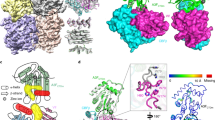
Abstract
HIV-1 Vif recruits host cullin-RING-E3 ubiquitin ligase and CBFβ to degrade the cellular APOBEC3 antiviral proteins through diverse interactions. Recent evidence has shown that Vif also degrades the regulatory subunits PPP2R5(A–E) of cellular protein phosphatase 2A to induce G2/M cell cycle arrest. As PPP2R5 proteins bear no functional or structural resemblance to A3s, it is unclear how Vif can recognize different sets of proteins. Here we report the cryogenic-electron microscopy structure of PPP2R5A in complex with HIV-1 Vif–CBFβ–elongin B–elongin C at 3.58 Å resolution. The structure shows PPP2R5A binds across the Vif molecule, with biochemical and cellular studies confirming a distinct Vif–PPP2R5A interface that partially overlaps with those for A3s. Vif also blocks a canonical PPP2R5A substrate-binding site, indicating that it suppresses the phosphatase activities through both degradation-dependent and degradation-independent mechanisms. Our work identifies critical Vif motifs regulating the recognition of diverse A3 and PPP2R5A substrates, whereby disruption of these host–virus protein interactions could serve as potential targets for HIV-1 therapeutics.





Similar content being viewed by others
Data availability
The model of the PPP2R5A–VCBC complex has been deposited in the Protein Data Bank (PDB) with accession code PDB 8SZK. The cryo-EM map of the PPP2R5A–VCBC complex has been deposited in the Electron Microscopy Data Bank (EMDB) with accession code EMD-40919. The structures used for model building of the PPP2R5A–VCBC complex and making figures could be downloaded from the PDB: VCBC (extracted from PDB 4N9F), PPP2R5A (extracted from PDB 6NTS), A3F (PDB 6NIL), A3G (PDB 8CX0) and Cul5CTD/Rbx2 (PDB 3DPL). Source data are provided with this paper.
References
Desimmie, B. A. et al. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J. Mol. Biol. 426, 1220–1245 (2014).
Hu, Y., Knecht, K. M., Shen, Q. & Xiong, Y. Multifaceted HIV-1 Vif interactions with human E3 ubiquitin ligase and APOBEC3s. FEBS J. 288, 3407–3417 (2021).
Salamango, D. J. & Harris, R. S. Demystifying cell cycle arrest by HIV-1 Vif. Trends Microbiol. 29, 381–384 (2021).
Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002).
Chaipan, C., Smith, J. L., Hu, W. S. & Pathak, V. K. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J. Virol. 87, 444–453 (2013).
Desimmie, B. A. et al. APOBEC3 proteins can copackage and comutate HIV-1 genomes. Nucleic Acids Res. 44, 7848–7865 (2016).
Hultquist, J. F. et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 85, 11220–11234 (2011).
Refsland, E. W., Hultquist, J. F. & Harris, R. S. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 8, e1002800 (2012).
Conticello, S. G. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9, 229 (2008).
Harris, R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 (2003).
Lecossier, D., Bouchonnet, F., Clavel, F. & Hance, A. J. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300, 1112 (2003).
Mangeat, B. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003).
Adolph, M. B., Webb, J. & Chelico, L. Retroviral restriction factor APOBEC3G delays the initiation of DNA synthesis by HIV-1 reverse transcriptase. PLoS ONE 8, e64196 (2013).
Bishop, K. N., Verma, M., Kim, E. Y., Wolinsky, S. M. & Malim, M. H. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4, e1000231 (2008).
Holmes, R. K., Koning, F. A., Bishop, K. N. & Malim, M. H. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282, 2587–2595 (2007).
Iwatani, Y. et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35, 7096–7108 (2007).
Li, X. Y., Guo, F., Zhang, L., Kleiman, L. & Cen, S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 282, 32065–32074 (2007).
Luo, K. et al. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 81, 7238–7248 (2007).
Mbisa, J. L. et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81, 7099–7110 (2007).
