Abstract
The quick Sequential Organ Failure Assessment (qSOFA) system identifies an individual’s risk to progress to poor sepsis-related outcomes using minimal variables. We used Support Vector Machine, Learning Using Concave and Convex Kernels, and Random Forest to predict an increase in qSOFA score using electronic health record (EHR) data, electrocardiograms (ECG), and arterial line signals. We structured physiological signals data in a tensor format and used Canonical Polyadic/Parallel Factors (CP) decomposition for feature reduction. Random Forests trained on ECG data show improved performance after tensor decomposition for predictions in a 6-h time frame (AUROC 0.67 ± 0.06 compared to 0.57 ± 0.08, \(p = 0.01\)). Adding arterial line features can also improve performance (AUROC 0.69 ± 0.07, \(p < 0.01\)), and benefit from tensor decomposition (AUROC 0.71 ± 0.07, \(p = 0.01\)). Adding EHR data features to a tensor-reduced signal model further improves performance (AUROC 0.77 ± 0.06, \(p < 0.01\)). Despite reduction in performance going from an EHR data-informed model to a tensor-reduced waveform data model, the signals-informed model offers distinct advantages. The first is that predictions can be made on a continuous basis in real-time, and second is that these predictions are not limited by the availability of EHR data. Additionally, structuring the waveform features as a tensor conserves structural and temporal information that would otherwise be lost if the data were presented as flat vectors.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Sepsis is a syndrome induced by an existing infection in the body that produces life-threatening organ dysfunction in a chain reaction. The clinical criteria for sepsis include suspected or documented infection and an increase in two or more Sequential Organ Failure Assessment (SOFA) points. Septic shock, a more severe subset, consists of substantially increased abnormalities1 and higher risk of mortality2. It is imperative to risk-stratify patients early in their course in order to appropriately direct critical, but potentially limited, resources and therapies.
Sepsis’ heterogeneity complicates its diagnosis and prognosis. Its current definition, based on SOFA score, requires measurement or collection of variables which may not be immediately available. The quick-SOFA (qSOFA) is a screening tool that can be performed at the bedside. It consists of three criteria—Glasgow Coma Scale of < 15 (indicating mental status change), respiratory rate \(\ge\) 22 breaths per minute, and systolic blood pressure \(\le\) 100 mmHg—where two of the three must be met1. It includes the poorly characterized variable mental status change, but it is a better predictor of organ dysfunction than systemic inflammatory response syndrome (SIRS), which is less sensitive3,4. SIRS is the body’s response to a stressor such as inflammation, trauma, surgery, or infection, while sepsis is specifically a response to infection; many septic patients have SIRS, but not all patients who meet SIRS criteria have an infection or experience septic organ failure. In comparison to qSOFA, SIRS has four criteria, three of which must be met to positively identify SIRS. These are: respiratory rate > 20 breaths per minute or partial pressure of CO2 < 32 mmHg; heart rate > 90 beats per minute; white blood cell count > 12,000/microliter or < 4000/microliter or bands > 10%; and temperature >38 \(^{\circ }\)C or < 36 \(^{\circ }\)C5. For each of these scoring systems, factors such as comorbidities, medication, and age may confound the phenotype in different patient groups. In previous work, SOFA score predicted sepsis onset upon ICU admission with AUROC of 0.73, qSOFA with AUROC of 0.77, and SIRS with AUROC of 0.616. Among patients with suspected infection in the ICU, SOFA score predicted in-hospital mortality with AUROC of 0.74, qSOFA with AUROC of 0.66, and SIRS with AUROC of 0.644.
A system of sepsis detection which is too strict or time-consuming can delay necessary care to patients, and criteria that are too broad can lead to over-treatment or inappropriate use of limited resources. For example, false positive sepsis prognoses can lead to patients receiving unnecessary care and antibiotics, which contribute to antibiotic resistance and emergence of “superbugs”7,8,9. Similarly, qSOFA is not recommended as a single screening tool for diagnosis of sepsis10, but it can be used as a method of predicting prolonged ICU stay or in-hospital mortality4. Predicting the trajectory of a patient with suspected infection may be a more efficient use of resources than detecting existing sepsis, and therefore trajectory prediction is the focus of this study.
Many models for detecting, monitoring, or predicting outcomes related to sepsis depend on Electronic Health Record (EHR) data, including SOFA score1, EPIC’s sepsis model11, and others12,13,14. EHR data can include static variables like demographics information, and dynamic variables such as vital signs or lab values. While useful for determining a patient’s status, EHR data are limited by time. Lab values require time for collection and processing, and continuous variables may be updated less than hourly or at irregular intervals. In contrast, physiological readings, such as those generated from electrocardiography, blood pressure monitoring, or pulse oximetry, are collected continuously. Our study examines the use of continuous physiological signals, namely electrocardiogram (ECG) and arterial line, in outcome prediction related to sepsis.
