Abstract
In our research, we studied the regulatory effect of miR-29c-3p on HNSCC progression. First, bioinformatics approach was utilized to find significantly differentially expressed genes in The Cancer Genome Atlas-HNSCC. Then the target miRNA and its target mRNA were screened out. Next, qRT-PCR was utilized to examine miR-29c-3p expression in HNSCC and the adjacent tissue. Western blot was introduced to test the protein level of C1QTNF6. Besides, the results of MTT, wound healing, transwell and angiogenesis assays verified the proliferation, migration, invasion and angiogenesis of HNSCC cells. Finally, the targeted relationship of miR-29c-3p and C1QTNF6 was confirmed through dual-luciferase assay. Our study found the negative correlation of miR-29c-3p and C1QTNF6 in HNSCC. Besides, overexpressed miR-29c-3p suppressed proliferation, migration, invasion, and angiogenesis of HNSCC cells. Additionally, overexpressed C1QTNF6 facilitated these biological functions of HNSCC cells while upregulating miR-29c-3p reversed such effect. Altogether, miR-29c-3p was proven to suppress the tumor-promoting effects and angiogenesis in HNSCC by targeting C1QTNF6. We also revealed a novel mechanism of HNSCC progression. MiR-29c-3p/C1QTNF6 might be a target in HNSCC treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As an invasive cancer, head and neck squamous cell carcinoma (HNSCC) generates from the larynx, oral cavity, and pharynx, which is associated with alcohol, smoking, or human papillomavirus exposure [1,2,3]. Although most HNSCC cases develop from the local invasion of cervical neural and vascular tissue, its mortality is mainly caused by the metastasis of distant organs and regional lymph nodes [4,5,6]. A critical event contributing to this metastasis is tumor-related angiogenesis [7]. Angiogenesis is increasingly considered as a key process to the solid tumors development because it can support tumor growth as well as the metastasis of malignant cells from primary tumor site to distant organs [8, 9]. However, the molecular regulatory mechanisms about the angiogenesis and cancer metastasis are complex. This study aimed to elucidate the mechanisms facilitates future monitoring and targeted therapy of HNSCC metastasis.
MicroRNAs (miRNAs) inhibit protein translation or induce mRNA degeneration. The aberrant expression of miRNAs is tightly related to cancers, including HNSCC [10]. Research showed that miRNAs like miR-34a [11], miR-96-5p [12] and miR-205 [13] are aberrantly expressed in HNSCC patients’ tissue. Besides, these miRNAs target-related genes to affect the cell progression as well as cell cycle in HNSCC. Thus, the modulation of specific miRNA activity represents a potential therapy strategy for cancers [14, 15]. MiR-29c-3p as a member of miR-29 family can affect the proliferation, invasion and migration of SAS cell line (primary lesion derived from tongue SCC) and the FaDu cell line (primary lesion derived from scaly cell carcinoma of the hypopharynx) [16]. Additionally, growing evidence proves that miR-29c-3p is involved in angiogenesis. For example, TUG1 reverses the inhibitory effect on endothelial progenitor tube formation caused by miR-mimic transfection [17]. Circ-001971 acts as a ceRNA and alleviates the inhibition of VEGFA by miR-29c-3p, thereby aggravating angiogenesis of colorectal cancer cells [18].
C1q and TNF-Related 6 (C1QTNF6), also known as C1qTNF-related protein-6 (CTRP6), belongs to the CTRP family and to functions in fatty acid metabolism [19]. C1QTNF6 endows with special molecular structure composed of short variable region, signal peptide, C-terminal globular domain and collagen-like region [20]. As an endogenous complement regulator, C1QTNF6 contributes to the effective treatment of arthritis [21]. In addition, C1QTNF6 can also exert effect on tumors. C1QTNF6 expression is high in gastric cancer tissue and it mediates cancer cell proliferation, migration and apoptosis [22]. Furthermore, C1QTNF6 is less expressed in serum of ovarian cancer patients, whereby suppressing the progression of the cancer cells by blocking IL-8/NEGF signaling pathway [23]. Yet no study has revealed whether C1QTNF6 affects HNSCC. Hence, our study unprecedentedly investigated the regulatory mechanism of C1QTNF6 in HNSCC cells.
