Abstract
Polygenic risk scores (PRSs), which often aggregate results from genome-wide association studies, can bridge the gap between initial discovery efforts and clinical applications for the estimation of disease risk using genetics. However, there is notable heterogeneity in the application and reporting of these risk scores, which hinders the translation of PRSs into clinical care. Here, in a collaboration between the Clinical Genome Resource (ClinGen) Complex Disease Working Group and the Polygenic Score (PGS) Catalog, we present the Polygenic Risk Score Reporting Standards (PRS-RS), in which we update the Genetic Risk Prediction Studies (GRIPS) Statement to reflect the present state of the field. Drawing on the input of experts in epidemiology, statistics, disease-specific applications, implementation and policy, this comprehensive reporting framework defines the minimal information that is needed to interpret and evaluate PRSs, especially with respect to downstream clinical applications. Items span detailed descriptions of study populations, statistical methods for the development and validation of PRSs and considerations for the potential limitations of these scores. In addition, we emphasize the need for data availability and transparency, and we encourage researchers to deposit and share PRSs through the PGS Catalog to facilitate reproducibility and comparative benchmarking. By providing these criteria in a structured format that builds on existing standards and ontologies, the use of this framework in publishing PRSs will facilitate translation into clinical care and progress towards defining best practice.
Similar content being viewed by others
Main
The predisposition to common diseases and traits arises from a complex interaction between genetic and non-genetic factors. In the past decade there has been enormous success at discovering disease-associated genetic variants, facilitated by many collaborative consortia and large cohorts of well-phenotyped individuals with matched genetic information1,2,3,4,5. In particular, genome-wide association studies (GWASs) have yielded summary statistics that describe the magnitude (effect size) and statistical significance of association between an allele and the outcome of interest4,6. GWASs have been applied to many complex human traits and diseases, including height, blood pressure, cardiovascular disease, cancer, obesity and Alzheimer’s disease.
The associations identified through GWASs can be combined to quantify genetic predisposition to a heritable trait, and this information can be used to conduct disease risk stratification or to predict prognostic outcomes and response to therapy7,8. Typically, information across many variants is combined by means of a weighted sum of allele counts, in which the weights reflect the relative magnitude of association between variant alleles and the trait or disease. These weighted sums can include millions of variants, and are frequently referred to as polygenic risk scores (PRSs), or genetic or genomic risk scores (GRSs), if they refer to risk estimates of disease outcomes; or, more generally, polygenic scores (PGSs) when referring to any outcome (Box 1). Although algorithms are actively being developed to decide how many and which variants to include and how to weigh them so as to maximize the proportion of variance explained or disease discrimination, there is an emerging consensus that inclusion of variants beyond those that meet stringent GWAS significance levels can boost predictive performance9,10. Methodological research has also established theoretical limits of the performance of PGSs and PRSs on the basis of the genetic architecture and heritability of the trait11,12,13,14,15.
In the last decade, the landscape of genetic prediction studies has transformed. Over 900 publications mention PGS or PRS, and there have been significant developments in how PRSs are constructed and evaluated, as well as many new proposed uses. The data available in the current era of biomedical research are larger and more consolidated than ever before. Biobanks and large-scale consortia have become dominant, yet frequently researchers have limited access to individual-level data. As individual data are often unavailable, most PRS models are developed from summary-level data (for example, GWAS summary statistics) in secondary datasets, each of which come with their own specific methodological considerations16,17,18. At the same time, there has been a push towards open data sharing as outlined in the FAIR (Findable, Accessible, Interoperable and Reusable) Data Principles3,19, with an emphasis on ensuring that research is reproducible by all.
The capacity of PRSs to quantify genetic predisposition for many clinically relevant traits and diseases has begun to be established, with many potential clinical uses in settings related to disease risk stratification as well as proposed prognostic uses (for example, predicting responses to intervention or treatment). Readiness for implementation varies by outcome, and mature PRSs with potential clinical utility are available for only a few diseases—for example, coronary heart disease (CHD) and breast cancer (Boxes 2 and 3, respectively). There has also been a rapid rise of direct-to-consumer assays and for-profit companies (23andMe, Color, MyHeritage, and so on) that provide PGS and PRS results to customers outside of the traditional patient–provider framework. These concomitant developments have resulted in healthcare systems developing new infrastructures to deliver genetic risk information20. Individually and combined, these advances have raised substantial challenges for PRS reporting standards—from the very basic (for example, reporting performance metrics on an external validation dataset) to the complicated (for example, making raw variant and weight information for a PRS available)—and necessitate the updating of existing standards for reporting genetic risk prediction studies to convey the increased scope of PRSs and the complexity of their clinical applications.
