Abstract
Naked carp (Gymnocypris przewalskii), endemic to the saline-alkaline Lake Qinghai, have the capacity to tolerate combinations of high salinity and alkalinity, but migrate to spawn in freshwater rivers each year. In this study, we measured the drinking rate over a 24 h period for naked carp exposed to saline-alkaline lake waters with salinities of 15 (L15) and 17 (L17). We also assessed the daily feed intakes of naked carp exposed to L15 and fresh water (FW). Additionally, we studied the daily expression of acid–base regulation and osmoregulation related genes and proteins in the intestine of naked carp exposed to saline-alkaline lake waters. Our results revealed that the drinking rate at night was significantly higher than in daytime when exposed to either L15 or L17, while feed intakes in daytime were significantly higher than at night. The relative expression of Na+/K+-ATPase α (NKA-α), solute carrier family members 26A6 (SLC26A6) and 4A4 (SLC4A4) in the intestine of naked carp exposed to L17 at night was higher than in daytime. Specifically, NKA-α mRNA expression at 4:00 was 7.22-fold and 5.63-fold higher than that at 10:00 and 16:00, respectively, and the expression at 22:00 was 11.29-fold and 8.80-fold higher than that at 10:00 and 16:00, respectively. Similarly, SLC26A6 mRNA expression was greatest at 22:00, exceeding that observed at 4:00, 10:00 and 16:00 by 3.59, 4.44 and 11.14-fold, respectively. Finally, the expression of NKA-α and SLC26A6 protein at the single cell level was also higher at night than during the day, which was 1.65-fold and 1.37-fold higher at 22:00 respectively compared to 16:00. Overall, the present findings revealed that naked carp drinks at night and feeds during the day, demonstrating that intestinal ion regulation exhibits a daily rhythm when exposed to high saline and alkaline lake water.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Gymnocypris przewalskii, also known as naked carp or the scale-less carp, is endemic to the extreme environment of Lake Qinghai1. Lake Qinghai has a low concentration of dissolved oxygen (4.16–6.08 mg L−1), strong alkalinity (carbonate alkalinity approximately 29 mmol L−1, pH 9.1–9.5), and a high salinity (13–15 ppt)2. Due to evaporation and agricultural irrigation, the salinity and alkalinity levels of Lake Qinghai are increasing by 7 and 0.5% per year, respectively3. Although historically rich in resources, the naked carp population decreased significantly because of climate change, overfishing and destruction of spawning habitat4.
Teleost fish living in seawater drink surrounding water to avoid dehydration. This plays an important role in their osmoregulation and presents the challenge of salts excretion and water absorption5. While not full strength seawater, the salinity of Lake Qinghai is nevertheless hyperosmotic to naked carp, thus necessitating the need for these fish to drink in order to effectively osmoregulate6.
The osmoregulatory mechanism of fish inhabiting such saline-alkaline water is unknown. Bergman et al.7 described the intestinal structure and physiology of the Lake Magadi tilapia (Alcolapia grahami) when feeding and drinking in highly alkaline water and found that they differ from other freshwater and marine teleosts. The esophagus, stomach and intestine form an unusual configuration ‘T’ that allows for the selective shunting of ingested food and water. The pyloric sphincter is open when the fish is drinking and closed when the fish is eating. This unique configuration allows water to almost completely bypass the stomach and enter directly into the intestine7. However, the naked carp has no stomach and thus cannot physically partition intestinal water absorption from food digestion. Moreover, additional research has shown that the digestive and drinking behavior of fish in general cannot be performed at the same time8. This led us to question, whether a stomachless fish, such as naked carp might partition these functions temporally rather than physically.
The mechanisms of daily rhythm influenced by the periodic changes of environmental factors such as temperature and illumination, and have a coordinated effect on life activities such as physiology and behavior9,10. The regulation mechanism of fish feeding rhythm integrates external signals with peripheral and central nervous system by brain-gut axis11. It was recently discovered that the intestinal rhythm is mainly related to light and food12. Feeding behavior of fish is limited by sensory ability. Fish, those ingest rely on vision, usually feed during the day. Fish with poor visual ability are more active at night13. In addition, the daily rhythm of some fish appears to be more flexible than that of higher vertebrates14. For example, in natural environments, rainbow trout (Oncorhynchus mykiss) swims to the bottom to feed during the day and rises to the surface for locomotor activity at night15, while in Nile tilapia (Oreochromis niloticus) the situation is reversed14. In addition, goldfish (Carassius auratus)16, Atlantic salmon (Salmo salar L.)17, silver carp (Hypophthalmichthys molitrix), bighead carp (Hypophthalmichthys nobilis)18 have obvious daytime feeding behaviors, while mackerels19 feed more intensely at night. On the other hand, European sea bass (Dicentrarchus labrax L.) exhibit continuous feeding characteristics over both diurnal and nocturnal scales20. While zebrafish (Danio rerio) developed a food anticipatory activity, displayed locomotor activity mostly during the daytime21.