ECG signal information has previously been used in the study of risk for sepsis and sepsis progression15,16,17. The advantage that continuous monitoring devices like ECG offer over EHR data is real-time, continuous assessment of a patient’s status. In addition, ECG is routinely collected in the intensive care unit (ICU), and is minimally invasive. In our analysis, we also include arterial line. Yearly, roughly eight million arterial catheters are placed in patients at hospitals in the United States, or roughly 10–12% of patients that undergo anesthesia18,19. We choose to include arterial line in the study as both SOFA and qSOFA use blood pressure to assess the status of a patient’s cardiovascular system1.
Given sepsis’ complexity and heterogeneity, it is necessary to incorporate multiple variables into a trajectory prediction method. Modeling data as a tensor provides the ability to observe changes in different variables with respect to time and to one another. The prognosis and severity assessment of sepsis rely on a large amount of heterogeneous data, including body temperature, arterial blood pressure, blood culture tests, and molecular assays. Treatment of sepsis does not rely on any individual variable, but on all of these measurements, which vary as a function of time. Because no individual feature is sufficient, integrating data across time and incorporating structure is necessary for improved sepsis prognosis, and therefore can better inform care decisions.
In this study, we use ECG and arterial line signals to predict an increase in an individual’s qSOFA score, where a qSOFA of \(\ge\) 2 indicates poor outcomes related to sepsis. The results of signal-trained models are then compared to models trained using both signals and EHR data. This is to (1) predict which individuals are at risk of septic shock, future organ failure, or other complications related to sepsis, rather than focusing on a sepsis diagnosis, and (2) assess the usefulness of continuous physiological signals in the event that EHR data are unavailable, such as the time between EHR data collection times. Outside of the hospital, one such scenario is the case of home monitoring, where after a patient is released from the hospital, their ECG is still being recorded in case of a cardiac event20.
Methods
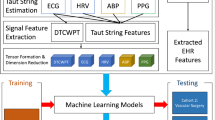
A schematic of the methods used in this paper is presented in Fig. 1.
Dataset
The retrospective dataset consisted of 1803 unique individuals age \(\ge\) 18 years with 3516 unique encounters between 2013 and 2018 at Michigan Medicine. Individuals’ characteristics are presented in Supplementary Table 1. The detailed inclusion/exclusion criteria for the dataset are provided in Supplementary Materials Sect. 1.3, but, briefly, inclusion criteria selected for inpatient encounters with: ECG lead II waveforms at least 15 min in length and ICD9/10 codes for pneumonia, cellulitis, or urinary tract infection (UTI), excluding UTIs associated with catheters. These are infections that have been documented in previous sepsis cases. Exclusion criteria included positive HIV status, solid organ or bone marrow transplant, and ongoing chemotherapy. These exclusion criteria were selected as individuals undergoing organ/bone marrow transplants are usually given immunosuppressant medications, and therefore react differently to infection than a typical patient who enters the ICU with an infection. Additionally, positive HIV status and chemotherapy treatments also affect the immune system, and therefore affect how these individuals react to infection. These criteria created a dataset that did not specifically select for sepsis diagnosis, but instead focused on patients with an infection who were at risk to develop sepsis and septic shock.
This dataset that we used was selected from an existing Michigan Medicine biobank, whose original data collection was approved by the institutional review board of University of Michigan’s medical school, IRBMED. The protocols of this retrospective study (accession number HUM00092309) were reviewed and approved by IRBMED. The protocols were carried out in accordance with applicable guidelines, state and federal regulations, and the University of Michigan’s Federalwide Assurance with the Department of Health and Human Services. Informed consent was waived, as this was a retrospective study of previously collected and de-identified data, without direct involvement of human subjects and therefore no chance of physical harm or discomfort to the individuals being studied. Individuals reported their own sex and race/ethnicity, from categories defined by Michigan Medicine, and this information is included in Supplementary Table 1 to provide information on the population of this study. The study performed in this paper only used the retrospective data previously collected by the existing biobank, and did not perform any new recruitment or data collection. Individuals’ data are not shared in this project’s publicly available code. The risk of re-identification from the de-identified dataset is low; (1) the key linking de-identified patients to their original patient records is not made available at any point of the machine learning stages, from feature extraction to model training or deployment, (2) dates of EHR data collected are obfuscated from the model, and instead, relative dates (e.g., time between collections) are used, so training data retained within the model cannot necessarily be linked back to exact dates within the EHR data.
This larger dataset was reduced by selecting for individuals who had EHR, ECG, and arterial line data available. In this study, EHR data included labs, medications, hourly fluid output, and vital signs. Because poor signal quality can result in false alarms21, the ECG signal was reviewed automatically using Pan-Tompkins to identify QRS complexes22,23. Upon collecting 10-min signals for feature extraction, signals determined to be 50% or more noise were discarded. The method for identifying noise in ECG has previously been used in studies of arrhythmia and atrial fibrillation24,25.
Change in qSOFA score was used to assign positive and negative classes for machine learning. Given an individual who meets one of the criteria for qSOFA, the model predicts whether the score will increase to \(\ge\) 2, which Sepsis-3 deems as “likely to have poor outcomes”1. This increase in qSOFA is considered the positive outcome in a learning context, because the patient meets at least 2 qSOFA criteria as defined by Sepsis-3 after the prediction gap. Thus, the negative outcome is qSOFA < 2 after the prediction gap.