Poorly designed and/or described studies call into question the validity of some PRSs to predict their target outcome21,22, and relatively few studies have externally benchmarked the performance of multiple scores. At present, there are no best practices for developing PRSs that are uniformly agreed upon, nor are there widely adopted standards or regulations that are sufficiently tailored to assess the eventual clinical readiness of a PRS. There are emerging applications of PRSs that further compound the heterogeneity in reporting; for example, using PRSs as tools for testing gene–environment interactions or shared aetiology between diseases23,24,25,26. The rapid evolution in both methodological development and applications of PRSs make it challenging to compare or reproduce claims about the predictive performance of a PRS for a specific outcome when studies are not properly documented. These deficiencies are barriers to PRSs being interpreted, compared, and reproduced, and must be addressed to enable the application of PRSs to improve clinical practice and public health.
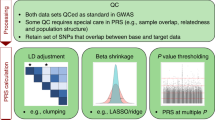
Frameworks have been developed to establish standards around the transparent, standardized, accurate, complete and meaningful reporting of scientific studies. In 2011, an international working group published the Genetic Risk Prediction Studies (GRIPS) Statement—a set of reporting guidelines for risk prediction models that include genetic variants, from genetic mutations to gene scores27. These guidelines are analogous to those developed for observational epidemiological studies (STROBE28) and genome-wide association studies (STREGA29), and are in line with the reporting guidelines for multivariate prediction models (TRIPOD30). Adherence to reporting statements has been low, and the same holds for GRIPS. One reason might be that researchers feel that the GRIPS Statement inadequately addresses PRSs. Researchers are frequently uncertain as to what precisely should be reported for a PRS study to be assessed as rigorous, reproducible and ultimately translatable, especially with the increased push for data availability and transparency. Most PRS studies follow a prototypical process (Fig. 1) that can be used as a template for standardizing reporting and benchmarking in the field.
The prototypical steps for PRS construction, risk model development and validation of performance are displayed with select aspects of the PRS-RS guidelines (labelled in bold). During PRS development, variants associated with an outcome of interest, typically identified from a GWAS, are combined as a weighted sum of allele counts. Methods for optimizing variant selection (PRS construction and estimation) are not shown. To predict the outcome of interest the PRS is added to a risk model and may be combined with non-genetic variables (for example, age, sex, ancestry or clinical variables; collectively referred to as risk model variables). After fitting procedures to select the best risk model, this model is validated in an independent sample. The PRS distribution should be described, and the performance of the risk model demonstrated in terms of its discrimination, predictive ability and calibration. Though not displayed in the figure, these same results should also be reported for the training sample for comparison to the validation sample. In both training and validation cohorts, the outcome of interest criteria, demographics, genotyping and non-genetic variables should be reported (Table 1) HLA, human leukocyte antigen; HR, hazard ratio; IDI, integrated discrimination improvement; IQR, interquartile range; NRI, net reclassification improvement; OR, odds ratio; β, effect estimate from linear regression.
Here, the Clinical Genome Resource (ClinGen) Complex Disease Working Group and the Polygenic Score (PGS) Catalog (Supplementary Note 1) jointly present the Polygenic Risk Score Reporting Standards (PRS-RS), an expanded and updated set of reporting standards for PRSs that addresses current research environments with advanced methodological developments to inform clinically meaningful reporting on the development and validation of PRSs in the literature, with an emphasis on reproducibility and transparency throughout the development process. Additional methods are detailed in Supplementary Note 2.
The PRS-RS
The PRS-RS is a set of standard items specifying the minimal criteria that need to be described in a manuscript to accurately interpret a PRS and reproduce results throughout the PRS development process31 (see Fig. 1 for a brief summary). It applies to PRS development and validation studies that aim to predict disease onset, diagnosis and prognosis, as well as response to therapies; however, other research uses of PRSs have overlapping steps that should be reported in a similar manner. Table 1 presents the full PRS-RS, with reporting items organized into key components along the developmental pipeline of PRSs for clear interpretation and to encourage their documentation from the inception of the study, well before publication.
