Abstract
It has been reported that MicroRNAs (miRNAs) play pivotal roles in the occurrence and progression of a variety of cancers. As reported, miR-4295 promotes cell growth and metastasis in a lot of cancers. Nonetheless, the role and molecular mechanism of miR-4295 in HNSCC still remain unknown. In this study, we discovered miR-4295 expression was significantly upregulated in HNSCC tissues and cell lines, which is also associated with the overall survival of patients. Additionally, suppression of miR-4295 significantly inhibited cell proliferation, migration and EMT process in HNSCC. Through Targetscan website, it was predicted that NPTX1 might be a direct target gene of miR-4295. Then, we verified that NPTX1 could directly interact with miR-4295 via luciferase reporter and RNA assays. What’s more, we discovered that there was a significantly negative correlation between NPTX1 and miR-4295 expression. It was indicated by further investigation that the effect of miR-4295 suppression on cell proliferation, migration and EMT process in HNSCC can be restored by knockdown of NPTX1 at the same time. Our results suggested that miR-4295 promoted the progression of HNSCC via regulating NPTX1 expression and miR-4295/NPTX1 axis, which may be a new therapeutic strategy for HNSCC.
Similar content being viewed by others
Introduction
Head and neck cancer (HNC) is the sixth most common malignant cancer worldwide and a heterogeneous collection of tumors originated from the upper aerodigestive tract [1, 2]. Head and neck squamous cell carcinoma (HNSCC) is the most common type of HNC, accounting for more than 90% of HNC and occurs in the pharynx, oral cavity, and larynx [3, 4]. Despite of intensive efforts for diagnosis and treatment of HNSCC, the overall survival rate has had no significant increase in the past few decades. Therefore, it is crucial to investigate the underlying molecular biological mechanism of HNSCC, and then seek for new therapeutic targets for HNSCC.
It is well known that microRNAs (miRNAs), a class of small noncoding RNA with a length of 19–24 nucleotides, can modulates downstream gene expression and then involved in the regulation of cancers [5,6,7]. As reported in many studies, miRNAs have oncogenic or tumor-suppressive function in cancers. For instance, miR-346 promotes HCC progression by suppressing breast cancer metastasis suppressor 1 expression [8]. MiR-9-5p promotes cell growth and metastasis in non-small cell lung cancer through the repression of TGFBR2 [9]. MiR-142-3p inhibits cancer cell proliferation by targeting CDC25C [10]. MiR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells [11]. In addition, there were many miRNAs involved in the regulation of HNC. For instance, microRNA-203 suppresses invasion and epithelial-mesenchymal transition induction via targeting NUAK1 in HNC [12]. Loss of miR-125b-1 contributes to HNC development by dysregulating TACSTD2 and MAPK pathway [13]. MiR-124 acts as a tumor suppressor by inhibiting the expression of sphingosine kinase 1 and its downstream signaling in HNSCC [14]. Furthermore, recent studies have found that miR-4295 is upregulated in multiple cancers and affects the tumor phenotypes. For example, miR-4295 promotes cell growth in bladder cancer by targeting BTG1 [15]. MiR-4295 promotes cell proliferation and invasion in anaplastic thyroid carcinoma via CDKN1A [16]. Still and all, there is no report on the molecular mechanism and biological function of miR-4295 in HNSCC.
The aim of this study was to investigate the biological function and potential regulatory mechanism of miR-4295 in HNSCC.
Materials and methods
Clinical samples
A total of 92 clinical tumor tissues and adjacent normal tissues were collected from patients with HNSCC who underwent surgery at Sichuan Cancer Hospital & Institute. All tissue samples were immediately stored at −80 °C for future experiments. None of the patients suffered from any tumor-specific treatment prior to surgery. Written informed consents were provided from all patients. The Ethics Committee of Sichuan Cancer Hospital & Institute approved the protocol of this study. The tissue samples were used for further experiments (92 samples and 92 controls).