The intestine has the function of both digestion and osmoregulation22. The food intake of fish is not only affected by external factors such as temperature and photoperiod, but also by internal factors such as gastrointestinal tract fullness and gastrointestinal tract emptying rate23. And the intestine digestion showed obvious daily rhythm. For example, the trypsin and chymotrypsin activities in foregut, midgut and hindgut of Nile tilapia showed significant daily rhythms. The enzyme activity in daytime was higher than that in night24. Moreover, the osmoregulatory role of the intestine is linked to acid–base balance in marine and euryhaline teleosts and its ability to effectively absorb fluid is essential to compensate for water loss in hypertonic environments5,25. Previous results have shown that Na+/K+-ATPase (NKA) expression and activity increase following transition from freshwater to seawater26. The majority of teleost fish regulate acid–base balance by adjusting plasma HCO3− levels through the differential regulation of H+ and HCO3− transport in exchange for Na+ and Cl−, respectively27, via exchangers such as solute carrier family members 26A6 (SLC26A6), solute carrier family members 4A4 (SLC4A4). However, an apparent exception is the Lake Magadi tilapia which showed more than 70% of intestinal flux occurred via Na+ and base co-transport, and less than 30% by Na+ and Cl− co-transport7.
In the end, the question of how stomach less fish living in a hypersaline environment, such as naked carp, are able to sufficiently balance water absorption and food digestion without a means of physical partitioning within the gut. We hypothesized that naked carp living in a high saline-alkaline water environment partition these functions temporally such that they drink at night and feed during the day. Studies on G. przewalskii have shown that the mRNA expression of SLC26 and SLC4 gene families were up-regulated in the intestine when exposed to different saline-alkaline waters. This was particularly evident under conditions of high salinity and alkalinity, where the up-regulation of expression was clear, indicating that under these conditions naked carp secrete and excrete the accumulated HCO3− in vivo through the intestine Cl−/HCO3− exchanger and Na+-HCO3− co-transporter6, respectively. Furthermore, a decrease in cytosolic carbonic anhydrase c expression in these conditions has been shown, likely reflecting a compensatory response to a respiratory alkalosis and as a means for aiding in osmoregulation during the transition from a freshwater river to a saline-alkaline lake3. With this background in mind, the focus of this study was to investigate the potential effect of a daily rhythm on osmoregulation, acid–base balance and feeding in naked carp living in the saline-alkaline water of Lake Qinghai. In this study, naked carp were placed in saline-alkaline lake waters with salinities of 15(L15) and 17(L17). We measured the drinking rate over a 24 h period in both treatments. Additionally, we measured the daily feed intakes of naked carp exposed to saline-alkaline lake water (L15) and fresh water (FW).We also studied the daily expression of genes and proteins, which related to acid–base regulation and osmoregulation, in the intestine of naked carp exposed to saline-alkaline lake waters.
Results
Drinking rate
Drinking rate of naked carp exposed to L15 and L17 were assessed (Fig. 1). In each treatment, drinking rate of naked carp at night (22:00–4:00) were significantly higher than that in the daytime (10:00–16:00) (P < 0.05). In the L15 treatment, the drinking rate at night was 1.77 uL g−1 h−1, which was significantly higher (p < 0.05) than that in the daytime. In the L17 treatment, the drinking rate at night was 2.09 uL g−1 h−1, which was significantly higher (p < 0.05) than that in the daytime.
Drinking rates of Gymnocypris przewalskii exposed to saline-alkaline lake water (L15 and L17) at different times (Day 10:00–16:00, Night 22:00–4:00) (n = 9 per treatment). Light–dark cycle was 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx;19:00–5:00 with light intensity of 0 lx); L15 and L17 refers to saline-alkaline lake water with a salinity of 15 or 17. Lowercase indicates significantly difference among time points in each treatment (P < 0.05).
Daily feed intake
Daily feed intakes of naked carp exposed to L15 and FW were assessed (Fig. 2). In each treatment, daily feed intakes in the daytime were significantly higher than those at night. In the L15, the daily feed intake in the daytime was 3.50 g kg−1 which was significantly higher (P < 0.05) than that at night. In FW, the daily feed intake in the daytime was 10.17 g kg−1 which was significantly higher (P < 0.05) than that at night.
Daily feed intakes of Gymnocypris przewalskii exposed to saline-alkaline lake water (L15) and fresh water (FW) at different times (Day 5:00–19:00, Night 19:00–5:00) (n = 3 per treatment). Light–dark cycle was 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx;19:00–5:00 with light intensity of 0 lx); L15 refers to saline-alkaline lake water with a salinity of 15; Lowercase indicates significantly difference between phases in each treatment (P < 0.05).
