions when compared to the other homoeologues and OsDWF4 (Suppl. Fig. 2). The first insertion is located between exons 1 and 2, and the second insertion is located between exons 5 and 6, relative to the predicted CDS in the B and D homoeologues. The genes on chromosome 4 showed the highest expression levels, in particular TaDWF4-4B, while low to no expression could be seen for any of the genes on chromosome 3 in the wheat expression.com databases39 (Suppl. Fig. 1a). The expression patterns of the identified homoeologues on chromosome 4 showed unbalanced expression with TaDWF4-4B dominating expression in the shoot (70.9%), and TaDWF4-4A dominating expression in the spike (56.6%). There was no dominant homeologue expressed in the roots as TaDWF4-4A and TaDWF4-4B showed similarly but the higher expression in the roots than TaDWF4-4D.
DWF4 gene duplications in wheat
In diploid plant species where the DWF4 gene family has been characterized (including Arabidopsis, rice, and maize (Zea mays), DWF4 is encoded by a single gene28,40,41. It is interesting that in hexaploid wheat, cv. Chinese Spring, seven putative orthologs could be identified, suggesting that there may have been one or possibly two duplication events: the first duplication event leading to the presence of DWF4 genes on both chromosome 3 and chromosome 4 groups, and the second duplication event leading to two DWF4 genes on chromosome 3D, as the three gene models are co-linear on chromosome 3D.
The DWF4 inter-chromosomal duplication event in wheat, which led to the presence of the DWF4 gene on both chromosome groups 3 and 4, cannot be found in crop or model species such as Arabidopsis, rice, maize, or brachypodium (Brachypodium distachyon) (Suppl. Fig. 1b). In rice, the closest related proteins to OsDWF4 are other cytochrome P450s more commonly known as CPD1 or CPD2 sharing 46.7 percent identity at the amino acid level (Ensembl v 46). In Arabidopsis, the closest protein to AtDWF4 (AT3G50660) is also a cytochrome P450, CYP72A1 sharing 36.1% identity at the amino acid level (Ensembl v 46). In maize and brachypodium the closest related proteins are ZmCYP724A1 (Zm00001d003349) with 50.5% identity and BdCYP724A1 (BRADI_5g12990v3) with 47.4% identity.
The inter-chromosomal duplication of the DWF4 genes can also be inferred for both T. urartu and Aegilops tauschii, two of the three wheat progenitor species (progenitors of the A and D genomes in T. aestivum), as well as in the tetraploid T. dicoccoides (Suppl. Figure 1b). An intra-chromosomal duplication in Aegilops tauschii on chromosome 3, supports the hypothesis that these copies arose before the progenitors hybridized. Therefore, one gene duplication event seems to have arisen before the progenitor species hybridized, and a second event after the hybridization occurred to produce modern bread wheat.
Overexpression of TaDWF4-4B in wheat leads to the upregulation of BR responsive genes
Given that TaDWF4-4B in Chinese spring showed the highest expression levels in the shoot tissues identified in public RNAseq databases, we selected this homeologue for overexpression in hexaploid wheat (Suppl. Fig. 1). A total of 40 independent T0 plants were generated in the cv. Fielder, with TaDWF4-4B CDS, expressed from the constitutive promoter, OsActin42. In six out of nine T0 plants with a single T-DNA insertion, TaDWF4 transcript levels (including both transgene and endogenous gene) were significantly increased from 1.6 to 4.9-fold compared to wild type (Suppl. Fig. 3a). In the four over-expressing (OE) lines showing the highest TaDWF4 transcript levels, we measured the transcript levels for EXORDIUM (EXO), DWF3/CPD/CYP90A, BZR2, and BES1, as these genes either have been shown in Arabidopsis to be upregulated by BR43 or involved in the BR pathway44,45,46. TaEXO and TaBES1 transcript levels were significantly higher than WT in the four OE lines tested (p < 0.05) and followed the same trend as those of TaDWF4 (Suppl. Fig. 3b). TaBZR2 and TaDWF3 did not show significantly increased expression in OE wheat lines and this contrasts with previous reports in other plant species of either DWF4 overexpression or the application of exogenous BL44,47.
Overexpression of TaDWF4-4B increases productivity under low to high N levels
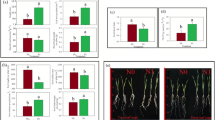
The four highest expressing transgenic lines (OE-1–OE-4) and a corresponding null segregant (WT) were grown in pots on a low fertility soil supplemented with NH4NO3 to reach field-equivalent N levels of 70, 140, and 210 kg/ha N. A low fertility soil with limited N was used as a substrate in order to demonstrate N deficiency symptoms. The lowest level (70 kg/ha N) corresponds to an N deficiency and the highest level (210 kg/ha N) to the agronomic level typical of current UK agronomic practice48. All four OE lines tested showed significant increases in yield per plant when compared to the WT at all 3 N levels (p = 0; Fig. 1a). There was also a large increase in above-ground biomass (p = 0), ranging from a 45 to 101% increase in the OE lines compared to WT (Fig. 1b). An overall increase in tiller number (p = 0; Fig. 1c), grain number per plant (p = 0; Fig. 1e) and grain weight measured as thousand grain weight (TGW) (p = 0; Fig. 1f) were also observed. The OE lines also showed a significant increase in harvest index (HI, defined as the proportion of biomass in the grain divided by the total biomass) relative to WT (p = 0; Fig. 1d). The significant change in yield per plant under all levels of N tested resulted in a significant increase (p = 0) in the NUE in OE lines relative to WT (Suppl. Fig. 5), as expected with yield increase. Full statistical comparisons are included in Suppl Tables 2–7.