We tested prediction gaps of 6 and 12 h. These gaps were chosen because, if a decompensation event is predicted six or more hours in advance, this gives ample time for healthcare providers to give the appropriate therapies or move the patient to appropriate facilities. For a 6-h gap, there were 199 negative and 59 positive cases. For a 12-h gap, there were 189 negative and 37 positive cases.
Signal processing
For every sample, we collected the 10 min of signal occurring directly before the prediction gap for processing. This 10-min signal was divided into 2 5-min windows, and then preprocessed according the relevant sections below.
Arterial line data
Arterial line signals were sampled at 120 Hz. We applied a third order Butterworth bandpass filter with cutoff frequencies 1.25 and 25 Hz to remove artifacts. These cutoff frequencies were selected from a previous study that used equipment from the same hospital26, and were determined to adequately capture the movement of the arterial waveform while also reducing noise and other artifacts. The BP_Annotate software package27 annotated the signal. Following previous methodology28, we extract features from the annotated signal: number of peaks, as well as the minimum, maximum, mean, median, and standard deviation (SD) of time between sequential systolic peaks, time between a systolic peak and its subsequent diastolic reading, relative amplitude between systolic peaks, and relative amplitude between a systolic peak and its subsequent diastolic reading.
Electrocardiogram data
ECG data consisted of four leads and signals were sampled at 240 Hz. We used lead II of the ECG, following previously established methods29. A second order Butterworth bandpass filter with the cutoff frequencies 0.5 and 40 Hz removed noise, and baseline wander. This follows from previous work26. Lead II was selected as it is commonly used for monitoring in the ICU, and therefore is clinically relevant.
Taut string
Peak-based and statistical features were calculated from the Taut String (TS) estimation30 of the ECG waveform. Others have previously used such features to detect hemodynamic instability29 and predict hemodynamic decompensation26,31. TS provides a piecewise linear estimation of an input signal at a specified level of “wiggle room”, \(\epsilon\). After applying TS, the resulting approximation looks like a piece of string pulled tight between the peaks and valleys of input signal \(\pm \epsilon\). An illustration of the Taut String approximation is provided in Fig. 2
TS estimation functions as follows. Given a discrete signal \(f = (f_0, f_1, ..., f_n)\) for a fixed value \(\epsilon > 0\), the TS estimate of f is the unique function g such that
and
is minimal, with D being the difference operator.
TS estimation was applied to the filtered ECG signal using the five values of the parameter \(\epsilon\): 0.0100, 0.1575, 0.3050, 0.4525, and 0.6000. These values were selected from previous work26. Six features were computed from each TS estimate of a 5-min window and value of \(\epsilon\). These features were: number of line segments, number of inflection segments, total variation of noise, total variation of denoised signal, power of denoised signal, and power of noise. This resulted in a tensor of size \(2 \times 5 \times 6\) for each signal, where the modes of the tensor were window, \(\epsilon\), feature.
Electronic health record data
We assigned an ordinal encoding to labs and cardiovascular infusions ranging from 0–4 or 0–3, respectively. A score of 1 indicates less severity and a score of 3 or 4, more severity. If a lab value had been recorded before the time of interest, this value was carried forward. This would be considered the most up-to-date assessment of a lab value and not be considered missing data. To differentiate from missing data, we assigned a score of 0 to represent a missing value with no previous recordings. The Supplementary Materials Sect. 1.2 provides tables detailing these assignments. Vital signs and urine output were included, but not given an ordinal encoding. If vital signs or urine output were not reported in the time of interest, we carried forward the most recent known value. As it cannot be guaranteed that missing urine output or vital signs were missed completely at random, carrying the last value forward has some risk of biasising the data32.
We added a retrospective component for lab values, cardiovascular infusions, and vital signs where, in addition to the 10 min occurring before the prediction gap, we include four look-back periods. For the prediction gap of 6 h, these look-back periods are increments of 4 h; for the prediction gap of 12 h, they are increments of 8 h. Look-back periods were developed in a previous study of postoperative cardiac decompensation events31. They allow for the inclusion of previous observations of data, before signal collection began for patients in the ICU.
Feature reduction with tensor methods
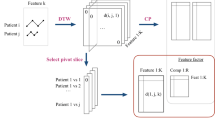
For each 10-min ECG signal, 60 features were computed and arranged as a tensor of size \(2 \times 5 \times 6\). For each 10-min arterial line signal, 42 features were arranged as a tensor of size \(2 \times 1\times 21\), where the second mode, TS parameter \(\epsilon\), was inflated to create a uniform presentation to the tensor reduction algorithms. The reasoning behind using a tensor structure was similar to envisioning the different incoming signals as an image. Similar to how the rows of pixels in an image have spatial relationships to one another33, the series of TS approximations of ECG have temporal relationships to one another. By separating these features into different vectors, that temporal relationship would be lost. As an example, flattening image data into a vector before feature reduction was found to be less effective than tensor reduction when training a model to detect changes in images34. See Fig. 3 for an illustration of how tensors are produced from the ECG signal.