Gene expression in response to saline-alkaline exposure
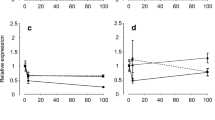
Expression of NKA-α, SLC26A6 and SLC4A4 in intestine of naked carp exposed to L17 were investigated (Fig. 3). In L17, NKA-α mRNA expression at 4:00 and 22:00 was significantly (P < 0.05) higher than other time points. The expression at 4:00 was 7.22-fold and 5.63-fold that of 10:00 and 16:00, respectively, and the expression at 22:00 was 11.29-fold and 8.80-fold that of 10:00 and 16:00, respectively; SLC26A6 mRNA expression at 22:00 was significantly (P < 0.05) higher than the other time points, which was 3.59-fold of 4:00, 4.44-fold of 10:00 and 11.14-fold of 16:00. The expression of SLC4A4 mRNA at 4:00 and 22:00 was significantly (P < 0.05) higher than at 10:00 and 16: 00. The expression at 4:00 was 5.34-fold of 10:00 and 5.75-fold of 16:00, whereas the expression at 22:00 was 5.32-fold of 10:00 and 5.73-fold of 16:00.
NKA-α, SLC26A6, SLC4A4 mRNA expression levels in the mid-intestine of Gymnocypris przewalskii at different time points (4:00, 10:00, 16:00 and 22:00) exposed to saline-alkaline lake water with a salinity of 17. Light–dark cycle was 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx;19:00–5:00 with light intensity of 0 lx); Lowercase indicates significantly difference among time points in each treatment (P < 0.05).
Intestinal single cell positivity rates for NKA-α and SLC26A6
Western blot analysis for NKA-α and SLC26A6 revealed distinct bands of ~ 113 kDa and ~ 50 kDa respectively (Fig. S1). Single cell positivity rates for NKA-α and SLC26A6 in naked carp in response to L17 acclimation from fresh water were investigated in mid-intestine. In L17, there was significant differences at each time point (P < 0.05) (Fig. 4). The expression of NKA-α and SLC26A6 at 22:00 was 1.65-fold and 1.37-fold of 16:00, respectively. Figure 5 shows the protein staining results of NKA-α and SLC26A6 at 16:00 and 22:00, respectively.
NKA-α, SLC26A6 positivity rate in the mid-intestine of Gymnocypris przewalskii at different time points exposed to saline-alkaline lake water with a salinity of 17 (L17). Light–dark cycle was 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx;19:00–5:00 with light intensity of 0 lx); Lowercase indicates significant difference among time points in each treatment (P < 0.05); Positive rate = protein staining/nuclear staining. Image J was used for measurement, analysis and calculation).
Discussion
Teleosts living within hyperosmotic environments are faced with the constant challenges of diffusive water loss and ion gain25. Thus, to maintain osmoregulatory homeostasis in these environments28, teleosts must actively drink the surrounding water, absorb salts and water across the gut and then actively secrete the added salt load across the gills29. The naked carp migrates annually between freshwater rivers and Lake Qinghai as part of their life history strategy30. Thus, migrating to lake water demands a transition in osmoregulatory strategy as the fish moves from a hypoosmotic environment to a hyperosmotic environment. Considering that the naked carp lacks a stomach for physically partitioning digestion and absorption, we hypothesized that these fish might partition these functions temporally when moving into the hyperosmotic lake water. To address this, we analyzed the daily changes in feeding, drinking and expression/activity of key proteins involved in digestion and osmoregulation of naked carp in saline-alkaline environments. Our results first time revealed that naked carp exhibit a daily rhythm whereby drinking is partitioned more to evening hours and feeding is partitioned to a greater extent during the day. This was supported by the observations that the drinking rate and expression of key osmoregulatory genes of naked carp at night were significantly higher than that observed during the day, while the feed intake was significantly lower at night than during the day.
Transporters involved in the regulation of intestinal epithelial ions mainly include NKA, Cl−/HCO3− exchangers, Na+-HCO3− co-transporter and others. When teleost fish are transferred to seawater, osmoregulation is maintained by increasing the expression of NKA mRNA and its activity in intestinal epithelial cells31,32. The SLC4 and SLC26 gene families are the main proteins involved in HCO3− transport in organisms, and play an important role in maintaining the pH and ion balance of body fluids33,34. When naked carp were transferred directly from fresh water to saline-alkaline water, the expression of NKA increased in the basolateral membrane of the intestine likely as a response to increase salt uptake and maintain the osmotic pressure gradient for water absorption. Yao et al.3 found that the naked carp transferred from river water to lake water resulted in a respiratory alkalosis and ion rise in the blood. The expression of SLC26A6 and SLC4A4, on the other hand, likely increased as a compensatory measure to aid in the secretion and excretion of accumulated bicarbonate concentrations6. Perhaps most notably, the drinking rate of naked carp was significantly higher at night than that during the day which aligned with the heightened expression of NKA-α, SLC26A6 and SLC4A4, further supporting that osmoregulation and acid–base balance appear to be partitioned to a greater extent during this period of the daily cycle.