Cell culture and transfection
HNSCC cell lines (FaDu, Hep2, SCC22B, and SCC154) and human normal oral epithelial cell line (iNOE) were investigated in this study, and they were all obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). They were recently authenticated by STR profiling and tested for mycoplasma contamination. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco/Invitrogen Inc., Carlsbad, CA, USA), phytomycin (100 μg/ml) and penicillin (100 U/ml) at 37 °C in a humidified atmosphere containing 5% CO2.
MiR-4295 inhibitor, miR-4295 mimic and the negative controls (NC inhibitor and NC mimic) were purchased from GenePharma (Shanghai, China). Short hairpin RNA (shRNA) targeting NPTX1 (sh-NPTX1) and a negative control (sh-NC) were also designed and synthesized by GenePharma. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect these above plasmids into FaDu or Hep2 cells according to manufacturer’s recommendations.
RNA extraction and quantitative real-time PCR
Trizol reagent (Takara, Otsu, Japan) was used to extract the total RNA from tissues and cells. The reverse transcription for RNAs into cDNA was performed by using TaqManTM Advanced miRNA cDNA Synthesis Kit (Waltham, MA, USA) or the reverse transcription kit (Takara, Otsu, Japan). The RT-qPCR was performed using SYBR Green PCR Kit (Takara, Otsu, Japan). GAPDH and U6 served as endogenous controls. Applied Biosystems Step One Plus Real-Time PCR System (Applied Biosystems, Foster city, USA) was used to perform RT-qPCR analysis, and relative quantification was assessed using the 2−ΔΔCt method. The primers used for RT-qPCR were as follows:
miR-4295: 5′-CAGTGCAATGTTTTCCTTGCC-3′ (Forward) and 5′-GTAGGAACAGTCTAGTTTCTTAGCC-3′ (Reverse);
NPTX1: 5′-CTCAGCCAACTCGGGCAAACTT-3′ (Forward) and 5′-ATCCTTGAGGCTGTTGGTCTGG-3′ (Reverse);
GAPDH: 5′-GAA GGT GAA GGT CGG AGT C-3′ (Forward) and 5′-GAA GAT GGT GAT GGG ATT TC-3′ (Reverse);
U6: 5′-ATT GGA ACG ATA CAG AGA AGA TT-3′ (Forward) and 5′-GGA ACG CTT CAC GAA TTT G-3′ (Reverse).
CCK-8 assay
Cell proliferation capability was assessed using Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies) assay. Firstly, the transfected cells (1 × 103 cells/well) were seeded in 96-well plates. Plates were incubated for 24, 48, 72, and 96 h. Each well was added with CCK-8 reagent, followed by subsequent incubation for another 4 h. A microplate reader (Molecular Devices, Sunnyvale, CA, USA) was used to detect the optical density of each well at 450 nm.
EdU immunofluorescence assay
Cell proliferation was assessed by using 5-ethynyl-2′-deoxyuridine (EdU) immunofluorescence assay. HNSCC cells (1 × 103 cells/well) were cultured into 96-well plates. After incubation for 48 h, new medium containing 50 μM EdU replaced the original medium. With another 2 h incubation, cells were fixed with 4% formaldehyde for 30 min and treated with 0.5% Triton X-100 for 20 min. After washing with PBS, cells were counterstained with anti-EdU reagents for 30 min. After incubation with 100 μL DAPI for 30 min, cells were visualized applying fluorescence microscopy.
Transwell assay
Transwell assay was performed to detect the migration ability of cells. The transwell membranes (8 µm pore size, Corning, NY, USA) without Matrigel were used for migration assay. The upper chambers were supplemented with 200 μl serum-free medium containing transfected cells (1 × 104 cells/well), while the lower chambers were supplemented with 600 μl DMEM medium containing 10% FBS. After incubation for 48 h, migrated cells that located in the lower membrane were fixed by menthanol and then stained by 0.5% crystal violet. Finally, all stained cells were counted under a light microscope (Olympus Corporation, Tokyo, Japan).