Data are shown as mean values (central line), lower and upper quartiles (box), minimum and maximum values (whiskers), and outliers as individual points. Fifteen plants were grown per replicate for each genotype by N level combination. The overall plant growth experiment was replicated twice; data from one replicate is shown. The statistical analysis was performed with ANOVA and post hoc Tukey test, asterisk denotes p val < 0.05. a yield (g) per plant; b above-ground biomass (g) per plant; c ears per plant; d harvest index; e seed number per plant; f thousand-grain weight per plant (TGW); g OE-1 (left) and WT (right) plants grown on 70 kg/ha equivalent; h OE-1 (left) and WT (right) plants grown on 210 kg/ha equivalent; i 10-day-old plants from OE-1 (left) and WT (right) grown in hydroponic solution under at 70 kg/ha N. Yellow bars in g and h = 10 cm, size standard in i = 40 mm diameter. Full statistical comparisons are included in Supplementary Data 2.
As anticipated, increasing N supply led to increased yield in both WT and OE lines. The WT line showed a defined N-dependent yield increase, with OE lines producing higher yield at all N levels and generally mirroring the gains seen by WT. The converse effect was seen for biomass where increased N levels decreased the differences seen in biomass; as N became less limiting the differences in biomass decreased (Fig. 1b). Overexpression of TaDWF4-B in cv. Fielder led to significantly increased growth compared to the WT even under low N conditions and also led to higher productivity per plant.
Nitrogen responsive genes in TaDWF4 OE lines show altered expression
At the whole plant level, OE lines appear to be highly responsive to low N levels, maintaining yield increases under N-deficient to replete conditions. The expression of four genes known to be differentially regulated under high or low N conditions in wheat was selected to test the N-responsiveness at the transcript level of an OE line (OE-1) relative to WT49,50. The genes selected encode the high and low-affinity N uptake transporters (NRT2.1 and NRT1), an N transporter involved in N translocation through the plant (NPF7.1), and glutamate dehydrogenase (GHD2), an enzyme involved in N remobilization in response to limiting carbon (C). All four genes tested show a clear increase in OE-1 and WT transcript levels under low N compared to replete N conditions in both shoots and roots of 10-day-old seedlings grown in a hydroponic system (Fig. 2).
Transcript levels for NRT1, NRT2.1, NPF7.1, and GDH2 genes in wheat are shown relative to the expression of TaUbi under low (LN) and high nitrogen (HN) in hydroponic solution in a shoot; and b root tissues. Data are shown as mean values (central line), lower and upper quartiles (box), minimum and maximum values (whiskers), and outliers as individual points. The statistical analysis was performed with ANOVA and post hoc Tukey test, letters correspond to significant differences between transcript levels of either line under either treatment (p < 0.05).
Transcript abundance in OE-1 shoot tissues under N limited conditions showed significantly lower expression of TaNRT1 (p = 0), TaNPF7 (p < 0.001) and TaGDH2 (p < 0.001), but not TaNRT2.1 (p = 0.07), compared with WT. Under replete conditions, no significant differences were found between OE-1 and WT. A similar pattern was seen in root tissues under N limited conditions with significantly lower expression of TaNRT1 (p = 0), TaNPF7 (p < 0.001), TaNRT2.1 (p < 0.001), and TaGDH2 (p = 0), in the OE-1 line compared with WT. Under replete conditions only TaNRT1 and TaNRT2.1 showed significantly lower expression in OE-1 compared with WT (p < 0.01, p < 0.05).
Given the putative role of DWF4 in N response we also measured the level of transcripts for TaDWF4 from chromosome 4 in the shoots and roots of WT Fielder plants grown under four N levels (0, 1/3, 2/3, and full as a proxy for 0, 70, 140, and 210 kg/ha fertilizer application rate). Transcript levels of TaDWF4 were higher in the shoots relative to the roots of wheat plants grown under the four N levels (p < 0.05, Suppl. Fig. 6). Increased N concentrations led to higher TaDWF4 transcript levels in the shoots and lower transcript levels in the roots (Suppl. Fig. 6, p < 0.05). The effect of N level on TaDWF4 transcript levels was driven by the difference between the lowest N levels. There were no significant differences in transcript levels observed amongst the 1/3N, 2/3N, and N full treatments. TaDWF4 expression, therefore, appears to be responsive to changes in N levels under very limiting N conditions promoting expression in the roots and decreasing expression in the shoots in WT Fielder.
Increases in yield and biomass in TaDWF4 OE lines are independent of N uptake and translocation
To further understand how the OE lines were driving an increase in yields, the flux of N was measured using 15N tracer studies. Uptake and translocation of N were measured between the four OE lines and the WT when grown under low N conditions for 2 weeks and supplied with15N to quantify short-term uptake and translocation from the root to the shoot. No significant differences were seen in the uptake or translocation of N between any of the lines tested (Fig. 3a, b) (ANOVA, n.s.).
a Uptake of 15N in roots of overexpression lines relative to WT roots. b Translocation of 15N from the soil to the shoot in 5 min of uptake in OE lines relative to WT. c Percentage N of flag leaves and grains are grown on 210 kg/ha at harvest. d N content of grain grown on three N levels. Data are shown as mean values (central line), lower and upper quartiles (box), minimum and maximum values (whiskers), and outliers as individual points. The statistical analysis was performed with ANOVA and post hoc Tukey test. Asterisks indicate a significant difference (p < 0.05) between WT and an OE line at the same N level.
To understand if the OE lines showed altered N content compared with WT over a longer plant growth cycle, total N was measured in the grain and senesced flag leaf from plants grown under three N levels to compare N taken up from the soil at the end of the life cycle. At harvest, no significant differences in flag leaf N content could be measured in plants grown on eq. 210 kg/ha N (Fig. 3c), while the grain N content was significantly lower in the OE compared to WT. The decrease in grain N content in OE lines compared to WT was also measured when plants were grown under three different N levels (Fig. 3d) (ANOVA p < 0.05).