Fish usually exhibit certain feeding rhythms that can be connected with external environment and internal factors. Generally, feeding rhythms represent the evolutionary trade-off between food availability, metabolic demand and predator risk35. During the experiment, the mode of self-feeding was closer to the survival conditions of natural environment than the mode of artificial feeding by schedules. The use of self-feeders can not only promote growth, improve feed conversion and reduce feed waste, but also find some behavioral characteristics related to self-feeding activities36. The study found that self-feeding intake of naked carp was higher in the daytime, indicating increased feeding behavior during the day. Naked carp triggered the self-feeder when they need to ingest feed. The daily feed intakes of L15 and FW in daytime was 3.50 g kg−1 and 10.17 g kg−1, respectively, significantly higher than that of 1.79 g kg−1 and 5.55 g kg−1 at night. This indicated that high feed intake in daytime could depend on surrounding lights, since visual feeder usually feed during the day13. In addition, the intestinal tract of fish can improve digestive physiological activities through rhythmic digestive enzyme activities, and then affect feeding activities37. Daily rhythms have evolved from long-term adaptation of life to daily changes of environmental factors on the Earth9,38. Studies represented by zebrafish have shown that fish exhibit distinct daily rhythms in physiology and behavior21, which include feeding rhythms.
In conclusion, naked carp appear to cope with the conflicting demands of feeding/digestion and drinking/osmoregulation by alternating these functions within a daily rhythm, whereby the former is largely achieved during the day and the latter by night. This was supported by the fact that the drinking rate of naked carp at night was significantly higher than that during the day while the feed intakes exhibited the opposite pattern. Moreover the relative expression of key osmoregulatory and acid–base proteins (i.e., NKA, SLC26A6 and SLC4A4) paralleled the daily changes in drinking rate. Given the high salinity/alkalinity environment of naked carp, we predict that the daily rhythm may therefore play an important role in osmoregulation and ion regulation of this fish.
Methods
Experimental animals
Gymnocypris przewalskii used in this study were obtained from the Rescue Center for Naked Carp of Lake Qinghai in Xining. Only healthy fish without visible body damage were used. Wet mass and body length were recorded before the fish were sampled. All fish were collected under permits issued by local and national authorities, and experimental procedures were in accordance with national animal care regulations. Experimental waters were prepared daily, and water qualities were measured before each experiment. The water temperature and salinity were measured using an YSI6600 multiprobe sensor (YSI Incorporated, Ohio, USA), and the carbonate alkalinity was determined by titration39. All fish (average body weight: 33.21 ± 2.74 g; average body length:14.81 ± 0.35 cm) were held in an indoor RAS system at a density of approximately 6.5 kg m−3. The holding and experimental water were filtered tap water (Canature/AC/KDF150-1–300) (salinity 0.16, pH 7.56, carbonate alkalinity 2.7 mmol L−1, temperature 17.1 ± 0.61 °C). Fish were fed daily with commercial feed. Fish husbandry and experimental procedures were approved by the Second Scientific Research Ethics Committee of East China Sea Fisheries Research Institute.
Experimental design
Fish were placed on a 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx; 19:00–5:00 with light intensity of 0 lx) photoperiod aquaculture system. To examine the effect of rhythm on osmoregulation and acid–base balance, this study measured four endpoints: drinking rate, self-feeding intake, mRNA expression and the single cell expression level of osmoregulation and acid–base regulation relevant proteins. Fish held in filtered tap water were transferred directly to saline-alkaline lake water with salinities of 15 (L15, salinity 14.83, pH 8.65, carbonate alkalinity 30.54 mmol L−1) and 17 (L17, salinity 16.80, pH 9.02, carbonate alkalinity 34.61 mmol L−1), which was prepared by adding the same ratio of NaCl, MgCl2.6H2O, Na2SO4, CaCl2, KCl, NaHCO3 and Na2CO3 as in Qinghai Lake. The experimental period was 4–5 days.