Rather than treating this information as 60 or 42 feature vectors, we preserved the underlying tensor structure by using a tensor-based dimensionality reduction method, inspired by previous work26 and described below.
First, each tensor’s underlying structure was determined. All \(2 \times 5 \times 6\) ECG-feature tensors in the training set were stacked along the fourth mode, generating a new tensor of size \(2 \times 5 \times 6 \times N\), where N was the number of observations in the training set. Similarly, all \(2 \times 1 \times 21\) arterial line-feature tensors were stacked along the fourth mode to generate a new tensor of size \(2 \times 1 \times 21 \times N\).
Tensor Toolbox’s35 Canonical Polyadic / Parallel Factors (CP) decomposition36 was used to obtain the underlying structure of the tensors. A CP decomposition breaks the initial tensor down into a sum of rank-1 tensors, so it can be considered an extension of singular value decomposition to a higher order. Similar to how performing a singular value decomposition on an image (matrix) can create a compressed and simplified image, the CP decomposition creates a compressed and simplified estimation of the original tensor.
In general, given a tensor
and a predetermined rank r, the CP decomposition gives a tensor
such that \(\Vert X - \hat{X}\Vert\) is minimized, where \(\otimes\) denotes the Kronecker product. The multiplication of vectors \(v_1, \dots v_d\) yields a component rank-1 tensor. Because the tensors used in this specific case are fourth-order, this can be written as:
The vectors \(a_1,\dots ,a_r\in \mathbb {R}^{n_1}\), and so on, can be combined to form factor matrices, such as \(A = [a_1, \dots , a_r]\in \mathbb {R}^{n_1\times r}\), and similarly for B, C, D. In this manner, each mode of the original tensor X can be approximated by the product of these factor matrices, such as:
where \(\odot\) denotes the Khatri-Rao product for the first mode. Because finding a CP decomposition is NP-hard37, we used the Alternating Least Squares (ALS) heuristic method, which is an iterative algorithm to find the best approximation of X for a given rank r36.
The dataset was divided into an 75/25 split 100 times, and tensor reduction was performed on each of those splits. A fit score, defined as
was calculated to determine how well the reduced tensor approximated the original. This CP-ALS process was repeated 15 times, with the selected reduction being the one with the highest fit, or the first reduction with fit score equal to one, whichever occurred first. CP-ALS was run using rank values of 1–4.
After applying CP-ALS to the training data, the resultant factor matrices A and B were retained, which related to the modes of the original tensor that were not the feature mode (C) or the patient encounter mode (D).
With this process completed, for any given individual’s third-order tensor T, a reduced set of features was extracted using the factor matrices computed from the training data. The feature vectors \(c_{T,1}, \dots , c_{T,r}\) were computed via a least squares problem, where
is minimal. After computing the individual vectors, they were concatenated to create \(C_T\), a feature matrix with a reduced set of features compared to matricization \(T_{(3)}\) of the original tensor T along the third mode.
Machine learning
When constructing training and test datasets, 75/25 splits were created based on individuals so that no individual would overlap between the training and test sets.
After extracting features, the three types of learning models used for training were linear Support Vector Machines (SVM)38, Random Forest (RF)39, and Learning Using Concave and Convex Kernels (LUCCK)40. We selected a linear kernel for SVM in this experiment because it would be less susceptible to overfitting when many features are present41 (such as in the case when no tensor reduction is used), and a linear kernel is both faster to train and more easily interpretable than a nonlinear kernel42. Additionally, datasets with many features can become linearly separable, making the linear kernel a good option both in terms of its transparency as well as its faster training time43. We opted not to test deep learning models because we wanted to offer transparency to the end user of the model and to patients who would receive care, as deep learning models are known for operating as a “black box”; a patient would trust a clinician who understands the “explainable” machine learning method that they use to assist in their decision-making (referred to as the AI-user dyad)44.
For all methods, the training phase consisted of threefold cross-validation (3FCV) on a 75/25 split of the data, where the test set was held and not used for training. The test set was presented to the three models generated from 3FCV to produce three sets of prediction scores. We computed the final prediction scores for the test set by taking the median of the three prediction scores, thus creating a voting system. This process was repeated 100 times to obtain mean and standard deviation values of model performance.
A grid search selected optimal hyperparameters for each model using the validation fold in 3FCV. For RF, these hyperparameters included: number of trees, minimum leaf size, fraction of maximum number of splits, and number of predictors to sample. For SVM, grid search selected the best box constraint C. Sequential minimal optimization45 was used for the optimization routine. For LUCCK, grid search selected optimal \(\Lambda\) and \(\Theta\) parameters. All grid searches used F1 score as the value to optimize.