TaDWF4-B overexpression maintains photosynthesis under N limitation
In rice, OE of DWF4 led to an increase in leaf-level photosynthetic C fixation30. Leaf level CO2 assimilation per unit area was measured on the fourth expanded leaf in the OE-1 and WT lines grown under both low (eq. 70 kg/ha) and high (eq. 210 kg/ha) N levels. No differences in CO2 assimilation could be measured when OE-1 and WT plants were supplied with adequate N and high light (Fig. 4a). A significant increase in CO2 assimilation over a range of CO2 concentrations in the OE-1 line was observed when plants were grown under low N conditions and high light (Fig. 4b). The increase in CO2 assimilation in OE-1 compared to WT under low N was not driven by a significant difference in chlorophyll content, a major sink for N in a plant, as both the OE-1 and WT line had similar levels of chlorophyll under both low and high N (Fig. 4c) (p = 0.34). Overall, this suggests that under low N conditions an increased photosynthetic capacity may drive the yield and biomass gains measured in the OE lines. However, this is not the case when plants are grown under N-replete conditions.
Plants were grown in a growth chamber with a light intensity of 1250 µmol m−2 s−1 and CO2 level of 400 ppm: a A/Ci curve of OE-1 and WT plants grown on under high N conditions (equivalent N 210 kg/ha) or b under low N conditions (equivalent N 70 kg/ha). c Chlorophyll content in the leaves of plants used for A/Ci measurements. In panels a and b the shading represents the 95% confidence interval. In panel c, data is shown as mean values (central line), lower and upper quartiles (box), minimum and maximum values (whiskers), and outliers as individual points. Data were collected from six individual plants measured on the fourth fully expanded leaf.
To understand if the plants were able to maintain photosynthesis at a higher level when grown under lower input conditions, plants were grown on the same low fertility soil as shown in Fig. 1 supplemented with either 70 or 210 kg/ha equivalent N. Spot measurements of CO2 assimilation were taken under growth chamber conditions, with light levels at 250 µmol m−2 s−1 and 400 ppm CO2. Under these conditions, a significant increase in CO2 assimilation could be measured in OE-1 compared to WT, under both N levels tested (Fig. 5a) (p < 0.05). Measurements of chlorophyll fluorescence were taken as a proxy for the operating efficiency of photosystem II ((Fm’–Fo’)/Fm’) on light-adapted leaves from plants grown under low (70 kg/ha) vs. high (210 kg/ha) N conditions. OE-1 plants had a greater proportion of PSII centers available to collect the saturating light pulse than that of WT plants under both low and high N conditions (Fig. 5b) (p = 0). While PSII efficiency declined in WT plants grown under low N conditions (p < 0.001), this was not the case for OE-1 plants.
a Spot measurements of C assimilation in OE-1 or WT undergrown in growth chamber conditions which include a light intensity of 250 µmol m−2 s−1 and CO2 level of 400 ppm. b Maximum efficiency of PSII under light (Fv’/Fm’) in light-adapted plants grown on two different N levels. Plants were grown on either (Low) 70 or (High) 210 kg/ha N equivalent and the fourth leaf was measured for PSII activity. The statistical analysis was performed with ANOVA and post hoc Tukey test. Data are shown as mean values (central line), lower and upper quartiles (box), minimum and maximum values (whiskers), and outliers as individual points. Data were collected from six individual plants measured on the fourth fully expanded leaf. Asterisk in panels a and b indicates a significant difference (p < 0.05) between WT and OE-1 at the same N level.
To understand the effect of continued photosynthesis on C content, flag leaves of fully senesced plants grown on 210 kg/ha equivalent N were measured. There was a significant increase (1–2%) in the flag leaf C content in OE-1 lines compared to WT (p < 0.01) (Suppl. Fig. 7). Soluble sugars were measured in 14-day-old seedlings grown in hydroponic solution under low (1/3, 70 kg/ha) or replete (Full, 210 kg/ha) conditions. Under replete N conditions, WT showed significantly higher levels of both soluble sugars measured (d-fructose, d-glucose) (p < 0.05) (Fig. 6a–d), led mostly by large differences in the roots of both sugars measured. Under low N conditions, there were similar levels of bother sugars per gram dry weight in the shoots of OE-1 plants relative to WT plants. Although the distribution in the two tissues tested of the sugars is different and significant differences were observed in both sugars in both tissues measured (p < 0.05).
Plants grown under hydroponic conditions lead to increased transcript levels of TaTOR and soluble sugars. Fructose (a, b), and glucose (c, d)) were extracted from wheat leaves of 14-day old plants grown High N (Full N) or Low N (0N). The transcript abundance of TaTOR was measured in the shoots (e) and roots (f) in both OE-1 and WT 14-day old wheat plants grown under High N or Low N, the expression shown is relative to TaUbi. n = 3 plants for each treatment and line testing. The statistical analysis in all panels was performed with ANOVA and post hoc Tukey test. Data in all panels are shown as mean values (central line), lower and upper quartiles (box), minimum and maximum values (whiskers), and outliers as individual points. Asterisk indicates a significant difference (p < 0.05) between WT and OE-1 at the same N level. Letters in panels e and f indicate significant differences (p < 0.05) amongst both the line and the treatment.
Next, we tested whether the effect of TaDWF4-B overexpression on photosynthesis and soluble sugar/C levels was mediated by TOR. Significantly higher transcript levels for TaTOR were measured in the roots of OE-1 relative to WT when plants were grown under both low and replate N in hydroponics. TaTOR transcripts levels were higher in roots than shoots in OE-1 (p < 0.05) and a significant difference in TOR expression was seen under both low and high N growth conditions (p < 0.05) (Fig. 6d, e). The effect of low N led to increased TaTOR expression in OE-1 shoots, however, the converse was seen in WT shoots where TaTOR transcripts levels were markedly reduced under low N. In roots OE-1 showed significantly higher transcript levels than WT under both high and low N (Fig. 6e).