Reporting on the background of risk scores
The development and validation of a PRS tests a specific hypothesis with a defined outcome and study population. Therefore, authors should define a priori (note that in the next few sections, inverted commas are used to refer to each of the Reporting Standards referred to in Table 1) the ‘study type’ (for example, development and/or validation), ‘risk model purpose’ (for example, risk prediction versus prognosis) and ‘predicted outcome’ (for example, CHD) in enough detail to understand why the study population and risk model selected are relevant (for example, the value for CHD risk stratification and primary prevention is highest in younger individuals compared to those over 80 with lifetime accumulated risk). As the PRS-RS is focused on clinical validity and implementation, authors must outline the study and appropriate outcomes to understand what risk is measured, what the purpose of measuring risk would be and why this purpose may be of clinical relevance. To establish the internal validity of a study, authors should use the appropriate data for the intended purpose (for example, prediction of incident disease versus prognosis), with adequate documentation of dataset characteristics to understand nuances in measured risk.
Reporting on study populations
The applicability of any risk prediction to an external target population (the who, where, and when) depends on its similarity to the original study populations that were used to derive the risk model. Therefore, authors need to define and characterize the details of their study population (‘study design and recruitment’), and describe study ‘participant demographics’ for key variables (most often age and sex) and ancestry. Notably, there are often inconsistent definitions and levels of detail associated with ancestry, and the transferability of genetic findings between different racial and ethnic groups can be limited1,9,32. It is therefore essential for authors to provide a detailed description of the genetic ‘ancestry’ of participants—including how ancestry was determined—using a common controlled vocabulary where possible (for example, the standardized framework developed by the NHGRI-EBI GWAS Catalog1). Authors should provide a sufficient level of detailed criteria for defining all of the factors relevant to the ‘outcome of interest’, including but not limited to those used in the risk model (‘non-genetic variables’). These details should accompany information about how the population was genotyped (‘genetic data’), including assays and all quality control measures.
Reporting on the development of risk models
At present there are several commonly used methods to select variants that constitute the PRS and fine-tune their weights7,16,17,18,31. Methods using GWAS summary statistics should clearly cite the relevant GWAS, preferably using unique and persistent study identifiers from the GWAS Catalog (GCSTs)33. As the performance and limitations of the combined risk model are dependent on methodological considerations, authors must provide complete details including the method used and how variants are combined into a single PRS (‘PRS construction and estimation’). Apart from genetic data, authors should also describe the defining criteria for other demographic and non-genetic predictors (‘non-genetic variables’) included in the model. Often authors will iterate through numerous models to find the optimal fit. In addition to the estimation methods, it is important to detail the ‘integrated risk model(s) fitting’ procedure, including the measures that were used for the selection of the final model. Translating the continuous PRS distribution to a risk estimate, whether absolute or relative, is highly dependent on assumptions and limitations that are inherent to the specific dataset used. When describing the ‘risk model type’, authors should detail the timescale used for prediction, or the study period and follow-up time for a relative hazard model. Furthermore, if relative risk is estimated, the reference group should be well described. These details should be described for the training set, as well as for validation and sub-group analyses.
Reporting on the evaluation of risk models
Authors should report estimates for all evaluated models (including the methods used to derive them) to equip readers with the information necessary to evaluate the relative value of an increase in performance against other trade-offs. We recommend that authors provide summary information of the ‘PRS distribution’ to aid in model interpretation. The ‘predictive ability’, ‘calibration’, and ‘discrimination’ of the risk model should also be assessed and detailed with common descriptions including the risk score effect size, variance explained (R2), reclassification indices and metrics like sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). The risk model ‘calibration’ and ‘discrimination’ should be described for all analyses, although their estimation and interpretation are most relevant for the PRS validation sample. It is imperative for the PRS and integrated risk models to be evaluated on a population that is external (independent, non-overlapping) to the individuals in the study population. The ability of the risk model to classify individuals of interest (‘risk model discrimination’) is commonly described and presented in terms of the area under the receiver operating characteristic (AUROC) or precision-recall curve (AUPRC), or the concordance statistic (C-index). Any differences in variable definitions or performance discrepancies between the training and validation sets should be described.