RNA pull-down assay
MiR-4295-Wt, miR-4295-Mut, and miR-NC were transcribed by TranscriptAid T7 High Yield Transcription Kit (ThermoFisher Scientific, USA). Biotin RNA labeling mix (Roche Diagnostics, Indianapolis, IN, USA) was used to produce Bio-miR-4295-Wt, Bio-miR-4295-Mut, and Bio-miR-NC. Two hundred micrograms cell lysates (FaDu and Hep2) and 50 pmol biotinylated RNA were mixed, and then incubated with 50 μl streptavidin agarose beads (Invitrogen, Carlsbad, CA, USA) at 4 °C for 1 h. After washing, RT-qPCR was used to measure the mRNA expression of eluted proteins.
Luciferase reporter assay
The 3′-UTR sequences of NPTX1 containing the binding sites of miR-4295 were cloned and constructed into the pGL3 vector (Promega, Madison, WI, USA) to produce the wild-type NPTX1 reporter (NPTX1-Wt). GeneArt™ Site-Directed Mutagenesis System (Thermo Fisher Scientific) was used to produce the mutant-type NPTX1 reporter (NPTX1-Mut). NPTX1-Wt or NPTX1-Mut was co-transfected with NC mimic, NC inhibitor, miR-4295 mimic or miR-4295 inhibitor into FaDu or Hep2 cells. After transfection for 48 h, the luciferase activity was determined with luciferase reporter assay system (Promega, Madison WI, USA).
Western blot
Total proteins from HNSCC cells were extracted using RIPA lysis buffer (Beyotime Biotechnology, China) containing protease inhibitors (Roche, China). The proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto the polyvinylidene difluoride (PVDF) membranes. Next, membranes were blocked by skim milk and probed with the primary antibodies overnight at 4 °C. The primary antibodies anti-NPTX1 (ab75917), anti-E-cadherin (ab15148), anti-N-cadherin (ab18203), anti-Vimentin (ab92547), and anti-GAPDH (ab8245) were both purchased from Abcam company (Abcam, Cambridge, UK). After that, the appropriate secondary antibody was added to membranes and then incubated for another 2 h at room temperature. Finally, ECL detection kits (Millipore, MA, USA) was used to visualize the proteins. GADPH served as a loading control.
Statistical analysis
Statistical analysis was carried out using SPSS 20.0 software (SPSS, Chicago, IL, USA). All data were expressed as the mean ± standard deviation (SD). Each experiment was repeated three times. The one-way ANOVA or Student’s t-test was used to analyze the differences between groups. All data was found to follow a normal distribution. The estimation of variance homogeneity was carried out. ANOVA with Dunnett post hoc test was applied for comparison between groups when there were no significant differences among variance. Otherwise, ANOVA with Dunnett’s T3 post hoc test was used. Overall survival analysis was performed using the Kaplan–Meier method and the log-rank test. Correlation analysis was performed using Spearman’s rank correlation test. P < 0.05 was considered statistically significant.
Results
MiR-4295 is upregulated in HNSCC tissues and cells, and correlated with the overall survival of patients
Compared with adjacent normal tissues, miR-4295 expression was significantly upregulated in HNSCC tissues (n = 92) (Fig. 1a). Likewise, miR-4295 expression was remarkably upregulated in HNSCC cells (FaDu, Hep2, SCC22B, and SCC154) compared with that in human normal oral epithelial cells (iNOE) (Fig. 1b). We also discovered that miR-4295 expression was significantly increased in advanced clinical stages (Fig. 1c). Next, we analyzed the correlation between the miR-4295 expression and the clinical characteristics of HNSCC patients, and therefore found that miR-4295 expression was significantly associated with tumor size, TNM stage, and lymph node metastasis (Table 1, P < 0.05). To explore the potential relationship between miR-4295 expression and the patients’ prognosis, the effects of miR-4295 expression on overall survival were evaluated via Kaplan–Meier analysis, which indicated that the over-all survival of the high miR-4295 expression group was notably shorter compared with that of the low miR-4295 group (Fig. 1d). Multivariate analysis showed that only miR-4295 expression (P = 0.030) and TNM stage (P = 0.009) were independent prognostic factors for HNSCC patients (Table 2, P < 0.05). These results suggested that miR-4295 may act as an oncogene in HNSCC.