Indirect selection has not increased DWF4 expression in modern wheat varieties
To understand if the increased photosynthesis observed in the OE lines has already been indirectly selected for in modern wheat cultivars, seven diverse spring wheat cultivars, as well as Fielder and OE-1, were tested for both their ability to maintain PSII operating efficiency under limiting N levels, and to determine if this could be a factor in determining NUE differences. The ability of the different varieties to maintain PSII electron transport could be seen in three different varieties including the former elite UK spring wheat cultivar Paragon (Fig. 7a) suggesting that this trait may have been indirectly fixed via breeding. TaDWF4 transcript levels were lower in all cultivars tested compared to OE-1, and there was no significant difference amongst cultivars (Fig. 7b). The lack of selection for higher DWF4 expression is further supported as lines that maintained PSII reaction centers open had both high and low NUE, where NUE is defined as the yield on low N (eq. 70 kg/ha) divided by yield on high N (eq. 210 kg/ha) (Suppl. Fig. 1).
a Operational PSII efficiency in light-adapted plants grown on two different N levels. Plants were grown on either 70 or 210 kg/ha N equivalent and the fourth leaf was measured for PSII activity. b Expression of TaDWF4 in the shoots of 10-day old wheat plants grown under low N (LN = 0 N) or high N (HN = Full N), the expression shown is relative to TaUbi in each cultivar. Data in panels a and b are shown as the mean values (central line), lower and upper quartiles (box), minimum and maximum values (whiskers), and outliers as individual points. Statistical analysis was performed with ANOVA and post hoc Tukey test, asterisk indicates a significant difference between low and high N levels (p < 0.05).
Discussion
In this study, we identified the functional orthologues of the OsDWF4 in wheat and generated OE lines to understand whether increased TaDWF4 expression could be used to increase wheat NUE and drive an increase in yield as seen in other crops with modified DWF4 expression. We identified seven putative orthologues in wheat, four on chromosome 3 plus three on chromosome 4, and have shown that the putative orthologues on chromosome 3 had low level to no expression, whereas the 4B homoeologue had the highest expression, based on data available from public wheat expression databases. Significant tissue type differences were seen in the expression of the three homoeologues, suggesting that specific regulatory elements may control modification of TaDWF4 expression in particular tissues to increase plant growth in wheat. The duplication of putative orthologues on the two different chromosome groups also appears to have happened before hexaploid bread wheat arose 5.5 Mya, as each of the progenitor species, durum wheat, and T. dicoccoides have the duplication24. Further support for the orthologues on chromosome 4 is similar to the previously characterized DWF4 is the high degree of synteny between wheat chromosome group 4, and rice chromosome 3, on which OsDWF4 resides, extending most of its length. In contrast, chromosome group 3 is the best conserved of all wheat chromosome groups, along both short and long arms, compared with rice chromosome 151.
Based on the bioinformatic analysis the homoeologue on 4B was selected for the creation of OE lines in order to understand the role of increased TaDWF4 expression on increasing yield and NUE. Overexpression of TaDWF4-B resulted in dramatic increases in several agronomic measures including yield and biomass across a range of N levels. Interestingly, total biomass levels of WT plants started to approach those of OE lines as N levels increased, however, this alone did not lead to a decrease in the difference in yields suggesting that biomass alone is a proxy and not a perfect measure of ultimate yield potential (Fig. 1a, b). The OE lines showed significant increases in yield at all tested N levels and seemed to respond as if N was not limiting growth under low N. Similar results have been recently reported in maize overexpressing ZmDWF4 grown under field conditions although different N levels were not tested52. Uptake rates of N as well as translocation rates did not differ between the OE or WT lines and similar levels of N were found in most tissues measured outside of the grain, suggesting that the OE lines did not demand an increased level of N to support the increased plant growth. The altered N sensing by higher DWF4 expression was supported by the expression of N regulated genes which showed a muted response to the removal of N in TaDWF4-B OE lines. This expression data suggests that TaDWF4 overexpression can mitigate nutrient limitations and that other signals such as C or soluble sugars are more important to drive yield gains while lowering the need for N input. This difference in growth was not due to a difference in the photosynthetic capacity, which has been previously suggested as the reason for increased yields30. Overall we observed assimilation rates and levels of chlorophyll were similar at high N levels (Fig. 4). The maintenance of C assimilation under limited N conditions may contribute to the increased biomass and yield. The increase in levels of soluble sugars observed in WT plants including glucose possibly suggests altered sensing of required nutrient signals to activate growth and development. This is supported by the higher expression of TaTOR under both N levels tested suggesting that TaDWF4 OE lines maintain growth, where WT would slow down. Further work is required to understand the exact regulation of soluble sugars and other nutrient signals which mediates the increase in growth in DWF4 overexpressing lines. As BR levels were not directly measured as part of this work it is difficult to determine whether BR levels increased, thereby allowing growth to continue due to altered levels of soluble sugars, or whether the lack of BR pathway inhibition allowed the photosynthetic pathways to produce additional sugars and fuel further growth. There may indeed be other mechanisms outside of the known BR pathway to explain how increased DWF4 expression allows for growth on otherwise limiting N levels.
Data collected here suggest that higher TaDWF4 expression in wheat can redirect normal feedback inhibition of N limitation supporting continued plant growth. It would be interesting to determine if similar effects are observed with other common nutrient limitations such as phosphate or potassium. Certainly, a further understanding of how increased DWF4 expression allows for increased growth and yields will be paramount for translation to other crops and non-GM methods for increasing DWF4 expression in field-grown crops.
Finally, it has been suggested that breeding for increased BR levels could improve overall yields and improve the tolerance of crop species to various abiotic and biotic stress28,30,35,41. Observations here also suggest that wheat breeders are not selecting for increased DWF4 expression as TaDWF4 expression was not significantly higher in the more modern spring wheat varieties tested (Fig. 7). There were significant differences in DWF4 expression between the varieties tested as well as in response to N levels suggesting that variation may exist allowing for selection for higher expression in wheat shoots.