Different signal feature based models were tested using tensor reduction. The first, using only ECG data and presented in Figs. 4 and 5, was the most restricted model, assuming that both EHR and arterial line data were unavailable. This would apply to patients recently admitted, who would not have lab values or other EHR data available, and is also minimally invasive compared to having an arterial line in place. Next, a model trained on both ECG and arterial line features, presented in Figs. 6 and 7, which was tested to determine if the invasive arterial line improved performance compared to only using ECG data. Lastly, a model trained on signal features alongside EHR data was built, presented in Figs. 8 and 9.
Results
RF, LUCCK, and SVM were trained on tensor-reduced ECG features, presented in Figs. 4 and 5. We compare these models to those trained on tensor-reduced ECG features and arterial line features, presented in Figs. 6 and 7. These figures display the mean F1 Score and AUROC over 100 iterations, with error bars indicating one SD. The x-axis indicates the rank selected for CP-ALS, with the rightmost columns, separated with a dashed line, representing the case where no tensor decomposition was applied. Figures 8 and 9 show the results of models trained on both the tensor-reduced signal features and EHR data.
Data can also be viewed in a table format. Tables 1 and 2 present ECG-trained models at 6- and 12-h prediction intervals, Tables 3 and 4 show results for models trained with ECG and arterial line, and Tables 5 and 6 for models trained with ECG, arterial line, and EHR data, and Table 7 is for models trained with only EHR data. In these tables, “Rank” indicates the rank selected for CP-ALS, and a rank of “None” that CP-ALS was not performed.
Discussion
After extracting features from the EHR and from physiological signals, RF, LUCCK, and SVM models were trained. The results from these models are presented in “Results” section as graphs and tables. RF and LUCCK models performed similarly across different experiments, both performing better than SVM when tensor reduction was applied to the dataset. RF’s strong performance across different levels of feature reduction could be due to its bagging and bootstrapping procedures, which work to prevent overfitting and ignore noise39,46. In its introductory paper, LUCCK was shown to perform well even when trained with few samples of signal data, in part due to its similarity function, which allows for noise or large deviations in some features to not overwhelm the model40. Although SVM is known to perform well when few training samples are available47, there are also cases where if the data is feature-dense, linear SVM will perform as well as SVM trained with a nonlinear kernel48, as a large number of features can make a dataset linearly separable43. This may be why the non-tensor-reduced datasets tended to have stronger performance than datasets with tensor reduction for SVM.
For RF and LUCCK, both F1 Score and AUROC tended to increase when moving from no tensor reduction to tensor reduction when using only ECG signal data. For example, for LUCCK in the 6-h dataset, mean F1 score increased from 0.43 to 0.48 with SD remaining similar (0.06 to 0.07, \(p < 0.01\)), while RF’s F1 score increased from 0.41 to 0.48 without a change in SD, \(p < 0.01\). Here, p-values were generated from t-tests. We observed a similar increase in mean AUROC for LUCCK (0.60 ± 0.07 to 0.65 ± 0.07, \(p < 0.001\)) and RF (0.57 ± 0.08 to 0.67 ± 0.06, \(p = 0.01\)) going from using no tensor reduction to using CP-ALS with rank 4. SVM does not follow this trend, however, and tends to increase in performance as more information is added to the model, with no tensor reduction performing the best. We see a similar trend in the 12-h dataset. While AUROC is not a justification in and of itself for these models to be used in clinical practice, AUROC offers a method of comparing the discriminatory ability of each of the models presented in this paper, with higher AUROCs indicating stronger ability to distinguish between the at-risk (positive) and not-at-risk (negative) groups49.
For 6-h data, including the arterial line features improved both mean F1 Score and mean AUROC across different CP-ALS ranks, as can be seen comparing Figs. 4 and 6. For RF, including arterial line features improved performance compared to only using ECG signals without tensor reduction (AUROC 0.69 ± 0.07, \(p < 0.01\)), and also showed improvement in AUROC from tensor decomposition (AUROC 0.71 ± 0.07, \(p = 0.01\)). Adding EHR data features to a tensor-reduced signal model further improves performance (AUROC 0.77 ± 0.06, \(p < 0.01\)). For 12-h data, RF and LUCCK results are mixed across the different ranks, but including both Arterial Line and ECG data decreased SVM’s performance when no tensor reduction took place. When CP-ALS was used with ranks 1–3 to reduce the feature space for SVM, there is an increase in performance in the ECG + Arterial Line scenario; this suggests that SVM may not be a reliable model for these scenarios.
Adding EHR data to the signal features, presented in Figs. 8 and 9, further improves performance for both the 6- and 12-h datasets, across all three model types. For example in RF with tensor reduction rank 4, AUROC increased to 0.77 ± 0.06, (\(p < 0.01\)) in the 6-hour prediction range.
We included results from models trained on EHR data only as a comparison in Table 7, which shows that EHR data on its own is very informative. RF, SVM, and LUCCK models had an average AUROC of greater than 0.6 across all models. The purpose of this study, however, is to observe the performance of models informed by physiological signals.
While the results of models trained on tensor-reduced signal features show consistent mean AUROC \(\ge\) 0.65 for both LUCCK and RF, it is noted that these experiments were trained on data from only one hospital, the availability of signals led to a small sample pool, and the datasets used do not feature strong racial and ethnic diversity. To ensure the reproducibility and generalizability of these results, it will be necessary to perform similar experiments on a larger and more diverse dataset in future iterations.