Drinking rate during high saline-alkaline transfer
Fish were placed on a 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx;19:00–5:00 with light intensity of 0 lx) photoperiod aquaculture system. In this experiment, feeding was stopped 48 h prior to the experiments. Fresh water fish were transferred directly to L15 and L17 PEG-4000 free water for up to 4 days. For drinking rate analysis, new tanks were prepared which contained 50L saline-alkaline lake water with salinity 15 or 17 and with PEG-4000 (final concentration was 1.00 g L−1) during the day (10:00–16:00) and night (4:00–22:00) on the fourth day respectively. Nine fish per treatment were individually transferred from PEG-4000 free water to each tank which contained 1.00 g L−1 PEG-4000 at 10:00 or 4:00. Water samples were collected at 15 min after the fish were transferred to each treatment group for the determination of PEG-4000 concentration. The fish were terminally anesthetized with MS-222 (0.40 g L−1) after 6 h. The intestines were then quickly dissected out from nine individual fish per treatment group and the intestinal fluid were collected and stored at 4 °C. All fish were weighed before sampling.
Self-feeding intake during high saline-alkaline transfer
Fish were placed on a 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx;19:00–5:00 with light intensity of 0 lx) photoperiod aquaculture system. In this experiment, fish were kept in freshwater (FW) or acclimated to L15 for more than 15 days before the experiment started. Six RAS glass tanks (95 cm × 60 cm × 60 cm), which belong to two circulatory systems (3 tanks for FW and 3 for L15), were used for self-feeding experiment. Each tank had 15 individuals.
Before the experiment, fish were trained by a custom-made self-feeding system (Fig. S2). Trained fish triggered the self-feeder when they want to feed. In the self-feeding system, the photoelectric sensor converts the change of optical signal into the change of electrical signal, and the feeder release feed by recognizing level fluctuation.
During the formal experiment, we collected feed data at 5:00 and 19:00, which were the time points of the light and dark transition. Feed intakes of naked carp were calculated by weighing the feed quantities at two time points. The experiment lasted 5 days.
mRNA expression of osmoregulation and acid–base regulation relevant proteins during high saline-alkaline transfer
Fish were placed on a 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx;19:00–5:00 with light intensity of 0 lx) photoperiod aquaculture system. In this experiment, feeding was stopped 48 h prior to the experiments. Fresh water fish were transferred directly to L17 for up to 4 days. There were 24 fish per tank in triplicate. At the fourth day, six fish per tank were individually removed and terminally anesthetized with MS-222 (0.40 g L−1) at 4:00, 10:00, 16:00 and 22:00, respectively. The mid-intestine was quickly dissected out from six individual fish at each time point. Mid-intestine tissues for mRNA expression analyses were immediately snap-frozen in liquid N2, and stored at − 80 °C until analysis.
Single cell positive rate of osmoregulation and acid–base regulation relevant proteins
Fish were placed on a 14:10-h light:dark (5:00–19:00 with light intensity of 600–1000 lx;19:00–5:00 with light intensity of 0 lx) photoperiod aquaculture system. In this experiment, feeding was stopped 48 h prior to the experiments. To analyze the single cell positive rate of acid–base relevant proteins, a separate experiment was conducted. Fresh water fish were transferred directly to L17 for up to 4 days. There were 3 tanks (6 fish per tank) in this experimental group. At the fourth day, three fish per tank were individually removed and terminally anesthetized with MS-222 (0.40 g L−1) at 16:00 and 22:00, respectively. The mid-intestine was quickly dissected out from nine individual fish and immediately prepared for single-cell suspensions.
Analytical techniques
Drinking rate analysis
The measurement of drinking rate was performed according to the study of Buxton et al.40. After weighing the collected intestinal fluid, it was centrifuged at 13,000g for 1 min, and 50 μL of the supernatant was taken, added dropwise to 350 μL of 72% pre-cooled (4 °C) acetone, and vortexed to mix. Samples were then centrifuged at 2000g for 10 min at 4 °C, the supernatant filtered with 0.45 μm filter paper, followed by addition of 100 μL of filtrate to 175 μL 25 mg L−1 gum arabic and vortexed to mix. Finally, 200 μL of TCA-CaCl2 (trichloroacetic acid-calcium chloride, 30% and 5% by mass) was added to the mixture and the reaction allowed to proceed at room temperature for 20 min. An Epoch microplate (Bio Tek) spectrophotometry unit was used to measure the absorbance at 650 nm. The remaining solution was weighed again after drying at 60 °C for 48 h, and the volume of intestinal fluid was determined (quantity of collected intestinal fluid-mass after drying). The same method as above was used to process the standard solution. Solute concentrations for standard curve were prepared as 0.00 g L−1, 0.10 g L−1, 0.20 g L−1, 0.40 g L−1, 0.60 g/L−1, 0.80 g L–1, 1 g L−1, and 2 g L−1 PEG-4000. The PEG-4000 concentration of intestinal fluid was calculated based on the standard curve. Drinking rate (μLg-1h-1) = 1000 × (CI × VI)/(CW × W × t), where CI is the concentration of PEG-4000 in the intestinal fluid (gL-1), VI is the volume of intestinal fluid (mL), CW is the concentration of PEG-4000 in experimental water (gL-1),W is the body weight of the fish (g), t is the duration of the experiment (h).