Overall, we have shown that OE of TaDWF4 in wheat leads to plants that grow as if they need less N, increasing overall NUE. The plants do not sense N as a limitation to growth and keep growing under lower N levels. Based on these results we propose that a doubling of TaDWF4 expression could allow farmers to reduce the application of N fertilizer by up to 70 kg/ha whilst still producing the same amount of yield per plant as currently grown at standard 210 kg/ha N input level. When this 70 kg/ha saving is multiplied by the estimated land in which wheat is grown in the UK (1.69 million ha in 2019) there is the potential to reduce CO2 released into the atmosphere from fertilizer production by over 100,000 t5,53,54. This is equivalent to the energy needed to power and heat more than 15,000 UK homes for one year55. Here we show that it is possible to increase levels of DWF4 in wheat through genetic modification but that there is also potential to exploit native variation via traditional breeding. This could potentially have a large impact on wheat crop management, reducing fertilizer demands whilst maintaining levels of productivity.
Materials and methods
Gene identification
The OsDWF4 coding sequence from NCBI (XP_015633105.1) was used as a query for BLAST searches of the wheat genome (IWGSC 2014) and expressed sequence tag (EST) databases in GenBank (http://www.ncbi.nlm.nih.gov/), Ensembl (http://plants.ensembl.org/index.html) and Komugi (https://shigen.nig.ac.jp/wheat/komugi/). Gene prediction was carried out using FGENESH (www.softberry.com). Protein domain prediction was found using InterProScan (https://www.ebi.ac.uk/interpro/). Identification of DWF4 like genes was based on sequences in Ensembl (version 42) using BLASTP and using an e value cut off of <1e−50.
Plasmid construction for genetic modification
TaDWF4-B was synthesized from the public sequence available for the wheat cultivar Chinese Spring with attL1 and attL2 sites for direct recombination into a binary gateway vector. All primers used in this study are listed in Supplementary Data 1. TaDWF4-B was then recombined into the binary vector pSc4ActR1R2 using a Gateway LR Clonase II Kit (Thermofisher) to create pMM90. TaDWF4-B was expressed in planta from the rice Actin promoter and transcripts terminated by the A. tumefaciens nopaline synthase terminator (tNOS)42.
pMM90 was verified by restriction digest and sequencing before being electro-transformed into A. tumefaciens strain EHA105 Plasmids were re-isolated from Agrobacterium cultures and verified by restriction digest prior to use in wheat transformation experiments56.
Plant materials
Wheat cv. Fielder (USA) was used for genetic transformation experiments. The spring wheat varieties Baj-1 (India), Cadenza (UK), CDC Landmark (Canada), CDC Stanley (Canada), Lancer (Australia), Paragon (UK), and Weebill (Mexico) were obtained from the Germplasm Resource Unit, UK (www.seedstor.ac.uk/search-browseaccessions.php?idCollection=35), and used in photosynthetic measurements.
Plant growth conditions
Wheat cv. Fielder plants were grown in controlled environment chambers (Conviron) at 20 °C day/15 °C night with a 16 h day photoperiod (approximately 400 μE m−2 s−1) for the harvest of immature embryos for the transformation experiments.
Transgenic lines and corresponding null segregants were grown on TS5 low fertility soil to control total nitrogen with a starting nitrogen level of 0.1 mg/l (Bourne Amenity, Kent, UK). Ammonium nitrate was then added to reach a final concentration in the pots which contained roughly 750 g of dry soil in each pot. Equivalent to field fertilizer application of 70, 140, or 210 kg/ha N which equates to 23.3, 46.6, or 70 mg/pot. Each pot also received 4.2 mg Ca, 2.7 mg K, 0.62 mg Mg, 0.04 mg P, 0.56 mg S, 0.008 mg B, 0.13 mg Fe, 0.015 mg Mn, 0.0012 mg Cu, 0.0024 Mo, 0.0045 Zn, 0.00 mg Na, and 0.63 mg Cl per pot. Plants were grown in a climate-controlled glasshouse with supplemental light for 16 h day and 20 °C/15 °C day–night temperatures.
For transcript abundance measurements and sugar content, wheat seedlings were grown for ten days in 2.2 L pots containing Magnavaca solution containing 3 µM KH2PO4, 3.52 mM Ca(NO3)2, 0.58 mM, KCl, 0.58 mM K2SO4, 0.56 mM KNO3, 0.86 mM Mg(NO3)2 0.13 mM H3BO3, 5 µM MnCl2, 0.4 µM Na2MoO4, 10 µM ZnSO4, 0.3 µM CuSO4, Fe(NO3)3 and 2 mM MES (pH 5.5), and supplemented with either 0, 0.4 mM, 0.8 mM, or 1.3 mM NH4NO3. Plants were harvested on the tenth day separating tissue in to roots and shoots for analysis.
Plants were grown on Magnavaca solution containing either 0 or 1.3 mM NH4NO3 for ten days in controlled environment chambers (Conviron) at 20 °C day/15 °C night with a 16 h day photoperiod (approximately 400 μE m−2 s−1).
Wheat transformation
Immature seeds were collected 14–20 days post-anthesis (dpa) and immature wheat embryos were isolated and co-cultivated with A. tumefaciens for 2 days in the dark57. Subsequent removal of the embryonic axis and tissue culture was performed as previously described58. Individual plantlets were hardened off the following transfer to Jiffy-7 pellets (LBS Horticulture), potted up into 9 cm plant pots containing M2 compost plus 5 g/l slow-release fertilizer (Osmocote Exact 15:9:9) and grown to maturity and seed harvest in controlled environment chambers, as above.
DNA analysis of transformed wheat plants
Plantlets that regenerated under G418 selection in tissue culture, were tested for the presence of the nptII gene using QPCR. The nptII copy number was assayed relative to a single copy wheat gene amplicon, GaMyb, normalized to a known single copy wheat line59. Primers and Taqman probes were used at a concentration of 10 µM in a 10 µl multiplex reaction using ABsolute Blue qPCR ROX mix (Thermofisher) with the standard run conditions for the ABI 7900 HT. The relative quantification, ΔΔCT, values were calculated to determine nptII copy number in the T0 and subsequent generations60. Homozygous and null transgenic lines were identified on the basis of nptII copy number and segregation analysis. WT Fielder plants were null segregates.