Reporting on interpretation
By explicitly describing the ‘risk model interpretation’ and outlining potential ‘limitations’ to the ‘generalizability’ of their model, authors will empower readers and the wider community to better understand the risk score and its relative merits. Authors should justify the clinical relevance and the ‘intended uses’ of the risk model, such as how the performance of their PRS compares to other commonly used risk models, or previously published PRSs. This may also include comparisons to other genetic predictors of disease (for example, mutations in high or moderate risk genes associated with Mendelian forms of the disease), family history, simple demographic models or conventional risk calculators (see Boxes 2 and 3 for disease-specific examples). What indicates a ‘good’ prediction can differ between outcomes and intended uses, but should be reported with similar metrics to those described in the evaluation section.
Reporting on model parameters
The underlying PRS (variant alleles and derived weights) should be made publicly available, preferably through direct submission to an indexed repository such as the PGS Catalog, to enable others to reuse existing models (with known validity) and to facilitate direct benchmarking between different PRSs for the same trait (thus promoting ‘data transparency and availability’). The current mathematical form of most PRSs—a linear combination of allele counts—facilitates clear model description and reproducibility. Future genomic risk models may have more complex forms; for example, allowing for explicit non-linear epistatic and gene–environment interactions, or deep neural networks of lesser clarity. It will be important to describe these models in sufficient detail to allow their implementation and evaluation by other researchers and clinical groups.
Supplementary Note 3 provides explanations of reporting considerations in addition to the minimal reporting framework in Table 1. Authors intending downstream clinical implementation should aim for the level of transparent and comprehensive reporting covered in both Table 1 and the Supplementary Notes, especially concerning points related to discussing the interpretation, limitations and generalizability of results. The proper reporting of the development and performance of PRSs can also have implications for seeking regulatory approval of the PRS as a clinical test. Although not a comprehensive list of regulatory requirements, we highlight aspects of the PRS-RS that would be considered evidence of analytical and clinical validity from the perspective of the College of American Pathologists (CAP) and the Clinical Laboratory Improvement Amendments (CLIA) (Supplementary Table 1). CAP and CLIA approvals are additional incentives for the reporting adherence of researchers who wish to translate their work to the clinic, as well as a caution for researchers who want to avoid their findings being put to unintended use. Finally, we reiterate the need for both methodological and data transparency, and we encourage the deposition of PRSs (variant-level information necessary to recalculate the genetic portion of the score) in the PGS Catalog (www.pgscatalog.org34), which provides an invaluable resource for the widespread adoption and distribution of a published PRS. The PGS Catalog provides access to PGSs and related metadata to support the FAIR principles of data stewardship19, enabling subsequent applications and assessments of PGS performance and best practices (see Supplementary Table 2 for a description of the metadata captured in the PGS Catalog and its overlap with the PRS-RS).
Improving PRS research and translation
We surveyed 30 publications (selecting for a range of disease domains, risk score categories and populations) to understand how the information in the PRS-RS is presented and displayed as part of the larger iterative process to clarify and improve minimal reporting item descriptions. For 10 of these publications, we provide detailed annotations using the final minimal reporting requirements (Supplementary Table 3) and use these annotations to illustrate the detail necessary for each PRS-RS item (further described in Supplementary Note 3). The heterogeneity in the PRS reporting we observed in this pilot highlights a series of challenges. Critical aspects of PRS studies—including ancestry, predictive ability, and transparency or availability of information needed to reproduce PRSs—were frequently absent or reported in insufficient detail. This underscores the need for the PRS-RS to clearly and specifically define meaningful aspects of PRS development, testing and intended clinical use. However, these deficits in reporting are not unique to PRSs; previous reports of underreporting have found that 77% of GWAS publications in 2017 did not share summary statistics35 and 4% of GWASs do not report any relevant ancestry information1. In line with the push towards a culture of reproducibility and open data in genomics, we as the ClinGen Complex Disease Working Group and PGS Catalog joined to create this set of reporting standards (Table 1), which is specifically tailored to PRS research and adapts the previous standards on the basis of the opinions of multidisciplinary and international experts.