Molecular biology
The known sequences of the NKA-α gene of naked carp were compared with the corresponding genes of other species in GenBank, and highly conserved regions were selected for primer design (Table 1). The reference gene EF1α was used according to Yao et al.3. Previously published primers were used for SLC26A6 and SLC4A4 genes6. After extracting total RNA with Trizol (Invitrogen), the integrity of RNA was detected by 1% agarose gel electrophoresis, and the concentration and purity of total RNA were determined by a Bio Tek Epoch microplate spectrophotometer. The Rever Tra Ace-α (TOYOBO) kit was used to reverse transcribe mRNA to cDNA. Fluorescence quantitative PCR analysis was performed using a QuantStudio™ Real-Time PCR (Thermo life) with the SYBR Premix Ex TaqIII (TaKaRa) kit: total reaction volume of 10 μL, including 5 μL SYBR Premix Ex Taq, 2 μL upstream primers, 2 μL downstream Primers, and 1 μL cDNA template. The amplification procedure was as follows: 95 °C 30 s, 1 cycle; 95 °C 5 s, 60 °C 20 s, 40 cycles. Three replicates were included for each sample, with EF1α as the internal reference gene. The relative expression of each gene was calculated using the 2−ΔΔCt method41. Melting curve analysis was performed following each reaction to confirm that there was only a single product and no primer-dimer artifacts. In addition, representative samples were electrophoresed to verify that only a single product was present. Negative control reactions were performed for representative samples using RNA that had not been reverse transcribed to control for the possible presence of genomic DNA contamination. No-template control reactions were also performed to verify the absence of contaminating DNA or primer-dimer amplification in the reactions.
Single cell staining analysis
The naked carp mid-intestine was isolated and transferred to HBSS on ice. The mid-intestine was washed by HBSS (Corning, 21-022-CV) and transferred to pre-warmed digestion medium containing 0.2 mg·mL−1 Collagenase I (Gibco, 17100-017), 0.06 mg mL−1 Collagenase II (Gibco, 17101-015) and 0.2 mg mL−1 Collagenase IV (Gibco, 17104-019), which was shaken vigorously for 30 s and further incubated at 37 °C for about 30 min in incubator with gentle shaking every 5 min to release cells. Cells were then collected by centrifuging at 300 × g for 5 min, and resuspended in D-PBS (BBI, E607009-0500). Then taken an appropriate amount of single cell suspension and dropped it on poly-L-lysine-coated slides where the experimental area was drawn with a hydrophobic marker to allow the single cells to settle freely. When the cell sedimentation density was moderate, aspirated the excess cell suspension, slides were fixed with 4% paraformaldehyde fix solution (BBI, E672002-0500) for 10 min, and blocked with 3% BSA (Sigma, B2064) for 1 h, three washes in D-PBS. Subsequently, slides were incubated in NKA-α or SLC26A6 (antibody dilution ratio was 1:100) for overnight at 4 °C. The NKA-α antibody was a commercial polyclonal rabbit Na+/K+-ATPase α antibody (Santa Cruz Biotechnology, sc-28800). The SLC26A6 antibody was a commercial polyclonal rabbit SLC26A6 antibody (Abcam, ab-172684). After the incubation, three washes in D-PBS. The secondary antibodies consisted of Alexa flour 568 goat anti-rabbit IgG (Thermo Fisher Scientific, A11036) (antibody dilution ratio was 1:400). Slides were incubated in room temperature for 1 h, followed by three washes in D-PBS. Finally, incubate with Hochest for 30 min. Cells were then photographed with a fluorescence microscope. For every fish, positive protein expression was counted using at least three pictures. Image J was used to analyze the fluorescence intensity and record the positivity rate.
Statistical analysis
The data was expressed as mean ± standard error (SE). Two-way ANOVA and One-way ANOVA with LSD multiple comparison were employed to compare drinking rate, food intake and relative gene expression among different treatments and time courses respectively. Differences in single cell positive rate between 16:00 and 22:00 in L17 were evaluated by chi-square test. Assumptions for all parametric models (normality and equal residuals) were assessed via diagnostic plots. Means were considered significantly different when P < 0.05. All statistical analyses were conducted with SPSS 11. The bar charts were created using Origin 8.6.
References
Xiong, F., Chen, D. Q. & Duan, X. B. Threatened fishes of the world: Gymnocypris przewalskii (Kessler, 1876) (Cyprinidae: Schizothoracinae). J. Environ. Biol. Fishes 87, 351–352 (2010).