Transcript level analysis
Total RNA was isolated from both roots and shoots for each nitrogen treatment using an RNeasy Kit (Qiagen). Following treatment with DNaseI (Thermofisher), cDNA synthesis was conducted on 500 ng of total RNA using Omniscript RT Kit (Qiagen). The cDNA was diluted 1:2 with water and 0.5 µL was used as a template in each RT-PCR reaction. Transcripts levels were quantified using SYBR Green JumpStartTaq ReadyMix (SIGMA) with the standard run conditions for the ABI 7900 HT. Three technical replicates were performed on each of the three biological replicates. Two reference genes TaUbiquitin and TaEF1α were used for the normalization using the ΔΔCT. The sequence of primers used in qPCR assays is shown in Supplementary Data 1.
Whole plant measurements
Total shoot dry weight, seed weight (yield per plant), seed number, seed size, and tiller number were measured at maturity and following two weeks of drying at 35 °C. Biological replicates each contained 15 plants per line and were grown until seed maturation and grown in two separate experiments. NUE was calculated as yield per plant divided by the amount of ammonium nitrate added.
C and N content determination, N uptake
Wheat tissues (leaf or grains) were dried at 75 °C for 48 h before grinding. Four grains were placed in 2 mL microfuge tubes with 2 × 5 mm diameter stainless steel beads and shaken in a genogrinder until a fine powder was obtained. Dried and ground samples were carefully weighed (0.5 mg) into tin capsules, sealed, and loaded into the auto-sampler. Samples were analyzed for percentage carbon, percentage nitrogen, 12C/13C (δ13C) and 14N/15N (δ15N) using a Costech Elemental Analyzer attached to a Thermo DELTA V mass spectrometer in continuous flow mode.
To measure N uptake, roots from 2-week-old seedlings were exposed to 15NH415NO3 for 5 min, then washed in 0.1 mM CaSO4 for 1 min, harvested, and dried at 70 °C for 48 h. N content and isotopic levels were analyzed as described above. The excess 15N was calculated based on measurements of δ15N and tissue N%. First, the absolute isotope ratio (R) was calculated for labeled samples and controls, using Rstandard (the absolute value of the natural abundance of 15N in atmospheric N2).
Then, molar fractional abundance (F) and mass-based fractional abundance (MF) were calculated
The excess 15N in mg in a total tissue was calculated as in
Chlorophyll measurements
Leaf chlorophyll content was determined using the method developed by Hiscox et al.61. Chlorophyll was extracted from 100 mg of fresh leaf tissue from six independent plants in 10 mL DMSO, mixed for 30 min, and then placed at 4 °C overnight. Extracts were diluted 1:2 with DMSO before absorbance measurements at 645 and 663 nm (spectrophotometer Jenway model 7315, Staffordshire, UK).
Leaf gas exchange
Leaf level photosynthesis and stomatal conductance were measured on the youngest fully expanded leaf from the main tiller of at least six wheat plants at the tillering stage (GS23) grown in a controlled environment chamber with 16 h day and 20 °C/15 °C day–night temperatures. Photosynthesis and stomatal conductance were measured using a portable infrared gas analyzer (Licor 6400XP, Licor Environmental). Gas exchange was measured at a PPFD of 1500 µmol m−2 s−1 (using the LI-COR 6400 LED light source), block temperature was maintained at 22 °C, and humidity was maintained close to 60%. Each leaf was clamped in the Licor chamber, and let to acclimate until CO2 concentrations in the chamber reached a steady-state for 5 min. Gas exchange measurements were recorded under a range of CO2 concentrations (400, 300, 200, 100, 400, 600, 800, 1000, 1200, and 400 ppm).
Chlorophyll fluorescence measurements and A/Ci curve analysis
An LI-6800 portable photosynthesis infrared gas analyzer system (LI-COR) equipped with a multiphase flash fluorimeter was used to assess physiological differences for photosynthetic parameters between transgenic and WT wheat plants. Measurements were taken on the fourth leaf of plants grown on TS5 low fertility soil to control total nitrogen with a starting nitrogen level of 0.1 mg/l (Bourne Amenity, Kent, UK). Ammonium nitrate was then added to reach a final concentration in the pots equivalent to field fertilizer application of 70 or 210 kg/ha N. Plants were grown in a climate-controlled chamber with supplemented light (250 μmol m−2 s−1) for a 16 h day and 20 °C/15 °C day–night temperatures. For chlorophyll fluorescence measurements (Fm’–Fo’)/Fm’) leaves from at least six plants were pulsed four times to acclimatize tissues and a steady-state reading was taken. For measurements of the A/Ci curve was measured on six plants for each treatment. Ca reference values were 400, 400, 300, 200, 100, 50, 25, 400, 400, 400, 600, 800, 1000, 1200, and 400 µL L−1, with a saturating rectangular pulse of 12,000 µmol m−2 s−1 at each reference point. All leaves were also normalized for the amount of area of the measuring disk. Measurements were carried out on consecutive days between 1 and 8 h post-dawn, measuring three plants total selected at random from each treatment per day.
Sugar measurements
Freeze-dried samples were ground to a fine powder with a pestle and mortar. A 50 mg sub-sample of leaf tissue and 25 mg of root tissue was extracted in 1 and 2 ml of ultra-pure water, (Elgastat UHQPS), respectively. The extracts were mixed for 2 min on a miximatic (Jencons) and incubated for 60 min in a water bath at 60 °C and mixed again. The solutions were centrifuged (Sigma 4–16 KS) at 4500 g for 20 min. Five hundred microlitres of the supernatant were pipetted into a Thomson 0.45 µm PTFE filter vial (Thames Restek).
