Abstract
Condensin, a structural maintenance of chromosomes (SMC) complex, has been shown to be a molecular motor protein that organizes chromosomes by extruding loops of DNA. In cells, such loop extrusion is challenged by many potential conflicts, for example, the torsional stresses that are generated by other DNA-processing enzymes. It has so far remained unclear how DNA supercoiling affects loop extrusion. Here, we use time-lapse single-molecule imaging to study condensin-driven DNA loop extrusion on supercoiled DNA. We find that condensin binding and DNA looping are stimulated by positively supercoiled DNA, and condensin preferentially binds near the tips of supercoiled plectonemes. Upon loop extrusion, condensin collects nearby plectonemes into a single supercoiled loop that is highly stable. Atomic force microscopy imaging shows that condensin generates supercoils in the presence of ATP. Our findings provide insight into the topology-regulated loading and formation of supercoiled loops by SMC complexes and clarify the interplay of loop extrusion and supercoiling.







Similar content being viewed by others
Data availability
Source data files are available for Figs. 1e,g,h; 2c,e; 3g; 4a,b; 5b–d and 6g,h and Extended Data Figs. 1; 3b; 4a,b; 5c and 10a,b. Original imaging data are available upon request. Source data are provided with this paper.
Code availability
The Python-based data analysis source code used for the analysis of the imaging data is available at https://github.com/biswajitSM/LEADS.
References
Yatskevich, S., Rhodes, J. & Nasmyth, K. Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet. 53, 445–482 (2019).
Ganji, M. et al. Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 (2018).
Davidson, I. F. et al. DNA loop extrusion by human cohesin. Science 366, 1338–1345 (2019).
Kim, Y., Shi, Z., Zhang, H., Finkelstein, I. J. & Yu, H. Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 (2019).
Golfier, S., Quail, T., Kimura, H. & Brugués, J. Cohesin and condensin extrude DNA loops in a cell-cycle dependent manner. eLife 9, e53885 (2020).
Kim, E., Kerssemakers, J., Shaltiel, I. A., Haering, C. H. & Dekker, C. DNA-loop extruding condensin complexes can traverse one another. Nature 579, 438–442 (2020).
Baxter, J. et al. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331, 1328–1332 (2011).
Sutani, T. et al. Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat. Commun. 6, 7815 (2015).
Minchell, N. E., Keszthelyi, A. & Baxter, J. Cohesin causes replicative DNA damage by trapping DNA topological stress. Mol. Cell 78, 739–751.e8 (2020).
Canela, A. et al. Genome organization drives chromosome fragility. Cell 170, 507–521.e18 (2017).
Kimura, K. & Hirano, T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90, 625–634 (1997).
Kimura, K., Rybenkov, V. V., Crisona, N. J., Hirano, T. & Cozzarelli, N. R. 13S condensin actively reconfigures DNA by introducing global positive writhe. Cell 98, 239–248 (1999).
Bazett-Jones, D. P., Kimura, K. & Hirano, T. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell 9, 1183–1190 (2002).
Kimura, K., Cuvier, O. & Hirano, T. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 276, 5417–5420 (2001).
Eeftens, J. M. et al. Real‐time detection of condensin‐driven DNA compaction reveals a multistep binding mechanism. EMBO J. 36, 3448–3457 (2017).
Gutierrez-Escribano, P. et al. Purified Smc5/6 complex exhibits DNA substrate recognition and compaction. Mol. Cell 80, 1039–1054.e6 (2020).
Sun, M., Nishino, T. & Marko, J. F. The SMC1-SMC3 cohesin heterodimer structures DNA through supercoiling-dependent loop formation. Nucleic Acids Res. 41, 6149–6160 (2013).
Ganji, M., Kim, S. H., van der Torre, J., Abbondanzieri, E. & Dekker, C. Intercalation-based single-molecule fluorescence assay to study DNA supercoil dynamics. Nano Lett. 16, 4699–4707 (2016).
Kim, S. H. et al. DNA sequence encodes the position of DNA supercoils. eLife 7, e36557 (2018).
van Loenhout, M. T. J., de Grunt, M. V. & Dekker, C. Dynamics of DNA supercoils. Science 338, 94–97 (2012).
Ganji, M. et al. Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 (2018).