Researchers using the PRS-RS may identify fringe cases that are inadequately captured by these reporting items, as we have modelled our guidelines on prototypical steps for PRS development (Fig. 1). Although we anticipate that the field may change further as novel methods and technologies are generated, the PRS-RS items can be expanded and adapted to encompass new considerations. By updating previous standards, drawing on the knowledge of leaders in the field and tailoring the framework to common barriers observed in recent literature, we aim to provide a comprehensive and pragmatic perspective on the topic. In line with previous standards, the PRS-RS includes elements related to understanding the clinical validity of PRSs and consequent risk models. Items such as ‘predicted outcome’ and ‘intended use’ bookend our guidelines with the intended clinical framing of PRS reporting. In addition, we have modelled the guidelines by steps in experimental design—from hypothesis to interpretation—to more clearly emphasize the importance of considering the risk model’s intended purpose in defining what needs to be reported and to inform documentation throughout the process. As a reference, we have included a guide to where PRS-RS items should be reported in a manuscript in Supplementary Table 4. These expansions will further facilitate the curation and expert annotation of published PRSs as we move towards widespread clinical use.
Although the scope of our work encompasses clinical validity, it does not address the additional requirements that are needed to establish the clinical or public health utility of a PRS, such as randomized trials with clinically meaningful outcomes, health economic evaluations or feasibility studies36. In addition, the translation of structured data elements into useful clinical parameters may not be direct. One example is that the case definitions used in training or validation in any particular PRS study may deviate (sometimes substantially) from those used in any specific health system. CHD symptoms commonly include angina (chest pain), whereas PRSs are frequently trained on stricter definitions excluding angina. Another example is that the definitions used for race or ancestry as outlined in the PGS Catalog and the GWAS Catalog1 may differ from structured terms used to document ancestry information in the clinic. Consistent mappings and potentially parallel analyses may be necessary to translate from genetically determined ancestries to those that are routinely used in clinical care. Such translation issues potentially limit generalizability to target populations and warrant further discussion, and we reiterate the need for authors to be mindful of their intended purpose and target audience when discussing their findings. Authors’ understanding of potential translational barriers can be aided by considering the current CAP and CLIA analytical and clinical validity evidence requirements of peer-reviewed literature to ensure that the PRS-RS has value in informing later steps of the clinical translation spectrum, including clinical utility (Supplementary Table 1). Finally, although the principles of this work are clear, its scope does not include the complex commercial restrictions—such as intellectual property—that may be placed on published studies with regard to the reporting or distribution of PGSs or the data that underlie them. We hope that our work will inform downstream regulation and transparency standards for PRS as a commercial clinical tool.
The coordinated efforts of the ClinGen Complex Disease Working Group and PGS Catalog provide a set of compatible resources for researchers to deposit PGS- and PRS-related information. The PGS Catalog (www.PGSCatalog.org) provides an informatics platform, with data integration and harmonization to other PGSs as well as the source GWAS study through its sister platform, the GWAS Catalog1,34. In addition, it provides a structured database of scores (variants and effect weights) that can be reused, along with metadata requested in the PRS-RS. With these tools, the PRS-RS can be mandated by leading peer-reviewed journals and, consequently, the quality and rigour of PRS research will be increased to a level that facilitates clinical implementation. We encourage readers to visit the ClinGen website (https://clinicalgenome.org/working-groups/complex-disease/) for any future changes or amendments to our reporting standards.
Although we have provided explicit recommendations on how to acknowledge study design limitations and their effects on the interpretation and generalizability of a PRS, future research should attempt to establish best practices to guide the field. Moving forward, supplementary frameworks should be developed for the reporting of new methods, such as deep learning, as well as requirements for clinical utility and readiness. Together, the PRS-RS enables the rapid development of PRSs as potentially powerful tools for the translation of genomic discoveries into clinical and public health benefits, and provides a framework for PRSs to transform multiple areas of research in human genetics.
References
Morales, J. et al. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol. 19, 21 (2018).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017).
Claussnitzer, M. et al. A brief history of human disease genetics. Nature 577, 179–189 (2020).
Visscher, P. M., Brown, M. A., McCarthy, M. I. & Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012).
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Pasaniuc, B. & Price, A. L. Dissecting the genetics of complex traits using summary association statistics. Nat. Rev. Genet. 18, 117–127 (2017).