By summarizing the results of previous studies and bioinformatics analysis, it was speculated that miR-29c-3p/C1QTNF6 axis affected the development and angiogenesis of HNSCC. Subsequently, miR-29c-3p hampered proliferation, migration, invasion and angiogenesis of HNSCC cells by targeting C1QTNF6. MiR-29c-3p/C1QTNF6 axis may work in targeted therapy of HNSCC, and this study will lay a foundation for the development of effective clinical treatment for HNSCC.
Materials and Methods
Bioinformatics Approach
Mature miRNA data (normal samples: 44, tumor samples: 520) and mRNA Counts data (normal samples: 44, tumor samples: 497) of The Cancer Genome Atlas (TCGA)-HNSC were obtained from TCGA database (https://portal.gdc.cancer.gov/). Differentially expressed miRNAs (DEmiRNAs) were screened by differential analysis. The survival analysis was performed using R package “survival” based on the data from TCGA. Besides, differentially expressed mRNAs (DEmRNAs) were screened using “EdgeR” package (|logFC|> 2, adj. pvalue < 0.05). Furthermore, the targets of miR-29c-3p were predicted by miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php), starbase (http://starbase.sysu.edu.cn/) and TargetScan (http://www.targetscan.org/vert_72/) databases. Finally, the identified target gene was subjected to differential expression analysis (T-test detection) and survival analysis.
Cell Culture
Human immortalized keratinocytes cells HaCaT (normal control) and four human HNSCC cell lines (SCC-9, SCC-4, SCC-25 and Detroit 562) were bought from American Type Culture Collection (ATCC). Detroit 562 and HaCaT were incubated in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA). SCC-9 SCC-4, and SCC-25 cells were introduced to the DMEM/F12 (1:1) medium (Gibco, USA). The media contained 10% fetal bovine serum (FBS) (Gibco, USA), streptomycin (100 μg/mL) and penicillin (100 units/mL). All cell lines were kept under conditions of 5% CO2 and 37 °C.
Cell Transfection
miR-29c-3p mimic (miR-mimic), mimic negative control (miR-NC) as well as pcDNA3.1-C1QTNF6 plasmid (oe-C1QTNF6) and pcDNA3.1 plasmid (oe-NC) that encode C1QTNF6 were designed by GenePharma company (Shanghai, China). Lipofectamine 2000 (Invitrogen, USA) was applied to transiently transfect target plasmids with Detroit 562 cell line. Then the cells were held in corresponding media with 5% CO2 at 37 °C for later use. Prior to transfection, cells were maintained in the media for at least 24 h. Also, phosphate buffer saline (PBS, pH 7.4) was employed for cell wash.
Real-Time Fluorescent Quantitative PCR (qRT-PCR)
The total RNA was separated from cells based on the TRIzol kit (Invitrogen, USA), with the RNA concentration evaluated by the NanoDrop 2000 system (Thermo Fisher Scientific, Inc., USA). Next, miRNAs and mRNAs were reversely transcribed into cDNA through the miRNA 1st Strand cDNA Synthesis Kit (Yeasen, China) and PrimeScript RT Master Mix (Takara Bio Inc., Japan), respectively. Afterwards, miScript SYBR Green PCR Kit (Qiagen, Germany) and SYBR ® Premix Ex Taq TM II (Takara Bio Inc., Japan) were used to examine miRNA and mRNA expression, respectively. Furthermore, qRT-PCR was conducted based on the Applied Biosystems ® 7500 Real-Time PCR Systems (Thermo Fisher Scientific, USA). U6 and GAPDH were the internal references. Primer sequences are manifested in Table 1.
Differences in miR-29c-3p and C1QTNF6 mRNAs levels were compared using 2−ΔΔCt value.