MiR-4295 is upregulated in HNSCC tissues and cell lines and relates to poor prognosis of patients. a Relative expression of miR-4295 in tumor tissues (n = 92) and adjacent non-tumor tissues (n = 92). b Relative expression of miR-4295 in HNSCC cell lines (FaDu, Hep2, SCC22B and SCC154) and human normal oral epithelial cell line (iNOE). c Relative expression of miR-4295 in different clinical stages (I–II and III–IV). d Kaplan–Meier analysis of overall survival in HNSCC patients with high miR-4295 level and low miR-4295 level. Results were expressed as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001
MiR-4295 suppression inhibits proliferation, migration and EMT process in HNSCC cells
To explore the biological function of miR-4295 in HNSCC, miR-4295 inhibitor and its scramble controls (NC mimic and NC inhibitor) were respectively transfected into FaDu and Hep2 cells for the subsequent experiments. Compared with scramble control, the expression of miR-4295 was significantly downregulated in FaDu and Hep2 cells transfected with miR-4295 inhibitor (Fig. 2a). It was indicated that miR-4295 suppression inhibited the proliferation of FaDu and Hep2 cells through CCK-8 and EdU immunofluorescence assays (Fig. 2b, c). Transwell assay showed a significant reduction of the migration ability of FaDu and Hep2 cells transfected with miR-4295 inhibitor (Fig. 2d). EMT process has been reported to be a key step of tumor metastasis [17, 18]. Therefore, western blot was applied to determine the expression of EMT markers, and the results showed that miR-4295 suppression decreased the expression of mesenchymal marker (N-cadherin and vimentin), but increased the expression of epithelial marker (E-cadherin) in FaDu and Hep2 cells (Fig. 2e). Conclusively, all these results indicated that miR-4295 suppression inhibits proliferation, migration, and EMT process in HNSCC cells.
Suppression of miR-4295 inhibited cell proliferation, migration and EMT process in HNSCC. a The suppression efficiency of miR-4295 was detected in FaDu and Hep2 cells via RT-qPCR. b CCK-8 assay showed that suppression of miR-4295 inhibited cell proliferation. c EdU immunofluorescence assay showed that suppression of miR-4295 inhibited cell proliferation. d The effects of miR-4295 suppression on cell migration were detected in FaDu and Hep2 cells by transwell assay. e The effects of miR-4295 suppression on EMT markers (E-cadherin, N-cadherin and vimentin) expression were detected in FaDu and Hep2 cells after different transfections by western blot. Results were expressed as the mean ± SD of three independent experiments. **P < 0.01 and ***P < 0.001
NPTX1 is a direct target gene of miR-4295 in HNSCC
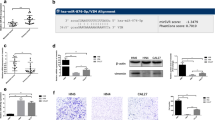
As has been reported before, miRNAs can regulate downstream gene expression in cancers [5,6,7]. In order to investigate the regulatory mechanism of miR-4295 in HNSCC, we used Targetscan to predict the potential target genes of miR-4295 and thereby noticed that Neuronal pentraxin 1 (NPTX1) had a binding site for miR-4295 (Fig. 3a). NPTX1 is reported to be associated with tumor progression in types of tumors [19, 20]. Therefore, we hypothesized that NPTX1 might be modulated by miR-4295 in HNSCC. Luciferase reporter assay was then performed to evaluate the interaction between miR-4295 and NPTX1, and the results showed that the luciferase activity of wild-type NPTX1 reporter vector was significantly decreased in cells transfected with miR-4295 mimic, whereas increased in cells transfected with miR-4295 inhibitor, indicating that NPTX1 could directly bind with miR-4295 (Fig. 3b). Nevertheless, no notable difference on the luciferase activity of mutation-type NPTX1 reporter vector was observed in cells transfected with miR-4295 mimic (or miR-4295 inhibitor) group and NC mimic (or NC inhibitor) group. RNA pull-down assay further manifested the direct interaction between miR-4295 and NPTX1 (Fig. 3c). Western blot assay showed that NPTX1 expression was downregulated in FaDu and Hep2 cells transfected with miR-4295 mimic, but upregulated in FaDu and Hep2 cells transfected with miR-4295 inhibitor (Fig. 3d). Moreover, NPTX1 expression was significantly upregulated in HNSCC tissues (Fig. 3e). Spearman’s correlation analysis indicated that a remarkably negative correlation was discovered between NPTX1 and miR-4295 expression in tumor tissues (r = −0.433, P < 0.001, Fig. 3f). To sum up, NPTX1 is a direct target gene of miR-4295.