Wang, Y. S. et al. Unusual physiology of scale-less carp, Gymnocypris przewalskii, in Lake Qinghai: A high altitude alkaline saline lake. Comp. Biochem. Physiol. Part A 134, 409–421 (2003).
Yao, Z. L. et al. Gymnocypris przewalskii decreases cytosolic carbonic anhydrase expression to compensate for respiratory alkalosis and osmoregulation in the saline-alkaline lake Qinghai. J. Comp. Physiol. B. 186, 83–95 (2016).
Wang, S. & Xie, Y. China Species Red List 258–259 (Higher Education Press, 2009).
Whittamore, J. M. Osmoregulation and epithelial water transport: Lessons from the intestine of marine teleost fish. J. Comp. Physiol. B 182, 1–39 (2012).
Wang, P. et al. Differential expressions of genes related to HCO3− secretion in the intestine of Gymnocypris przewalskiii during saline-alkaline water transfer. Mar. Fish. 37, 341–348 (2015).
Bergman, A. N. et al. Physiological adaptations of the gut in the Lake Magadi tilapia, Alcolapia grahami, an alkaline- and saline-adapted teleost fish. J. Comp. Biochem. Physiol. Part A 136, 701–715 (2003).
Lin, H. Fish Physiology (Zhongshan University Press, 2011).
Broussard, J. L. et al. Circadian rhythms versus daily patterns in human physiology and behaviour. In Biological Timekeeping: Clocks, Rhythms and Behaviour (ed. Kumar, V.) 279–295 (Springer, 2017).
Cowan, M., Azpeleta, C. & López-Olmeda, J. F. Rhythms in the endocrine system of fish: A review. J. Comp. Physiol. B. 187, 1057–1089 (2017).
Bonacic, K. et al. Dietary fatty acid composition affects food intake and gut-brain satiety signaling in Senegalese sole (Solea senegalensis, Kaup 1858) larvae and post-larvae. Gen. Comp. Endocrinol. 228, 79–94 (2016).
Xiong, Y. et al. Research progress of interaction between circadian clock and gut microbiota in mammals. Chin. Bull. Life Sci. 32, 551–557 (2020).
Kulczykowska, E. & Sánchez-Vázquez, F. J. Neurohormonal regulation of feed intake and response to nutrients in fish: aspects of feeding rhythm and stress. Aquaculture Research 41 (2010).
Fortes-Silva, R., Martínez, F. J., Villarroel, M. & Sánchez-Vázquez, F. J. Daily rhythms of locomotor activity, feeding behavior and dietary selection in Nile tilapia (Oreochromis niloticus). J. Comp. Biochem. Physiol. Part A 156, 445–450 (2010).
Sánchez-Vázquez, F. J. & Tabata, M. Circadian rhythms of demand-feeding and locomotor activity in rainbow trout. J. Fish Biol. 52, 255–267 (1998).
Sánchez-Vázquez, F. J., Madrid, J. A., Zamora, S., Iigo, M. & Tabata, M. Demand feeding and locomotor circadian rhythms in the goldfish, Carassius auratus: Dual and independent phasing. J. Physiol. Behav. 60, 665–674 (1996).
Kadri, S., Metcalfe, N. B., Huntingford, F. A. & Thorpe, J. E. Daily feeding rhythms in Atlantic salmon I: Feeding and aggression in parr under ambient environmental conditions. J. Fish Biol. 50, 267–272 (1997).
Li, S. F., Yang, H. Q. & Lu, W. M. Preliminary research on diurnal feeding rhythm and the daily ration for Silver Carp, Bighead Carp and Grass Carp. J. Fish. China 4, 275–283 (1980).
Wang, X. A., Ma, A. J., Zhang, X. M., Zhuang, Z. M. & Yu, H. A review on feeding behavior ecology of early developmentstage of marine fish. J. Mar. Sci. 30, 69–74 (2006).
Sánchez-Vázquez, F. J., Madrid, J. A. & Zamora, S. Circadian rhythms of feeding activity in Sea Bass, Dicentrarchus labrax L.: Dual phasing capacity of diel demand-feeding pattern. J. Biol. Rhythms 10, 256–266 (1995).
Sanchez, J. A. & Sanchez-Vazquez, F. J. Feeding entrainment of daily rhythms of locomotor activity and clock gene expression in zebrafish brain. Chronobiol. Int. 26, 1120–1135 (2009).
Grosell, M., Farrell, A. P. & Brauner, C. J. The multifunctional gut of fish. J. Fish Physiol. 30, 1–426 (2011).
Ivar, R. et al. Appetite-controlling endocrine systems in teleosts. Front. Endocrinol. 8, 73 (2017).
Oliveira, C. G. D., López-Olmeda, J. F., Costa, L. S., Santo, A. H. D. E. & Ribeiro, P. A. P. Gastrointestinal emptying and daily patterns of activity of proteinolytic enzymes in Nile tilapia (Oreochromis niloticus). Aquaculture 546, 737338 (2021).