Conclusion
In this study, predictions of increase in qSOFA score were created using tensor-reduced signal features and EHR data. It is possible to make a prediction of increase in qSOFA score using ECG data alone (for RF, AUROC 0.67 ± 0.06; for LUCCK, 0.65 ± 0.07), and results can be improved if tensor-reduced arterial line features are added (for RF, AUROC 0.71 ± 0.07; for LUCCK, 0.71 ± 0.07), but results are mixed when signal features are directly added without tensor reduction (for RF, AUROC 0.69 ± 0.07; for LUCCK, 0.69 ± 0.07). This may be because the models are overwhelmed with information, whereas tensor reduction improves performance because only pertinent information is given and noise is removed.
The previous experiments simulate the scenario when EHR data are completely unavailable. When EHR data are available and CP-ALS is used to reduce the feature space of the signal data, results can be further improved (for RF, AUROC 0.77 ± 0.06; for LUCCK, 0.73 ± 0.07). This indicates that ECG signal features, Arterial Line signal features, and EHR data features can all contribute to sepsis prognosis.
That said, we wish to draw attention to the first scenario, with signals information alone used for model training. The advantage of a signal features-based model is that predictions can be made in the ICU on a continuous basis in real-time; this model would not be limited by the wait times or availability of EHR data variables. From a clinical standpoint, further developing an ECG-only model would be advantageous as, (1) it is minimally invasive compared to an arterial line, and (2) it is possible to monitor ECG remotely outside the hospital. Devices such as Holter monitors and Zio patches could be used so that a patient with initially low qSOFA could be monitored at home, with a 6-h window to predict an increased risk for poor outcomes. Six hours would be adequate time for warning and arrival to the emergency department to seek appropriate treatment. Although at-home monitoring is more likely to be affected by movement than an in-hospital setting, Holter monitors are the current gold standard compared to other wearable technologies, which are more susceptible to motion artifacts50,51.
We stress that, while it may not achieve F1 or AUROC scores as high as the model including EHR data, our signal features-only model offers an advantage in that it is not prone to issues such as availability or inaccuracies of EHR data. Furthermore, it is continuously collected allowing for real-time evaluation and assessment. For future work, we recommend (1) the combination of EHR, tensor-reduced ECG, and tensor-reduced arterial line for use in the hospital or ICU and (2) tensor-reduced ECG only for use in home monitoring. Additionally, we can further study the use of interpretable deep learning models52, which can be coupled with tensor decomposition for feature reduction.
Data availability
Data from the electronic health records and physiological signals belong to the University of Michigan, and cannot be publicly distributed due to reasons of patient privacy. Data are located in controlled access storage at the University of Michigan. Access to patient-level data requires an agreement with University of Michigan. Any requests regarding data for this study can be sent to Drew Bennett (andbenne@umich.edu) of University of Michigan Innovation Partnerships and the corresponding author.
Code availability
The underlying code for this study is available on GitHub and can be accessed via this link: https://github.com/kayvanlabs/public-tensor-bigdata-qsofa. Our repository does not provide libraries created by others as to not violate their licenses. Additionally, the source code of LUCCK and Taut String methods cannot be shared due to proprietary reasons, but standalone Matlab executables are made available in the repository.
References
Singer, M. et al. The Third International Consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315(8), 801–810. https://doi.org/10.1001/jama.2016.0287 (2016).
Paoli, C. J., Reynolds, M. A., Sinha, M., Gitlin, M. & Crouser, E. Epidemiology and costs of sepsis in the united states—An analysis based on timing of diagnosis and severity level. Crit. Care Med. 46(12), 1889–1897. https://doi.org/10.1097/CCM.0000000000003342 (2018).
Bone, R. C. et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest 101(6), 1644–1655. https://doi.org/10.1378/chest.101.6.1644 (1992).
Seymour, C. W. et al. Assessment of clinical criteria for sepsis: For the Third International Consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315(8), 762. https://doi.org/10.1001/jama.2016.0288 (2016).
Chakraborty, R. K. & Burns, B. Systemic Inflammatory Response Syndrome (StatPearls Publishing, 2022).
Desautels, T. et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: A machine learning approach. JMIR Med. Inform. 4(3), 28. https://doi.org/10.2196/medinform.5909 (2016).
VanEpps, J. S. Reducing exposure to broad-spectrum antibiotics for bloodstream infection. J. Lab. Precis. Med. 3, 100. https://doi.org/10.21037/jlpm.2018.12.02 (2018).
Prestinaci, F., Pezzotti, P. & Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 109(7), 309–318. https://doi.org/10.1179/2047773215Y.0000000030 (2015).
Chokshi, A., Cennimo, D., Horng, H. & Sifri, Z. Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 11(1), 36. https://doi.org/10.4103/jgid.jgid_110_18 (2019).