Thundat, T. et al. Atomic force microscopy of DNA on mica and chemically modified mica. Scanning Microsc. 6, 2 (1992).
Balke, V. L. & Gralla, J. D. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J. Bacteriol. 169, 4499–4506 (1987).
Freeman, L., Aragon-Alcaide, L. & Strunnikov, A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149, 811–824 (2000).
Brandão, H. B. et al. RNA polymerases as moving barriers to condensin loop extrusion. Proc. Natl Acad. Sci. 116, 20489–20499 (2019).
Johzuka, K., Terasawa, M., Ogawa, H., Ogawa, T. & Horiuchi, T. Condensin loaded onto the replication fork barrier site in the rRNA gene repeats during S phase in a FOB1-dependent fashion to prevent contraction of a long repetitive array in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 2226–2236 (2006).
Jeppsson, K. et al. The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet. 10, e1004680 (2014).
Guo, M. S., Kawamura, R., Littlehale, M. L., Marko, J. F. & Laub, M. T. High-resolution, genome-wide mapping of positive supercoiling in chromosomes. eLife 10, e67236 (2021).
Marko, J. F. & Neukirch, S. Competition between curls and plectonemes near the buckling transition of stretched supercoiled DNA. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 85, 011908 (2012).
Racko, D., Benedetti, F., Dorier, J. & Stasiak, A. Transcription-induced supercoiling as the driving force of chromatin loop extrusion during formation of TADs in interphase chromosomes. Nucleic Acids Res. 46, 1648–1660 (2018).
Naughton, C. et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 20, 387–395 (2013).
Achar, Y. J., Adhil, M., Choudhary, R., Gilbert, N. & Foiani, M. Negative supercoil at gene boundaries modulates gene topology. Nature 577, 701–705 (2020).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Benedetti, F., Dorier, J. & Stasiak, A. Effects of supercoiling on enhancer–promoter contacts. Nucleic Acids Res. 42, 10425–10432 (2014).
Nolivos, S. et al. MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nat. Commun. 7, 10466 (2016).
Sen, N. et al. Physical proximity of sister chromatids promotes Top2-dependent intertwining. Mol. Cell 64, 134–147 (2016).
Japaridze, A. et al. Hyperplectonemes: a higher order compact and dynamic DNA self-organization. Nano Lett. 17, 1938–1948 (2017).
Krull, A., Buchholz, T.-O. & Jug, F. Noise2Void—learning denoising from single noisy images. In Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. 2124–2132 (IEEE, 2019).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Lyubchenko, Y. L. & Shlyakhtenko, L. S. AFM for analysis of structure and dynamics of DNA and protein–DNA complexes. Methods 47, 206–213 (2009).
Acknowledgements
We thank E. van der Sluis and A. van den Berg for protein purification, and J. Kerssemakers, J.-K. Ryu, A. Katan, R. Barth, M. Tisma and L. van Eendenburg for discussions. C.D. was supported by the European Research Council (ERC) Advanced Grant 883684 (DNA looping), Netherlands Organization for Scientific Research (NWO) grant OCENW.GROOT.2019.012, and the NanoFront and BaSyC programs.
Author information
Authors and Affiliations
Contributions
E.K. and C.D. designed the single-molecule imaging experiments, E.K. performed the imaging experiments and analyzed the imaging data, A.M.G. performed the AFM experiments and analyzed the AFM data, B.P. contributed in image analyses, J.v.d.T. prepared the DNA construct, C.D. supervised the work, and all authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors declare that they have no competing interests.
Peer review
Peer review information
Nature Structural and Molecular Biology thanks Alice Pyne, Anders Hansen, and Jan Lipfert for their contribution to the peer review of this work. Primary Handling Editor: Carolina Perdigoto, in collaboration with the Nature Structural and Molecular Biology team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Validation of our supercoiling generation assay.
To confirm the handedness of supercoiling, we introduced Topoisomerase I from E. coli, which only relaxes negative supercoils, to our DNA molecules on the surface that were generated to have positive/negative supercoils via SxO intercalation reaction. For negative supercoils, all the DNA molecules relaxed within 5 minutes of incubation with Topo1, while in the case of positive supercoils, the amount of supercoiled molecules only decreased by 25. Since, independent of Topo1, DNA molecules can be also nicked by exposure of excitation laser (which would resolve the supercoiling), the actual fraction of positive supercoiled DNA in our assay should be well above 75%. These data confirm the handedness of the supercoiling generated via SxO intercalation.