Carvalho, E. S. M., Gregório, S. F., Power, D. M., Canário, A. V. M. & Fuentes, J. Water absorption and bicarbonate secretion in the intestine of the sea bream are regulated by transmembrane and soluble adenylyl cyclase stimulation. J. Comp. Physiol. B 182, 1069–1080 (2012).
Evans, A. N. & Lambert, F. N. Na+/K+-ATPase α1 mRNA expression in the gill and rectal gland of the Atlantic stingray, Dasyatis sabina, following acclimation to increased salinity. J. BMC Res. Notes 8, 219 (2015).
Gilmour, K. M. & Perry, S. F. Carbonic anhydrase and acid-base regulation in fish. J. Exp. Biol. 212, 1647–1661 (2009).
Fuentes, J. & Eddy, F. B. Effect of manipulation of the renin-angiotensin system in control of drinking in juvenile Atlantic salmon (Salmo salar L.) in fresh water and after transfer to sea water. J. Comp. Physiol. B. 167, 438–443 (1997).
Whittamore, J. M., Cooper, C. A. & Wilson, R. W. HCO3− secretion and CaCO3 precipitation play major roles in intestinal water absorption in marine teleost fish in vivo. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R877–R886 (2010).
Wood, C. M. et al. Przewalski’s naked carp (Gymnocypris przewalskii): An endangered species taking a metabolic holiday in Lake Qinghai, China. Physiol. Biochem. Zool. 80, 59–77 (2007).
Cutler, C. P. et al. Expression of a duplicate Na, K-ATPase beta1-isoform in the European eel (Anguila anguilla). Am. J. Physiol. 279, 222–229 (2000).
Seidelin, M., Madsen, S., Blenstrup, H. & Tipsmark, C. Time-course changes in the expression of Na+, K+-ATPase in gills and pyloric caeca of brown trout (Salmo trutta) during acclimation to seawater. Physiol. Biochem. Zool. 73, 446–453 (2000).
David, B. M. & Michaelf, F. R. The SLC26 gene family of multifunctional anion exchangers. Pflügers Archiv. Eur. J. Physiol. 447, 710–721 (2004).
Pushkin, A. & Kurtz, I. SLC4 base (HCO3−, CO32−) transporters: Classification, function, structure, genetic diseases, and knockout models. Am. J. Physiol. Renal Physiol. 290, F580-599 (2006).
Flood, M. J. et al. Examining the daily feeding rhythms of amago Oncorhynchus masou masou using self-feeding systems. Aquaculture 318, 244–247 (2011).
Benhaïm, D. et al. Self-feeding behaviour and personality traits in tilapia: A comparative study between Oreochromis niloticus and Sarotherodon melanotheron. Appl. Anim. Behav. Sci. 187, 85–92 (2017).
Mata-Sotres, A. et al. Daily rhythms of digestive enzyme activity and gene expression in gilthead seabream (Sparus aurata) during ontogeny. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 197, 43–51 (2016).
Dunlap, J. C. Molecular bases for circadian clocks. Cell 96, 271–290 (1999).
Boyd, C. E. & Tucker, C. S. Water Quality and Pond Soil Analysis for Aquaculture (Auburn University, 1992).
Buxton, T. B., Crockett, J. K., Moore, W. L. & Rissing, J. P. Protein precipitation by acetone for the analysis of polyethylene glycol in intestinal perfusion fluid. J. Gastroenterol. 76, 820–824 (1979).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101 (2008).
Acknowledgements
We would like to thank Rescue Center for Naked Carp of Lake Qinghai for providing experimental materials.
Funding
This research was funded by National key R&D program of China (No. 2019YFD0900404) and Special Scientific Research Funds for Central Non-profit Institutes (No. 2021XT0401).
Author information
Authors and Affiliations
Contributions
Z.Y., F.W., and L.Z. conception and design of the research; F.W., L.Z., Z.Y., Y.W., P.G., Y.L, K.Z., Q.L., Z.S. performed the experiments; F.W., L.Z., Y.W., and Z.Y. analyzed the data; Z.Y., F.W., and L.Z. interpreted the results of the experiments; F.W., L.Z., Y.W., prepared the figures; F.W., L.Z. and Z.Y. drafted the manuscript; F.W., L.Z., Z.Y., Q.L, edited and revised the manuscript; Z.Y. approved the final version of manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, F., Zhu, L., Wei, Y. et al. Intestinal ion regulation exhibits a daily rhythm in Gymnocypris przewalskii exposed to high saline and alkaline water. Sci Rep 12, 807 (2022). https://doi.org/10.1038/s41598-021-04472-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-04472-5
- Springer Nature Limited