MiR-4295 directly targets NPTX1 and is negatively correlated to NPTX1 expression. a Bioinformatical predication showed the predicted binding sites of NPTX1 and miR-4295. b Luciferase reporter assay was performed to evaluate the interaction of NPTX1 and miR-4295. c RNA pull down assay was performed to further evaluate the binding ability between NPTX1 and miR-4295. d Western blot assay showed that miR-4295 could negative regulate the expression of NPTX1. e RT-qPCR showed NPTX1 expression was higher in tumor tissues than in adjacent non-tumor tissues. f The expression of miR-4295 was inversely correlated with the expression level of NPTX1 in HNSCC tissues. Results were expressed as the mean ± SD of three independent experiments. **P < 0.01 and ***P < 0.001
MiR-4295 promotes proliferation and migration in HNSCC by regulating NPTX1 expression
To explore whether miR-4295 promotes cell proliferation and metastasis in HNSCC via regulating NPTX1, rescue assays were performed by using sh-NPTX1 plasmid. As shown in Fig. 4a, NPTX1 expression was decreased in cells transfected with sh-NPTX1, but restored in cells co-transfected with miR-4295 inhibitor and sh-NPTX1. The inhibitory roles of miR-4295 inhibitor in the proliferation of HNSCC cells can be restored in cells co-transfected with sh-NPTX1 (Fig. 4b, c). Transwell assay showed that cells transfected with miR-4295 inhibitor decreased the migration ability of HNSCC cells, whereas transfected with miR-4295 inhibitor + sh-NPTX1 partly restored the migration ability of HNSCC cells (Fig. 4d). In addition, compared with cells transfected with miR-4295 inhibitor, the expression of EMT markers (E-cadherin, N-cadherin, and vimentin) was also restored in cells co-transfected with miR-4295 inhibitor + sh-NPTX1 (Fig. 4e). Thus, the conclusion can be drawn that miR-4295 promotes cell proliferation and metastasis in HNSCC progression by suppressing NPTX1 expression.
MiR-4295 promoted cell proliferation and metastasis in HNSCC by regulating NPTX1. a RT-qPCR assay showed that NPTX1 expression was decreased in cells transfected with sh-NPTX1, but restored in cells co-transfected with miR-4295 inhibitor and sh-NPTX1. b CCK-8 assay was showed that knockdown of NPTX1 weakened miR-4295 suppression-mediated effects on cell proliferation. c EdU immunofluorescence assay was showed that knockdown of NPTX1 weakened miR-4295 suppression-mediated effects on cell proliferation. d Transwell assay was showed that knockdown of NPTX1 weakened miR-4295 suppression-mediated effects on the migration of HNSCC cells. e Western blot assay presented that knockdown of NPTX1 weakened miR-4295 suppression-mediated effects on the expression of EMT markers. Results were expressed as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001
Discussion
Recently, it has been proved by mounting evidences that miRNAs play a variety of roles in development and progression of human cancers, such as breast cancer, bladder cancer as well as HNC. MiR-218 and miR-129 regulate breast cancer progression by targeting Lamins [21]. MicroRNA-124 inhibits cell proliferation, invasion, and migration by targeting CAV1 in bladder cancer [22]. MicroRNA-363 targets myosin 1B to reduce cellular migration in HNC [23]. Recent studies demonstrated that miR-4295 is frequently dysregulated in multiple tumors and affects the tumor phenotypes [15, 16]. Nonetheless, the biological function and molecular mechanism of miR-4295 in HNSCC are still unknown. In this study, we discovered that miR-4295 expression were significantly upregulated in HNSCC tissues and cell lines. In addition, suppression of miR-4295 inhibited the proliferation and migration in HNSCC. Moreover, it was also indicated that suppression of miR-4295 decreased the expression of mesenchymal marker (N-cadherin and vimentin), but increased the expression of epithelial marker (E-cadherin). To conclude, these results indicated that miR-4295 promoted cell proliferation and metastasis in HNSCC.