Five microliters of supernatant were injected into a Waters Alliance 2695 HPLC. Sugars were separated on an Ultra amino 100 Á 5 µm 250 × 4.6 mm column (Thames Restek, UK) and detected with a Waters 2414 refractive index detector. The mobile phase was [80:20] [acetonitrile: water] with a flow rate of 1 ml min−1, the column was heated to 35 °C. Standards of known amounts of the sugars were injected into the HPLC and EmpowerTM 3 software was used to produce linear calibration curves in the range of 0.625–10 µg for fructose, glucose, all curves had r2 greater than 0.998. These calibration curves were used to determine the concentration of the sugars found in the samples. Sugar standards were analar grade (Sigma Aldrich UK), solvents were HPLC grade (Fisher Scientific).
Statistics and reproducibility
Normal distribution of the data and equality of variance was verified using Shapiro and Levene tests (Lawstat package62), respectively. Analysis of variance (ANOVAs) or Wilcox Tests was run using the aov and TukeyHSD functions or Wilcox.test function in the R environment with the null hypothesis of no difference between lines63. Tukey’s post hoc test was added to identify each significant interaction between the lines tested. Data were plotted using R ggplot264.
Phylogenic trees
Trees were made using MEGA X65. Amino acid sequences were obtained from Ensembl. Amino acid sequences were aligned using the MUSCLE algorithm as part of MEGA X and the tree was constructed using the maximum likelihood method, Jones–Taylor–Thornton model, with 500 bootstrap replications.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Seed is available upon request from the corresponding author. Seed materials will be transferred under MTA. Data used to make figures and supplemental figures are deposited in figshare, 10.6084/m9.figshare.1907805866.
References
Shewry, P. R. & Hey, S. J. The contribution of wheat to human diet and health. Food Energy Secur. 4, 178–202 (2015).
Voss-Fels, K. P. et al. Breeding improves wheat productivity under contrasting agrochemical input levels. Nat. Plants 5, 706–714 (2019).
Ascott, M. J. et al. Global patterns of nitrate storage in the vadose zone. Nat. Commun. 8, 1–7 (2017).
Delgado, J. A., Del Grosso, S. J. & Ogle, S. M. 15N isotopic crop residue cycling studies and modeling suggest that IPCC methodologies to assess residue contributions to N2O-N emissions should be reevaluated. Nutr. Cycl. Agroecosyst. 86, 383–390 (2010).
Brentrup, F., Hoxha, A. & Christensen, B. Carbon footprint analysis of mineral fertilizer production in Europe and other world regions. (2016).
Swarbreck, S. M. et al. A roadmap for lowering crop nitrogen requirement. Trends Plant Sci. 24, 892–904 (2019).
Raun, W. R. & Johnson, G. V. Improving nitrogen use efficiency for cereal production. Agron. J. 91, 357–363 (1999).
Simkin, A. J., López-Calcagno, P. E. & Raines, C. A. Feeding the world: improving photosynthetic efficiency for sustainable crop production. J. Exp. Bot. 70, 1119–1140 (2019).
Ort, D. R. et al. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl Acad. Sci. USA 112, 8529–8536 (2015).
Long, S. P., Marshall-Colon, A. & Zhu, X. G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66 (2015).
South, P. F., Cavanagh A. P., Liu H. W. & Ort D. R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363, eaat9077 (2019).
Kromdijk, J. et al. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354, 857–861 (2016).
Raun, W. R. & Johnson, G. V. Improving nitrogen use efficiency for cereal production. Agron. J. 91, 357 (1999).
Renny-Byfield, S. & Wendel, J. F. Doubling down on genomes: polyploidy and crop plants. Am. J. Bot. 101, 1711–1725 (2014).
Chen, Z. J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58, 377–406 (2007).
Van De Peer, Y., Maere, S. & Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–732 (2009).
Huang, S. et al. Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 48, 805–820 (2002).
Mcfadden, E. S. & Sears, E. R. The origin of triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37, 107–116 (1946).
Matsuoka, Y. Evolution of polyploid triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 52, 750–764 (2011).
Cheng, H. et al. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 20, 136 (2019).
Pumphrey, M., Bai, J., Laudencia-Chingcuanco, D., Anderson, O. & Gill, B. S. Nonadditive expression of homoeologoes genes is established upon polyploidization in hexaploid wheat. Genetics 181, 1147–1157 (2009).
Akhunova, A. R., Matniyazov, R. T., Liang, H. & Akhunov, E. D. Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11, 505 (2010).
Feldman, M., Levy, A. A., Fahima, T. & Korol, A. Genomic asymmetry in allopolyploid plants: Wheat as a model. J. Exp. Bot. 63, 5045–5059 (2012).
Marcussen, T. et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092 (2014).
Clavijo, B. J. et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27, 885–896 (2017).
Ahammed, G. J., Li, X., Liu, A. & Chen, S. Brassinosteroids in plant tolerance to abiotic stress. J. Plant Growth Regul. 39, 1451–1464 (2020).
Nolan, T. M., Vukasinović, N., Liu, D., Russinova, E. & Yin, Y. Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. In Plant Cell 32, 298–318 (American Society of Plant Biologists, 2020).
Sakamoto, T. et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24, 105–109 (2006).
Morinaka, Y. et al. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141, 924–931 (2006).
Wu, C. et al. Brassinosteroids regulate grain filling in rice. Plant Cell 20, 2130–2145 (2008).
Sahni, S. et al. Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 6, 28298 (2016).
Eremina, M. et al. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl Acad. Sci. USA 113, E5982–E5991 (2016).
Li, X.-J. et al. Overexpression of a brassinosteroid biosynthetic gene Dwarf enhances photosynthetic capacity through activation of Calvin cycle enzymes in tomato. BMC Plant Biol. 16, 33 (2016).
Nie, S. et al. Enhancing Brassinosteroid Signaling via Overexpression of Tomato (Solanum lycopersicum) SlBRI1 Improves Major Agronomic Traits. Front. Plant Sci. 8, 1386 (2017).
Li, X.-J. J. et al. DWARF overexpression induces alteration in phytohormone homeostasis, development, architecture and carotenoid accumulation in tomato. Plant Biotechnol. J. 14, 1021–1033 (2016).