Lambert, S. A., Abraham, G. & Inouye, M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 28 (R2), R133–R142 (2019).
Torkamani, A., Wineinger, N. E. & Topol, E. J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018). References 7 and 8 are useful reviews outlining the potential benefits and clinical uses of polygenic risk scores.
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019).
Chatterjee, N., Shi, J. & García-Closas, M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 17, 392–406 (2016).
Wray, N. R., Yang, J., Goddard, M. E. & Visscher, P. M. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 6, e1000864 (2010).
Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013).
Pharoah, P. D. P. et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat. Genet. 31, 33–36 (2002).
Zhang, Y., Qi, G., Park, J.-H. & Chatterjee, N. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat. Genet. 50, 1318–1326 (2018).
Chatterjee, N. et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat. Genet. 45, 400–405 (2013).
Mak, T. S. H., Porsch, R. M., Choi, S. W., Zhou, X. & Sham, P. C. Polygenic scores via penalized regression on summary statistics. Genet. Epidemiol. 41, 469–480 (2017).
Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. GigaScience 8, giz082 (2019).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Ganguly, P. NIH funds centers to improve the role of genomics in assessing and managing disease risk. National Human Genome Research Institute https://www.genome.gov/news/news-release/NIH-funds-centers-to-improve-role-of-genomics-in-assessing-and-managing-disease-risk (2020).
Martens, F. K., Tonk, E. C. M. & Janssens, A. C. J. W. Evaluation of polygenic risk models using multiple performance measures: a critical assessment of discordant results. Genet. Med. 21, 391–397 (2019).
Janssens, A. C. J. W. Validity of polygenic risk scores: are we measuring what we think we are? Hum. Mol. Genet. 28, R143–R150 (2019).
Warrier, V. et al. Polygenic scores for intelligence, educational attainment and schizophrenia are differentially associated with core autism features, IQ, and adaptive behaviour in autistic individuals. Preprint at https://doi.org/10.1101/2020.07.21.20159228 (2020).
Matoba, N., Love, M. I. & Stein, J. L. Evaluating brain structure traits as endophenotypes using polygenicity and discoverability. Hum. Brain Mapp. https://doi.org/10.1002/hbm.25257 (2020).
Coleman, J. R. I. et al. Genome-wide gene-environment analyses of major depressive disorder and reported lifetime traumatic experiences in UK Biobank. Mol. Psychiatry 25, 1430–1446 (2020).
Kraft, P. & Aschard, H. Finding the missing gene–environment interactions. Eur. J. Epidemiol. 30, 353–355 (2015).
Janssens, A. C. J. W. et al. Strengthening the reporting of genetic risk prediction studies (GRIPS): explanation and elaboration. Eur. J. Hum. Genet. 19, 615 (2011). GRIPS was the first to propose standards for the reporting of risk prediction studies using genetics, and forms the basis of our updated PRS-RS.
von Elm, E. et al. Strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Br. Med. J. 335, 806–808 (2007).
Little, J. et al. STrengthening the REporting of Genetic Association Studies (STREGA): an extension of the STROBE statement. PLoS Med. 6, e1000022 (2009).
Moons, K. G. M. et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann. Intern. Med. 162, W1–W73 (2015).
Choi, S. W., Mak, T. S.-H. & O’Reilly, P. F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protocols 15, 2759–2772 (2020).
Wojcik, G. L. et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Lambert, S. A. et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. https://doi.org/10.1038/s41588-021-00783-5 (2021). This paper describes the development and contents of the PGS Catalog (www.PGSCatalog.org), an open database of polygenic scores.
Thelwall, M. et al. Is useful research data usually shared? An investigation of genome-wide association study summary statistics. PLoS One 15, e0229578 (2020).
Burke, W. & Zimmern, R. Moving Beyond ACCE: An Expanded Framework for Genetic Test Evaluation (PHG Foundation, 2007).
Huo, D. et al. Comparison of breast cancer molecular features and survival by African and European ancestry in The Cancer Genome Atlas. JAMA Oncol. 3, 1654–1662 (2017).
Choi, J. et al. The associations between immunity-related genes and breast cancer prognosis in Korean women. PLoS One 9, e103593 (2014).