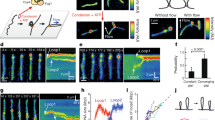
Extended Data Fig. 2 Fluorescence-intensity kymographs showing additional examples of loop extrusion events on supercoiled DNA.
(a) Examples showing condensin collects all the plectonemes upon loop extrusion and stabilizes them at its location. Yellow arrows show the location of the start of the gradually growing peak during loop extrusion. (b) Examples showing cases where condensin loop extrusion did not lead to absorption of all the plectonemes. In these events (11% of the total cases, Ntot=36), condensin was bound relatively close to the tethered end of DNA and the DNA loop extrusion was limited by reaching the end position of the DNA. Yellow arrows show the location of the start of the growing DNA loop.
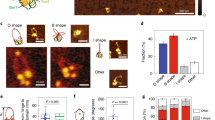
Extended Data Fig. 3 Estimation of condensin-binding position on supercoiled DNA measured with AFM.
a) Representative AFM images showing condensin binding near a plectoneme body (left) and near the plectoneme tip (right). Data represent 4 independent experiments. b) Statistics showing enrichment of condensin as observed outside of plectoneme, at plectoneme body, and at apical loops. In order to avoid possible supercoiling induced by condensin, the plasmids were incubated with AMP-PNP.
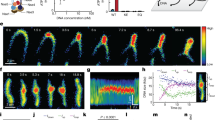
Extended Data Fig. 4 Additional examples showing that condensin initially loads near a plectoneme tip and moves downwards during plectoneme loop extrusion.
a) Snapshots showing condensin moving from the plectoneme tip (1st) towards the stem (till 3rd snapshot). Simultaneously the plectoneme becomes compacted and moves together with the condensin during extrusion while it grows back its length at later times (3rd – 5th frame). Data represent 3 independent experiments. b) Snapshot (left) and kymograph (right) showing condensin moving from the plectoneme tip towards the stem. Data represent 3 independent experiments. (c) Change in position for condensins that bound at the tip and moved along the length of DNA plectonemes extracted from the kymographs as in panel b for N = 6 molecules. The red colored trace corresponds to the kymograph in panel b.
Extended Data Fig. 5 Condensin-mediated loop extrusion on supercoiled DNA is strictly asymmetric.
(a) Examples of the DNA length calculated from the integrated fluorescence intensity kymographs of DNA loop extrusion by a single condensin on supercoiled DNA, showing asymmetric loop extrusion. (b) Same for an event where multiple condensins underlie the DNA loop, showing symmetric loop extrusion. (c) Statistics showing the ratios of the asymmetric and symmetric loop extrusion events observed for a single condensin (1) and multiple condensins (1 + ).
Extended Data Fig. 6 While plectonemes connected to the anchor side of condensin disappear, the corresponding DNA amount does not decrease.
(a) Example kymograph (top) and the DNA peaks (bottom) detected during condensin-mediated loop extrusion on supercoiled DNA. (b) Corresponding DNA lengths calculated from the kymograph. During the loop growth (blue; 45 s- 60 s) the length of DNA connected to the anchor side of condensin (pink) did not decrease, but instead DNA was accumulated from the motor side of condensin (green). This indicates that the disappearance of plectoneme at the anchor side is not due to the absorbance of plectoneme into the loop but rather to the stretching of the DNA.
Extended Data Fig. 7 Additional examples of condensin location along the plectoneme.
Condensin was found to be located at (a) the stem or (b) the middle after extrusion of a supercoiled loop in the case of single condensin. (c) Additional examples where multiple condensins are located along the supercoiled loop. Data in (a-c) represent 5 independent experiments.
Extended Data Fig. 8 AFM analysis of supercoiled DNA molecules.