Mbisa, J. L., Bu, W. & Pathak, V. K. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84, 5250–5259 (2010).
Newman, E. N. et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15, 166–170 (2005).
Pollpeter, D. et al. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat. Microbiol. 3, 220–233 (2018).
Dang, Y., Wang, X., Esselman, W. J. & Zheng, Y. H. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80, 10522–10533 (2006).
Marin, M., Rose, K. M., Kozak, S. L. & Kabat, D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9, 1398–1403 (2003).
Mehle, A. et al. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279, 7792–7798 (2004).
OhAinle, M., Kerns, J. A., Malik, H. S. & Emerman, M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 80, 3853–3862 (2006).
Sheehy, A. M., Gaddis, N. C. & Malim, M. H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9, 1404–1407 (2003).
Shirakawa, K. et al. Ubiquitination of APOBEC3 proteins by the Vif-Cullin5-ElonginB-ElonginC complex. Virology 344, 263–266 (2006).
Stopak, K., de Noronha, C., Yonemoto, W. & Greene, W. C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12, 591–601 (2003).
Wiegand, H. L., Doehle, B. P., Bogerd, H. P. & Cullen, B. R. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23, 2451–2458 (2004).
Yu, X. et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 (2003).
Zheng, Y. H. et al. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78, 6073–6076 (2004).
Jager, S. et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375 (2011).
Zhang, W., Du, J., Evans, S. L., Yu, Y. & Yu, X. F. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379 (2011).
Sakai, K., Dimas, J. & Lenardo, M. J. The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc. Natl Acad. Sci. USA 103, 3369–3374 (2006).
Wang, J. et al. The Vif accessory protein alters the cell cycle of human immunodeficiency virus type 1 infected cells. Virology 359, 243–252 (2007).
Greenwood, E. J. et al. Temporal proteomic analysis of HIV infection reveals remodelling of the host phosphoproteome by lentiviral Vif variants. eLife https://doi.org/10.7554/eLife.18296 (2016).
Naamati, A. et al. Functional proteomic atlas of HIV infection in primary human CD4+ T cells. eLife https://doi.org/10.7554/eLife.41431 (2019).
DeHart, J. L., Bosque, A., Harris, R. S. & Planelles, V. Human immunodeficiency virus type 1 Vif induces cell cycle delay via recruitment of the same E3 ubiquitin ligase complex that targets APOBEC3 proteins for degradation. J. Virol. 82, 9265–9272 (2008).
Du, J. et al. Vif-CBFbeta interaction is essential for Vif-induced cell cycle arrest. Biochem. Biophys. Res. Commun. 511, 910–915 (2019).
Kruse, T. et al. The Ebola virus nucleoprotein recruits the host PP2A-B56 phosphatase to activate transcriptional support activity of VP30. Mol. Cell 69, 136–145 e136 (2018).
Bhatt, V. et al. Structural basis of host protein hijacking in human T-cell leukemia virus integration. Nat. Commun. 11, 3121 (2020).
Maertens, G. N. B’-protein phosphatase 2A is a functional binding partner of delta-retroviral integrase. Nucleic Acids Res. 44, 364–376 (2016).
Moura, M. & Conde, C. Phosphatases in mitosis: roles and regulation. Biomolecules https://doi.org/10.3390/biom9020055 (2019).
Nilsson, J. Protein phosphatases in the regulation of mitosis. J. Cell Biol. 218, 395–409 (2019).
McCright, B., Rivers, A. M., Audlin, S. & Virshup, D. M. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J. Biol. Chem. 271, 22081–22089 (1996).
Wang, J. et al. Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization. Protein Cell 7, 516–526 (2016).
Janssens, V. & Goris, J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353, 417–439 (2001).
Marelli, S. et al. Antagonism of PP2A is an independent and conserved function of HIV-1 Vif and causes cell cycle arrest. eLife https://doi.org/10.7554/eLife.53036 (2020).