Marston, N. A. et al. Predicting benefit from evolocumab therapy in patients with atherosclerotic disease using a genetic risk score: results from the FOURIER trial. Circulation 141, 616–623 (2020).
Nikpay, M. et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 47, 1121–1130 (2015).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
Mars, N. et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat. Med. 26, 549–557 (2020).
Elliott, J. et al. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. J. Am. Med. Assoc. 323, 636–645 (2020).
Inouye, M. et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J. Am. Coll. Cardiol. 72, 1883–1893 (2018).
Abraham, G. et al. Genomic prediction of coronary heart disease. Eur. Heart J. 37, 3267–3278 (2016).
Ganna, A. et al. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler. Thromb. Vasc. Biol. 33, 2267–2272 (2013).
Kullo, I. J. et al. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES clinical trial). Circulation 133, 1181–1188 (2016).
Mavaddat, N. et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 104, 21–34 (2019).
Mars, N. et al. The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat. Commun. 11, 6383 (2020).
Zhang, H. et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat. Genet. 52, 572–581 (2020).
Zhang, X. et al. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: a nested case-control study. PLoS Med. 15, e1002644 (2018).
Lakeman, I. M. M. et al. Addition of a 161-SNP polygenic risk score to family history-based risk prediction: impact on clinical management in non-BRCA1/2 breast cancer families. J. Med. Genet. 56, 581–589 (2019).
Wilcox, A. N. et al. Prospective evaluation of a breast cancer risk model integrating classical risk factors and polygenic risk in 15 cohorts from six countries. Preprint at https://doi.org/10.1101/19011171 (2019).
Pal Choudhury, P. et al. Comparative validation of the BOADICEA and Tyrer-Cuzick breast cancer risk models incorporating classical risk factors and polygenic risk in a population-based prospective cohort. Preprint at https://doi.org/10.1101/2020.04.27.20081265 (2020).
Maas, P. et al. Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncol. 2, 1295–1302 (2016).
Lee, A. et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet. Med. 21, 1708–1718 (2019).
Garcia-Closas, M., Gunsoy, N. B. & Chatterjee, N. Combined associations of genetic and environmental risk factors: implications for prevention of breast cancer. J. Natl. Cancer Inst. 106, dju305 (2014).
Kapoor, P. M. et al. Combined associations of a polygenic risk score and classical risk factors with breast cancer risk. J. Natl. Cancer Inst. 113, djaa056 (2020).
Easton, D. F. et al. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 372, 2243–2257 (2015).
Muranen, T. A. et al. Genetic modifiers of CHEK2*1100delC-associated breast cancer risk. Genet. Med. 19, 599–603 (2017).
Kuchenbaecker, K. B. et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J. Natl. Cancer Inst. 109, djw302 (2017).
Fahed, A. C. et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat. Commun. 11, 3635 (2020.
Barnes, D. R. et al. Polygenic risk scores and breast and epithelial ovarian cancer risks for carriers of BRCA1 and BRCA2 pathogenic variants. Genet. Med. 22, 1653–1666 (2020).
van den Broek, J. J. et al. Personalizing breast cancer screening based on polygenic risk and family history. J. Natl. Cancer Inst. 00, jdaa127 (2020).
Esserman, L. J., the WISDOM Study and Athena Investigators. The WISDOM Study: breaking the deadlock in the breast cancer screening debate. NPJ Breast Cancer 3, 34 (2017).
Acknowledgements
We acknowledge the input of the ClinGen Complex Disease Working Group members, including C. D. Bustamante, M. Meyer, F. Harrell, D. Kent, P. Visscher, T. Assimes, S. Plon and J. Berg. We also thank D. Durham for her editorial support and A. Paolucci for her administrative support in the preparation and submission of this manuscript. ClinGen is primarily funded by the National Human Genome Research Institute (NHGRI), through the following three grants: U41HG006834, U41HG009649 and U41HG009650. ClinGen also receives support for content curation from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), through the following three grants: U24HD093483, U24HD093486 and U24HD093487. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH). In addition, the views expressed in this article are those of the author(s) and not necessarily those of the National Health Service (NHS), the National Institute for Health Research (NIHR) or the Department of Health. Research reported in this publication was supported by the NHGRI under award number U41HG007823 (EBI-NHGRI GWAS Catalog, PGS Catalog). We also acknowledge funding from the European Molecular Biology Laboratory (EMBL). Individuals were funded from the following sources: M.I.M. was a Wellcome Investigator and an NIHR Senior Investigator with funding from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; U01-DK105535) and Wellcome (090532, 098381, 106130, 203141 and 212259); M.I. is supported by the Munz Chair of Cardiovascular Prediction and Prevention at the Baker Heart and Diabetes Institute; M.I., S.A.L. and J.N.D. were supported by core funding from: the UK Medical Research Council (MR/L003120/1), the British Heart Foundation (RG/13/13/30194 and RG/18/13/33946) and the NIHR (Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust); S.A.L. is supported by a Canadian Institutes of Health Research postdoctoral fellowship (MFE-171279); and J.N.D. holds a British Heart Foundation Personal Chair and a NIH Research Senior Investigator Award. This work was also supported by Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome.