(a) AFM image of a non-entangled dsDNA molecule. (b) Same image in an expanded blue-red color scale to highlight the entangled regions of dsDNA (red) versus the non-entangled regions (light blue) and the mica (dark blue). Inset shows the Z color scale. Data in (a,b) represent 3 independent experiments. (c) Height profile along the molecule in a/b, showing an average height of 1.25 nm. (d) AFM image of a dsDNA molecule with one crossover. (e) Same image in an expanded blue-red color scale. The crossover point is denoted with a yellow arrow. Data in (d,e) represent 5 independent experiments. (f) Height profile along the molecule shown on d/e. A locally increased height of > 2 nm is observed at the crossing point (arrows). The arrows denote the two peaks that correspond to the crossing point – which is encountered twice upon tracking the contour of the DNA along this ‘figure-8’ profile. (g) AFM image of entangled dsDNA molecules. Yellow arrows signal the molecules of which the profiles are shown in panels h and i. Data represent 3 independent experiments. (h) Height profile along a molecule where the entangled patches are denoted in red. (i) Same for the molecule indicated by i in panel g. Notably, this molecule was almost entirely entangled along its full contour length. Scale bars are 200 nm.
Extended Data Fig. 9 Examples of bleaching time traces of fluorescence intensities of individual ATTO647N-labelled condensin complexes at the location of supercoiled loops.
(a) Bleaching occurred in a single step-wise manner for condensin after a loop extrusion event. (b) Bleaching occurred in a double step-wise manner after an event where two condensins had merged into a single supercoiled DNA loop. (c) Bleaching occurred in a single step-wise manner for condensin after a loop extrusion event. Additional short binding events occurred at the plectoneme loop.
Extended Data Fig. 10 Condensin binding is length and supercoiling dependent.
(a) Observation probability of condensin on nicked DNA with two different lengths (21 kb and 48.5 kb), estimated by summing the total number of detected condensin peaks over all time points, and dividing this by the DNA length and measurement time. N = 8 molecules for 21 kb, N = 22 molecules for 48 kb length, respectively. (b) Condensin enrichment on supercoiled loops and relaxed loops, estimated by summing up the total intensity of the detected condensin peaks within the loop region with similar lengths (~60 % of its contour length). N = 25 molecules for supercoiled loops and N = 20 for relaxed loops, respectively. Data represent mean ± standard deviation.
Supplementary information
Supplementary Information
Legends for Supplementary Videos 1 and 2.
Supplementary Video 1
Video showing condensin-mediated DNA loop extrusion on supercoiled DNA. A single DNA intensity peak appeared among the multiple plectoneme peaks upon loop extrusion, which gradually increased in size as it moved along the DNA.
Supplementary Video 2
Video showing that condensin initially bound to a plectoneme gradually reels in DNA while simultaneously localizing all of the nearby plectonemes. After the loop was formed, we applied a buffer flow and monitored the location of the condensin along the supercoiled DNA loop. This revealed that condensin was localized at the stem of the plectonemic loop.
Source data
Source Data Fig. 1
Statistical source data of Fig. 1e,g,h.
Source Data Fig. 2
Statistical source data of Fig. 2c,e.
Source Data Fig. 3
Statistical source data of Fig. 3g.
Source Data Fig. 4
Statistical source data of Fig. 4a,b.
Source Data Fig. 5
Statistical source data of Fig. 5b–d.
Source Data Fig. 6
Statistical source data of Fig. 6g,h.
Source Data Extended Data Fig. 1
Statistical source data of Extended Data Fig. 1.
Source Data Extended Data Fig. 3
Statistical source data of Extended Data Fig. 3b.
Source Data Extended Data Fig. 5
Statistical source data of Extended Data Fig. 5c.
Source Data Extended Data Fig. 10
Statistical source data of Extended Data Fig. 10a,b.
Rights and permissions
About this article
Cite this article
Kim, E., Gonzalez, A.M., Pradhan, B. et al. Condensin-driven loop extrusion on supercoiled DNA. Nat Struct Mol Biol 29, 719–727 (2022). https://doi.org/10.1038/s41594-022-00802-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-022-00802-x
- Springer Nature America, Inc.
This article is cited by
-
Topoisomerase-modulated genome-wide DNA supercoiling domains colocalize with nuclear compartments and regulate human gene expression
Nature Structural & Molecular Biology (2024)
-
The nucleolar shell provides anchoring sites for DNA untwisting
Communications Biology (2024)
-
DNA packaging by molecular motors: from bacteriophage to human chromosomes
Nature Reviews Genetics (2024)
-
Looking back at 30 years of Nature Structural & Molecular Biology
Nature Structural & Molecular Biology (2024)
-
Reeling it in: how DNA topology drives loop extrusion by condensin
Nature Structural & Molecular Biology (2022)