As has been reported, miRNAs can regulate the expression of target genes to modulate the progression of cancers. For instance, miR-361 targets Yes-associated protein (YAP) mRNA to suppress cell proliferation in lung cancer [24]. MiR-519d-3p suppresses breast cancer cell growth and motility via targeting LIM domain kinase 1 [25]. MiR-4326 promotes lung cancer cell proliferation through targeting tumor suppressor APC2 [26]. In consequence, we searched for downstream target genes of miR-4295 through Targetscan website. NPTX1 was predicted to have a binding site for miR-4295. It has been reported that NPTX1 is associated with tumor progression in some types of tumors. For example, NPTX1 inhibits colon cancer cell proliferation through down-regulating cyclin A2 and CDK2 expression [19]. NPTX1 is a novel epigenetic regulation gene and associated with prognosis in lung cancer [20]. In this study, it is indicated that NPTX1 expression could be negatively regulated by miR-4295. Consequent findings delineated that konckdown of NPTX1 partially restored miR-4295 suppression-mediated effects on cell proliferation, migration and EMT process in HNSCC. This work provided comprehensive evidence to show that miR-4295 facilitated cell proliferation and metastasis in HNSCC by targeting NPTX1. That is to say, miR-4295/NPTX1 axis might be a new diagnostic/therapeutic approach for HNSCC. This might be the first study to report that miR-4295 functions as an oncogenic miRNA for HNSCC tumorigenesis through targeting NPTX1. Additionally, other regulatory mechanisms of miR-4295 in HNSCC should be explored in the future so as to justify further evaluation of miR-4295 as a potential diagnostic/therapeutic target for HNSCC patients.
References
Guo T, Califano JA. Molecular biology and immunology of head and neck cancer. Surg Oncol Clin N Am. 2015;24:397–407.
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29.
Cramer JD, Speedy SE, Ferris RL, Rademaker AW, Patel UA, Samant S. National evaluation of multidisciplinary quality metrics for head and neck cancer. Cancer. 2017;123:4372–81.
Jou A, Hess J. Epidemiology and molecular biology of head and neck cancer. Oncol Res Treat. 2017;40:328–32.
Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–9.
Srivastava SK, Bhardwaj A, Leavesley SJ, Grizzle WE, Singh S, Singh AP. MicroRNAs as potential clinical biomarkers: emerging approaches for their detection. Biotech Histochem. 2013;88:373–87.
Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14.
Guo Z, Li J, Sun J, Sun L, Zhou Y, Yu Z. miR-346 Promotes HCC Progression by Suppressing Breast Cancer Metastasis Suppressor 1 Expression. Oncol Res. 2018;26:1073–81.
Li G, Wu F, Yang H, Deng X, Yuan Y. MiR-9-5p promotes cell growth and metastasis in non-small cell lung cancer through the repression of TGFBR2. Biomed Pharm. 2017;96:1170–8.
Cao XC, Yu Y, Hou LK, Sun XH, Ge J, Zhang B, et al. miR-142-3p inhibits cancer cell proliferation by targeting CDC25C. Cell Prolif. 2016;49:58–68.