Nagata, K., Shindo, K., Matsui, Y., Shirakawa, K. & Takaori-Kondo, A. Critical role of PP2A-B56 family protein degradation in HIV-1 Vif mediated G2 cell cycle arrest. Biochem. Biophys. Res. Commun. 527, 257–263 (2020).
Salamango, D. J. et al. HIV-1 Vif triggers cell cycle arrest by degrading cellular PPP2R5 phospho-regulators. Cell Rep. 29, 1057–1065 e1054 (2019).
Salamango, D. J. et al. Functional and structural insights into a Vif/PPP2R5 complex elucidated using patient HIV-1 isolates and computational modeling. J. Virol. https://doi.org/10.1128/JVI.00631-20 (2020).
Izumi, T. et al. HIV-1 viral infectivity factor interacts with TP53 to induce G2 cell cycle arrest and positively regulate viral replication. Proc. Natl Acad. Sci. USA 107, 20798–20803 (2010).
Hertz, E. P. T. et al. A conserved motif provides binding specificity to the PP2A-B56 phosphatase. Mol. Cell 63, 686–695 (2016).
Foley, E. A., Maldonado, M. & Kapoor, T. M. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 13, 1265–1271 (2011).
Lee, S. J., Rodriguez-Bravo, V., Kim, H., Datta, S. & Foley, E. A. The PP2A(B56) phosphatase promotes the association of Cdc20 with APC/C in mitosis. J. Cell Sci. 130, 1760–1771 (2017).
Hu, Y. et al. Structural basis of antagonism of human APOBEC3F by HIV-1 Vif. Nat. Struct. Mol. Biol. 26, 1176–1183 (2019).
Ito, F. et al. Structural basis for HIV-1 antagonism of host APOBEC3G via Cullin E3 ligase. Sci. Adv. 9, eade3168 (2023).
Li, Y. L. et al. The structural basis for HIV-1 Vif antagonism of human APOBEC3G. Nature 615, 728–733 (2023).
Russell, R. A., Smith, J., Barr, R., Bhattacharyya, D. & Pathak, V. K. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J. Virol. 83, 1992–2003 (2009).
Russell, R. A. & Pathak, V. K. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81, 8201–8210 (2007).
Letko, M., Booiman, T., Kootstra, N., Simon, V. & Ooms, M. Identification of the HIV-1 Vif and human APOBEC3G protein interface. Cell Rep. 13, 1789–1799 (2015).
Guo, Y. et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505, 229–233 (2014).
Wang, X., Bajaj, R., Bollen, M., Peti, W. & Page, R. Expanding the PP2A interactome by defining a B56-specific SLiM. Structure 24, 2174–2181 (2016).
Wu, C. G. et al. PP2A-B’ holoenzyme substrate recognition, regulation and role in cytokinesis. Cell Discov. 3, 17027 (2017).
Leonard, D. et al. Selective PP2A enhancement through biased heterotrimer stabilization. Cell 181, 688–701 e616 (2020).
Cho, U. S. & Xu, W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445, 53–57 (2007).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Strack, S., Cribbs, J. T. & Gomez, L. Critical role for protein phosphatase 2A heterotrimers in mammalian cell survival. J. Biol. Chem. 279, 47732–47739 (2004).
Flegg, C. P. et al. Nuclear export and centrosome targeting of the protein phosphatase 2A subunit B56alpha: role of B56alpha in nuclear export of the catalytic subunit. J. Biol. Chem. 285, 18144–18154 (2010).
Zhao, K. et al. Evolutionarily conserved pressure for the existence of distinct G2/M cell cycle arrest and A3H inactivation functions in HIV-1 Vif. Cell Cycle 14, 838–847 (2015).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Xiao, X., Li, S. X., Yang, H. & Chen, X. S. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat. Commun. 7, 12193 (2016).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D 66, 486–501 (2010).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. Sect. D 74, 814–840 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D 66, 213–221 (2010).