Xiong, Y. et al. Glc-TOR signalling leads transcriptome reprogramming and meristem activation. Nature 496, 181 (2013).
Zhang, Z. et al. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol. 26, 1854–1860 (2016).
Deprost, D. et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8, 864–870 (2007).
Borrill, P., Ramirez-Gonzalez, R. & Uauy, C. expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol. 170, 2172–2186 (2016).
Choe, S. et al. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243 (1998).
Liu, T. et al. Expression and functional analysis of ZmDWF4, an ortholog of Arabidopsis DWF4 from maize (Zea mays L). Plant Cell Rep. 26, 2091–2099 (2007).
McElroy, D., Zhang, W., Cao, J. & Wu, R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2, 163–171 (1990).
Coll-Garcia, D., Mazuch, J., Altmann, T. & Müssig, C. EXORDIUM regulates brassinosteroid-responsive genes. FEBS Lett. 563, 82–86 (2004).
Choe, S. et al. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26, 573–582 (2001).
Chung, Y. & Choe, S. The regulation of brassinosteroid biosynthesis in Arabidopsis. CRC Crit. Rev. Plant Sci. 32, 396–410 (2013).
Cui, X.-Y. et al. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180, 605 (2019).
Wang, Z. Y., Seto, H., Fujioka, S., Yoshida, S. & Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383 (2001).
Nitrogen for winter wheat-management guidelines THE N MANAGEMENT CYCLE 2 Nitrogen for winter wheat-management guidelines. (2009).
Buchner, P. & Hawkesford, M. J. Complex phylogeny and gene expression patterns of members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) in wheat. J. Exp. Bot. 65, 5697–5710 (2014).
Curci, P. L. et al. Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 7, 1176 (2017).
La Rota, M. & Sorrells, M. E. Comparative DNA sequence analysis of mapped wheat ESTs reveals the complexity of genome relationships between rice and wheat. Funct. Integr. Genomics 4, 34–46 (2004).
Liu, N. et al. Overexpression of ZmDWF4 improves major agronomic traits and enhances yield in maize. Mol. Breed. 40, 1–12 (2020).
(No Title). https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/868936/structure-jun2019provcrops-eng-28feb20.pdf. Accessed 26th August 2020.
Farming Statistics Provisional crop areas, yields and livestock populations At June 2019—United Kingdom. Office of National Statistics 1–24 (2019). https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/868943/structure-jun2019prov-UK-28feb20.pdf. Accessed 15th September 2021.
US EPA, O. Greenhouse Gas Equivalencies Calculator.
Bates, R., Craze, M. & Wallington, E. J. Agrobacterium-mediated transformation of oilseed rape (Brassica napus). In Current Protocols in Plant Biology 287–298 (2017). https://doi.org/10.1002/cppb.20060
Ishida, Y., Tsunashima, M., Hiei, Y. & Komari, T. Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 1223, 189–198 (2015).
Risacher, T., Craze, M., Bowden, S., Paul, W. & Barsby, T. Highly efficient agrobacterium-mediated transformation of wheat via in planta inoculation. Methods Mol. Biol. 478, 115–124 (2009).
Milner, M. J. et al. A PSTOL-like gene, TaPSTOL, controls a number of agronomically important traits in wheat. BMC Plant Biol. 18, 115 (2018).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Hiscox, J. D. & Israelstam, G. F. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 57, 1332–1334 (1979).
Gastwirth, J. L., Gel, Y. R., Hui, W., Lyubchich, V., & Miao, W. Tools for Biostatistics, Public Policy and Law. 1–44 (2017).
Computing, R. F. for S. R Core Team (2018). R: A language and environment for statistical computing. https://www.r-project.org/. (2018).
Wickham, H. Introduction. in ggplot2 (Springer New York, 2009). https://doi.org/10.1007/978-0-387-98141-3_1
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. https://doi.org/10.1093/molbev/msy096
Milner, M. DWF4 files. Figshare https://doi.org/10.6084/m9.figshare.19078058
Acknowledgements
This study was supported by BBSRC and the Newton–Bhabha Fund, under the Virtual Joint Centres in Agricultural Nitrogen Use initiative (BB/N013441/1: Cambridge—India Network in Translational Nitrogen (CINTRIN). We thank Dr. Gregory Reeves and Prof. Julian Hibberd (University of Cambridge) for assistance with photosynthetic measurements and CINTRIN members for additional discussions.
Author information
Authors and Affiliations
Contributions
M.C. and S.B. generated the transgenic wheat plants. M.M. performed molecular and biochemical analysis, plus growth, and gas-exchange experiments. S.M.S. performed 15N uptake experiments. M.M. and S.M.S. performed data analysis on their contributions. M.M. designed the experiments and wrote the paper with input and contributions from S.M.S., H.G., A.R.B., and E.J.W. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Golam Ahammed and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editor: Caitlin Karniski. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Milner, M.J., Swarbreck, S.M., Craze, M. et al. Over-expression of TaDWF4 increases wheat productivity under low and sufficient nitrogen through enhanced carbon assimilation. Commun Biol 5, 193 (2022). https://doi.org/10.1038/s42003-022-03139-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-022-03139-9
- Springer Nature Limited
This article is cited by
-
Bioinformatics and Functional Analysis of OsASMT1 Gene in Response to Abiotic Stress
Biochemical Genetics (2024)
-
Nitrogen deficiency tolerance conferred by introgression of a QTL derived from wild emmer into bread wheat
Theoretical and Applied Genetics (2024)
-
Precision breeding in agriculture and food systems in the United Kingdom
Transgenic Research (2024)
-
Identification of structural variations related to drought tolerance in wheat (Triticum aestivum L.)
Theoretical and Applied Genetics (2023)
-
The brassinosteroid biosynthesis gene TaD11-2A controls grain size and its elite haplotype improves wheat grain yields
Theoretical and Applied Genetics (2022)