Evans, L. et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock 2021. Crit. Care Med. 49(11), 1063–1143. https://doi.org/10.1097/CCM.0000000000005337 (2021).
Wong, A. et al. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern. Med. 181(8), 1065. https://doi.org/10.1001/jamainternmed.2021.2626 (2021).
Nesaragi, N., Patidar, S. & Thangaraj, V. A correlation matrix-based tensor decomposition method for early prediction of sepsis from clinical data. Biocybern. Biomed. Eng. 41(3), 1013–1024. https://doi.org/10.1016/j.bbe.2021.06.009 (2021).
Morrill, J. et al. The signature-based model for early detection of sepsis from electronic health records in the intensive care unit. In 2019 Computing in Cardiology (CinC) 1–4. https://doi.org/10.23919/CinC49843.2019.9005805 (2019).
Taylor, R. A. et al. Prediction of in-hospital mortality in emergency department patients with sepsis: A local big data-driven, machine learning approach. Acad. Emerg. Med. 23(3), 269–278. https://doi.org/10.1111/acem.12876 (2016).
Berger, T. et al. Shock index and early recognition of sepsis in the emergency department: Pilot study. West. J. Emerg. Med. 14(2), 168–174. https://doi.org/10.5811/westjem.2012.8.11546 (2013).
Moorman, J. R. et al. Cardiovascular oscillations at the bedside: Early diagnosis of neonatal sepsis using heart rate characteristics monitoring. Physiol. Meas. 32(11), 1821–1832. https://doi.org/10.1088/0967-3334/32/11/s08 (2011).
Nemati, S. et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit. Care Med. 46(4), 547–553. https://doi.org/10.1097/CCM.0000000000002936 (2018).
O’Horo, J. C., Maki, D. G., Krupp, A. E. & Safdar, N. Arterial catheters as a source of bloodstream infection: A systematic review and meta-analysis. Crit. Care Med. 42(6), 1334–1339 (2014).
Kim, S.-H. et al. Accuracy and precision of continuous noninvasive arterial pressure monitoring compared with invasive arterial pressure: A systematic review and meta-analysis. Anesthesiology 120(5), 1080–1097 (2014).
Faruk, N. et al. A comprehensive survey on low-cost ecg acquisition systems: Advances on design specifications, challenges and future direction. Biocybern. Biomed. Eng. 41(2), 474–502. https://doi.org/10.1016/j.bbe.2021.02.007 (2021).
Gambarotta, N., Aletti, F., Baselli, G. & Ferrario, M. A review of methods for the signal quality assessment to improve reliability of heart rate and blood pressures derived parameters. Med. Biol. Eng. Comput. 54(7), 1025–1035. https://doi.org/10.1007/s11517-016-1453-5 (2016).
Pan, J. & Tompkins, W. J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 32(3), 230–236. https://doi.org/10.1109/TBME.1985.325532 (1985).
Sedghamiz, H. Matlab Implementation of Pan Tompkins ECG QRS detector. MathWorks File Exchange. https://www.mathworks.com/matlabcentral/fileexchange/45840-complete-pan-tompkins-implementation-ecg-qrs-detector, https://doi.org/10.13140/RG.2.2.14202.59841 (2014).
Zhang, W. et al. Evaluation of capacitive ecg for unobtrusive atrial fibrillation monitoring. IEEE Sens. Lett. 7(10), 1–4. https://doi.org/10.1109/LSENS.2023.3315223 (2023).
Li, Z. et al. Prediction of cardiac arrhythmia using deterministic probabilistic finite-state automata. Biomed. Signal Process. Control 63, 102200. https://doi.org/10.1016/j.bspc.2020.102200 (2021).
Hernandez, L. et al. Multimodal tensor-based method for integrative and continuous patient monitoring during postoperative cardiac care. Artif. Intell. Med. 113, 102032. https://doi.org/10.1016/j.artmed.2021.102032 (2021).
Laurin, A. BP_annotate. https://www.mathworks.com/matlabcentral/fileexchange/60172-bp_annotate (Accessed 22 July 2022) (2017).
Luo, Y. The Severity of Stages Estimation During Hemorrhage Using Error Correcting Output Codes Method. PhD thesis, VCU Libraries. Publication Title: VCU Theses and Dissertations. https://doi.org/10.25772/MMMX-AF85, https://scholarscompass.vcu.edu/etd/406 (Accessed 05 August 2022) (2012).
Belle, A. et al. A signal processing approach for detection of hemodynamic instability before decompensation. PLoS ONE 11(2), 0148544. https://doi.org/10.1371/journal.pone.0148544 (2016).
Davies, P. L. & Kovack, A. Local extremes, runs, strings and multiresolution. Ann. Stat. 29, 1–65. https://doi.org/10.1214/aos/996986501 (2001).
Kim, R. B. et al. Prediction of postoperative cardiac events in multiple surgical cohorts using a multimodal and integrative decision support system. Sci. Rep. 12(1), 11347. https://doi.org/10.1038/s41598-022-15496-w (2022).
Lachin, J. M. Fallacies of last observation carried forward analyses. Clin. Trials 13(2), 161–168. https://doi.org/10.1177/1740774515602688 (2016).