DeLano, W. The PyMOL Molecular Graphics System (DeLano Scientific, 2002).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
We thank G. Hu, J. Kaminsky and L. Wang at the Brookhaven Laboratory Cryo-EM facility for assistance with data collection. We thank other Xiong laboratory members for discussions. This work was supported by National Institutes of Health (NIH) grant no. R37AI116313 (Y.X.). This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and by the Innovation Award, Office of AIDS Research, NIH (V.K.P.).
Author information
Authors and Affiliations
Contributions
Y.X., V.K.P., Y.H. and K.A.D.-F. designed the experiments. Y.H. and C.W. performed the biophysical and biochemical experiments. K.A.D.-F. performed the virological experiments. Data were analyzed by Y.X., V.K.P., Y.H., K.A.D.-F. and C.W. F.A. contributed to experiments and discussions. Y.X., V.K.P., Y.H. and K.A.D.-F. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Christopher Hill and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The screening and optimization of PPP2R5A complex with HIV-1 Vif/CBFβ/Cul5 E3 ligase.
a, Size exclusion chromatography (SEC) profiles and SDS-PAGE analysis of individual PPP2R5A, Vif/CBFβ/Cul5 E3 and their complexes (repeated twice independently). The cryo-EM reconstructions (surface with fitted models in cartoon representation) of the fully assembled Vif/CBFβ/Cul5 E3/PPP2R5A showed no presence of PPP2R5A. b, The SEC profile and SDS-PAGE analysis of PPP2R5A/VCBC complex crosslinked by Bissulfosuccinimidyl suberate (BS3). The large scale crosslinking of the complex has been repeated 7 times independently. The OD280 and OD260 are shown in blue and red, respectively. The protein bands were detected by Coomassie blue stain.
Extended Data Fig. 2 The superposition of the VCBC complex structure with and without A3F (left) or A3G (right) bound.
The Vif flexible loop that shows the largest local conformational changes upon A3F or A3G binding is highlighted by dashed circles and with details illustrated in insets. PDBs used for the superpositions: A3FCTD alone: 3WUS; Vif/CBFβ: 4N9F; Vif/CBFβ/A3FCTD: 6NIL; VCBC/A3G: 8CX1. The individual Vif/CBFβ structures are shown in gray, the A3F or A3G bound Vif/CBFβ are shown in magenta/cyan.
Extended Data Fig. 3 Conservation of PPP2R5 family protein sequences at the Vif interface.
a, Alignment of PPP2R5A (amino acids 82 to 397) to PPP2R5B-E with the amino acids at the PPP2R5A:Vif interface and corresponding conserved amino acids in PPP2R5B-E highlighted in cyan (patch 3), yellow (patch 1), and magenta (patch 2). NCBI reference protein sequence (NP accession numbers): PPP2R5A (NP_006234); PPPP2R5B (NP_006235); PPP2R5C (NP_001339842); PPP2R5D (NP_006236); PPP2R5E (NP_006237). b, Structural modeling of PPP2R5-containing PP2As and their complexes with the Vif/CBFβ/Cul5 E3 ligase. Left: top, overlay of PPP2R5A (gray, PDB ID 6nts) and PPP2R5C (colored, PDB 2npp)-containing PP2A complex; bottom, overlay of different PPP2R5 family members (PPP2R5A, gray, PDB 6nts; PPP2R5B, purple, AlphaFold model; PPP2R5C, yellow, PDB 2npp; PPP2R5D, green, AlphaFold model; PPP2R5E, blue, AlphaFold model). Right: structural model of the HIV-1 Vif recruitment of PPP2R5C-containing PP2A onto the Cul5 E3 ligase complex.
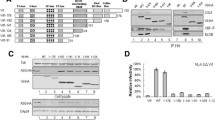
Extended Data Fig. 4 Mutational validation of the PPP2R5A-Vif interactions by western blot analysis.