Author information
Authors and Affiliations
Contributions
H.W., C.T., M.A.I., J.S.D., I.J.K., C.S., R.R., K.V., K.K., K.A.B.G., P.K. and G.L.W. conducted the literature review of reporting practices. H.W., S.A.L., C.T., M.A.I., J.W.O., I.J.K., R.R., D.B., E.V., M.I.M., A.C.A., D.F.E., R.A.H., A.V.K., N.C., C.K., K.E., E.M.R., M.C.R., K.E.O., M.J.K., A.C.J.W.J., K.A.B.G., P.K., J.A.L.M., M.I. and G.L.W. provided feedback on the reporting framework. H.W., S.A.L., C.T., M.A.I., J.W.O., J.S.D., I.J.K., A.V.K., J.N.D., H.P., E.M.R., M.C.R., A.C.J.W.J., K.A.B.G., P.K., J.A.L.M., M.I. and G.L.W. wrote and edited the manuscript text and items.
Corresponding author
Ethics declarations
Competing interests
M.I.M. is on the advisory panels for Pfizer, Novo Nordisk and Zoe Global; honoraria: Merck, Pfizer, Novo Nordisk and Eli Lilly; research funding: Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, Novo Nordisk, Pfizer, Roche, Sanofi Aventis, Servier and Takeda. As of June 2019, he is an employee of Genentech with stock and stock options in Roche. No other authors have competing interests to declare.
Additional information
Peer review information Nature thanks Guillaume Lettre, Paul O’Reilly and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
This file contains Supplementary Notes, including additional Supplementary Methods detailing the development and revisions of the Polygenic Risk Score Reporting Statement (PRS-RS), as well as additional considerations for each of the reporting items.
Supplementary Table 1
Considerations for clinical translation. This table outlines considerations to inform downstream stakeholder perspectives regarding the translational spectrum of lab test development.
Supplementary Table 2
Mappings between the PGS Catalog and PRS-RS fields. As the PGS Catalog and PRS-RS do not completely align in their fields, we have provided a mapping between the two frameworks to assist in reporting of published PRS.
Supplementary Table 3
Example curations of 10 manuscripts using PRSRS. We have detailed information from ten PRS manuscripts using the fields outlined in PRS-RS as an example for the research community.
Supplementary Table 4
Introduction, Methods, Results and Discussion (IMRAD) Mappings for PRS-RS. Additionally, we have provided mappings between PRS-RS reporting items and a traditional IMRAD structure to aid authors in the reporting of items in manuscript preparation.
Rights and permissions
About this article
Cite this article
Wand, H., Lambert, S.A., Tamburro, C. et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature 591, 211–219 (2021). https://doi.org/10.1038/s41586-021-03243-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03243-6
- Springer Nature Limited
This article is cited by
-
Inclusion of the severe and enduring anorexia nervosa phenotype in genetics research: a scoping review
Journal of Eating Disorders (2024)
-
Genetic distance and ancestry proportion modify the association between maternal genetic risk score of type 2 diabetes and fetal growth
Human Genomics (2024)
-
Generalizability of polygenic prediction models: how is the R2 defined on test data?
BMC Medical Genomics (2024)
-
Recent advances in polygenic scores: translation, equitability, methods and FAIR tools
Genome Medicine (2024)
-
Clinical data mining: challenges, opportunities, and recommendations for translational applications
Journal of Translational Medicine (2024)