Sripada L, Singh K, Lipatova AV, Singh A, Prajapati P, Tomar D, et al. hsa-miR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells. J Mol Med. 2017;95:641–51.
Obayashi M, Yoshida M, Tsunematsu T, Ogawa I. microRNA-203 suppresses invasion and epithelial-mesenchymal transition induction via targeting NUAK1 in head and neck cancer. Oncotarget. 2016;7:8223–39.
Nakanishi H, Taccioli C, Palatini J, Fernandez-Cymering C, Cui R, Kim T, et al. Loss of miR-125b-1 contributes to head and neck cancer development by dysregulating TACSTD2 and MAPK pathway. Oncogene. 2014;33:702–12.
Zhao Y, Ling Z, Hao Y, Pang X. MiR-124 acts as a tumor suppressor by inhibiting the expression of sphingosine kinase 1 and its downstream signaling in head and neck squamous cell carcinoma. Oncotarget. 2017;8:25005–20.
Nan Y-H, Wang J, Wang Y, Sun P-H. MiR-4295 promotes cell growth in bladder cancer by targeting BTG1. Am J Transl Res. 2016;8:4892.
Shao M, Geng Y, Lu P, Xi Y, Wei S, Wang L, et al. miR-4295 promotes cell proliferation and invasion in anaplastic thyroid carcinoma via CDKN1A. Biochem Biophys Res Commun. 2015;464:1309–13.
Huang R, Zong X. Aberrant cancer metabolism in epithelial-mesenchymal transition and cancer metastasis: mechanisms in cancer progression. Crit Rev Oncol Hematol. 2017;115:13–22.
Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13.
Peng X, Pan K, Zhao W, Zhang J. NPTX1 inhibits colon cancer cell proliferation through down-regulating cyclin A2 and CDK2 expression. Cell Biol Int. 2018;42:589–597.
Zhou C, Qin Y, Xie Z, Zhang J, Yang M, Li S, et al. NPTX1 is a novel epigenetic regulation gene and associated with prognosis in lung cancer. Biochem Biophys Res Commun. 2015;458:381–6.
Setijono SR, Park M, Kim G, Kim Y, Cho KW, Song SJ. miR-218 and miR-129 regulate breast cancer progression by targeting Lamins. Biochem Biophys Res Commun. 2018;496:826–33.
Zhou W, He L, Dai Y, Zhang Y, Wang J, Liu B. MicroRNA-124 inhibits cell proliferation, invasion and migration by targeting CAV1 in bladder cancer. Exp Ther Med. 2018;16:2811–20.
Chapman BV, Wald AI, Akhtar P, Munko AC, Xu J, Gibson SP, et al. MicroRNA-363 targets myosin 1B to reduce cellular migration in head and neck cancer. BMC Cancer. 2015;15:861.
Zhang S, Liu Z, Wu L, Wang Y. MiR-361 targets Yes-associated protein (YAP) mRNA to suppress cell proliferation in lung cancer. Biochem Biophys Res Commun. 2017;492:468–73.
Li D, Song H, Wu T, Xie D, Hu J, Zhao J, et al. MiR-519d-3p suppresses breast cancer cell growth and motility via targeting LIM domain kinase 1. Mol Cell Biochem. 2018;444:169–78.
Xu G, Zhang Z, Zhang L, Chen Y, Li N, Lv Y, et al. miR-4326 promotes lung cancer cell proliferation through targeting tumor suppressor APC2. Mol Cell Biochem. 2018;443:151–7.
Acknowledgements
The authors appreciate all laboratory members. This study was supported by Grant agency of the Science and Technology Department of Sichuan Province (No. 2017HH0096).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lu, S., Zhou, C., Zou, B. et al. MiR-4295 facilitates cell proliferation and metastasis in head and neck squamous cell carcinoma by targeting NPTX1. Genes Immun 21, 4–12 (2020). https://doi.org/10.1038/s41435-019-0081-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-019-0081-0
- Springer Nature Limited