The in vitro binding assay was performed using MBP-tagged Vif/CBFβ/EloB/EloC variants to pull down SUMO-PPP2R5A variants. The SUMO-PPP2R5A bands were recognized by anti-SUMO antibody, and the His-CBFβ bands were detected by Anti-His antibody (upper panel). The ratio of band intensities (SUMO-PPP2R5A/His-CBFβ) was quantified by mean ± sem; n = 2 biologically independent samples for R127E:WT, WT:E106K, R127E:E106K, n = 3 biologically independent samples for WT:Y294A, 273031AAA:WT, 273031AAA:Y294A, R33D:WT, WT:D205R/E251K, R33D:D205R/E251K, WT:Y373A, H43A/Y44A:WT, H43A/Y44A:Y373A, n = 4 biologically independent samples for K22E:WT, R23E:WT, K22E/R23E:WT, n = 7 biologically independent samples for WT:WT, with individual data points shown as dots (lower panel).
Extended Data Fig. 5 The observed structure differs from that predicted based on mutational analysis52.
The observed (upper panel) and computational predicted (lower panel) structures are shown in three different views to demonstrate the differences between the observed and predicted binding modes of PPP2R5A.
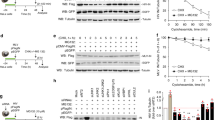
Extended Data Fig. 6 Structural analysis of Vif/CBFβ binding interfaces for different host targets.
Comparison of the distinct Vif/CBFβ interfaces (marked with ovals of different colors) for A3FCTD (bottom left), A3C (bottom middle, PDB 3VOW), A3DCTD (bottom right, homology model built from A3FCTD [PDB 3WUS]), A3G (middle left, [PDBs 8E40 and 8CX0]), A3H hapII (middle right, PDB 6BBO), and PPP2R5A (top). Critical residues observed are highlighted in red at the CBFβ-A3F/G interface and in yellow at the Vif-A3/PPP2R5A interfaces. A summary of the diverse locations of the Vif/CBFβ-A3/PPP2R5A interfaces is illustrated at middle center, with ovals of corresponding colors. Reproduced from: Fig. 3, Multifaceted HIV-1 Vif interactions with human E3 ubiquitin ligase and APOBEC3s, Yong Xiong, The FEBS Journal 288 3407–3417, Copyright © [2020], Federation of European Biochemical Societies, Wiley.
Extended Data Fig. 7 Cryo-EM image processing workflow for the Vif/CBFβ/EloB/EloC/PPP2R5A complex.
Tilted and untilted particles were combined and first cleaned by 3D and 2D classifications, then fractions of particles in two dominated views were discarded randomly to alleviate the preferred orientation issue. The remaining particles were further cleaned up by iterative heterogeneous refinement, and a final round of 3D classification identified a class of 500,511 particles with more balanced orientation distribution which was able to generate a 3D reconstruction of good quality. The resolution of the final reconstruction was further improved by non-uniform 3D refinement, local refinement and DeepEMhancer72.
Extended Data Fig. 8 Resolving the preferred orientation of the crosslinked PPP2R5A/VCBC complex.
a, An example of the raw image of the complex (left) and the particle orientation distribution of the untilted dataset (right). b, Top 2D class averages of the combined untilted and tilted datasets. From a total of 516,465 particles in the classes showing preferred orientations (boxed in red), 456,465 particles (~88%) were removed and excluded randomly from the subsequent analysis. c, The final particle set showed a more balanced orientation distribution.
Extended Data Fig. 9 Quality of the cryo-EM reconstruction of the crosslinked PPP2R5A/VCBC complex.
a, The Fourier shell correlation (FSC) curves of the cryo-EM reconstruction. b, Local resolution estimate of the cryo-EM map. c, The model-to-map FSC.
Supplementary information
Source data
Source Data Fig. 2
Unprocessed blue stain gels.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Unprocessed western blots and gels.
Source Data Extended Data Fig. 1
Unprocessed gels.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, Y., Delviks-Frankenberry, K.A., Wu, C. et al. Structural insights into PPP2R5A degradation by HIV-1 Vif. Nat Struct Mol Biol (2024). https://doi.org/10.1038/s41594-024-01314-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41594-024-01314-6
- Springer Nature America, Inc.