Wolf, L., Jhuang, H. & Hazan, T. Modeling appearances with low-rank svm. In 2007 IEEE Conference on Computer Vision and Pattern Recognition 1–6 (IEEE, 2007).
Yan, H., Paynabar, K. & Shi, J. Image-based process monitoring using low-rank tensor decomposition. IEEE Trans. Autom. Sci. Eng. 12(1), 216–227. https://doi.org/10.1109/TASE.2014.2327029 (2015).
Bader, B. W. & Kolda, T. G. Matlab Tensor Toolbox. www.tensortoolbox.org (2017).
Kolda, T. G. & Bader, B. W. Tensor decompositions and applications. SIAM Rev. 51(3), 455–500. https://doi.org/10.1137/07070111X (2009).
Hillar, C. J. & Lim, L.-H. Most tensor problems are NP-hard. J. ACM 60(6), 1–39. https://doi.org/10.1145/2512329 (2013).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20(3), 273–297. https://doi.org/10.1007/BF00994018 (1995).
Breiman, L. Random forests. Mach. Learn. 45(1), 5–32. https://doi.org/10.1023/A:1010933404324 (2001).
Sabeti, E. et al. Learning using concave and convex Kernels: Applications in predicting quality of sleep and level of fatigue in fibromyalgia. Entropy 21(5), 442. https://doi.org/10.3390/e21050442 (2019).
Han, H. & Jiang, X. Overcome support vector machine diagnosis overfitting. Cancer Inform. 13, 13875. https://doi.org/10.4137/CIN.S13875 (2014).
Huang, S. et al. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genom. Proteom. 15(1), 41–51 (2018).
Cervantes, J., Garcia-Lamont, F., Rodríguez-Mazahua, L. & Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 408, 189–215. https://doi.org/10.1016/j.neucom.2019.10.118 (2020).
Ferrario, A. & Loi, M. How explainability contributes to trust in AI. In 2022 ACM Conference on Fairness, Accountability, and Transparency 1457–1466. https://doi.org/10.1145/3531146.3533202 (ACM, 2022).
Fan, R.-E., Chen, P.-H. & Lin, C.-J. Working set selection using second order information for training support vector machines. J. Mach. Learn. Res. 6(63), 1889–1918 (2005).
Qi, Y. Random forest for bioinformatics. In Ensemble Machine Learning (eds Zhang, C. & Ma, Y.) 307–323 (Springer, 2012).
Gholami, R. & Fakhari, N. Support vector machine: Principles, parameters, and applications. In Handbook of Neural Computation (eds Samui, P. et al.) 515–535 (Elsevier, 2017).
Hsieh, C.-J., Chang, K.-W., Lin, C.-J., Keerthi, S. S. & Sundararajan, S. A dual coordinate descent method for large-scale linear SVM. In Proc. 25th International Conference on Machine Learning—ICML ’08 408–415. https://doi.org/10.1145/1390156.1390208 (ACM Press, 2008).
Hond, A. A., Steyerberg, E. W. & Calster, B. Interpreting area under the receiver operating characteristic curve. Lancet Dig. Health 4(12), 853–855 (2022).
Akintola, A. A., Pol, V., Bimmel, D., Maan, A. C. & Van Heemst, D. Comparative analysis of the equivital eq02 lifemonitor with holter ambulatory ecg device for continuous measurement of ecg, heart rate, and heart rate variability: A validation study for precision and accuracy. Front. Physiol. 7, 213365 (2016).
Van Voorhees, E. E. et al. Ambulatory heart rate variability monitoring: Comparisons between the empatica e4 wristband and holter electrocardiogram. Psychosom. Med. 84(2), 210–214 (2022).
Zhang, D. et al. An interpretable deep-learning model for early prediction of sepsis in the emergency department. Patterns 2, 2 (2021).
Funding
All aspects of this research were supported by the NSF Grant No. 1837985. The NIH training Grant No. T32GM070449 supported OPA and WZ. We also acknowledge NIH Grants 3-P30-ES-017885-11-S1 and 3-U24-CA-271037-02-S1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
JG, JSV, KN, and OPA conceptualised and designed the study. HD developed the code and algorithms for performing peak detection. HD, SC, and JP developed the method and code for tensor methods and dimensionality reduction. JG confirmed access to and verified the dataset used in this study. JSV and GO provided clinical knowledge. OPA performed feature extraction and developed the models. WZ developed the algorithm and code to perform the quality check of electrocardiogram data. All authors have discussed the results, and reviewed and contributed to the final manuscript.
Corresponding author
Ethics declarations
Competing interests
KN, JG, and HD have intellectual property with University of Michigan’s Office of Technology Transfer related to the content of this paper. All other authors do not hold any competing interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alge, O.P., Pickard, J., Zhang, W. et al. Continuous sepsis trajectory prediction using tensor-reduced physiological signals. Sci Rep 14, 18155 (2024). https://doi.org/10.1038/s41598-024-68901-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68901-x
- Springer Nature Limited