MTT Assay
Transfected HNSCC cell line Detroit 562 was digested and suspended, followed by inoculation into 96-well plate (5 × 103 cells/well). After 24, 48, 72 and 96 h, 10 μL MTT kit (5 mg/mL, Sigma-Aldrich, USA) was introduced to each well, followed by cell incubation at 37 °C for 4 h. Subsequently, the supernatant was removed and the precipitation was dissolved in 200 μL dimethyl sulfoxide (DMSO). Finally, the optical density (OD) was measured at 595 nm at present time.
Wound Healing Assay
HNSCC cell line Detroit 562 was seeded into 6-well plates. When the confluency was 80%, the single cell layer was scraped by a sterile pipette tip (200 μL). Thereafter, the separated cells were subjected to two washes. The remaining cells were supplemented with fresh serum-free media and incubated for 24 h. In the end, migrated cells at 0 and 24 h were photographed under a microscope.
Transwell Invasion Assay
Matrigel (BD, USA) was dissolved and diluted using serum-free medium in a ratio of 1:3. Next, it was coated on the upper chamber (50 μL/well) and equilibrated in an incubator for 30 min. Thereafter, cells were added to the upper chamber with a concentration of 1 × 105/mL. DMEM containing 10% FBS was supplemented to the lower chamber. After 48-h incubation, non-invading cells were removed using a cotton swab. Furthermore, invading cells were treated with 0.1% crystal violet staining. The number of invading cells was used as an indicator of invasive ability. Afterwards, 4 random fields under the microscope were selected for counting and photographing the invading cells. Finally, the average number of the cells in four fields was calculated.
Western Blot
The cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, USA) and the total protein concentration was evaluated utilizing bicinchoninic acid (BCA, Sigma-Aldrich, USA). Thereafter, the proteins were isolated using 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by transferring to a polyvinylidene difluoride membrane (Sigma-Aldrich, USA). The membrane was blocked with 5% skimmed milk for 1 h at standardized temperature, followed by the incubation with primary antibodies overnight. The primary antibodies were rabbit anti-C1QTNF6 (1:1000, ab36900, Abcam, UK) and rabbit anti-GAPDH (1:5000, ab16891, Abcam, UK). Afterwards, the membrane was subjected to 3 washes with phosphate-buffered saline + Tween-20 (PBST) buffer, 10 min each. The secondary antibody goat anti-rabbit IgG H&L (ab205718, Abcam, UK) was used for 1 h of incubation at routine temperature. In the end, the membrane was developed using electrochemiluminescence (ECL) reagent (Solarbio, China). Western blot was analyzed with gel imaging software.
Dual-Luciferase Assay
We transfected psiCHECK luciferase reporter plasmids (Sangon Co., China) containing C1QTNF6 wild type (wt) and mutant type (mut) 3′UTR. Afterwards, HNSCC cells Detroit 562 were seeded into 48-well plates for 24-h culture at 37 °C. Thereafter, miR-mimic/mimic NC and C1QTNF6-psiCHECK wt/mut plasmids were co-transfected with Detroit 562 cells. Finally, the luciferase activity was measured using luciferase detection kit (Promega, Fitchburg, WI, USA).
Angiogenesis Assay
The human umbilical vein endothelial cell line (HUVEC) was bought from ATCC and placed in 24-well plates coated with Matrigel (90 μL). After 30-min incubation at 37 °C, different Detroit 562 cell-conditioned media (CM) were introduced to the wells. After incubating the 24-well plates for 6 h, the tube formation was imaged with an inverted microscope at ×200 magnification.
Statistical Analysis
GraphPad Prism 6.0 (GraphPad Software, USA) was applied to analyze all data and each assay was repeated 3 times. The results were displayed as mean ± standard deviation and subjected to t-test for inter-group comparison. P < 0.05 denoted a remarkable difference in statistics.
Results
MiR-29c-3p is Less Expressed in HNSCC Cells
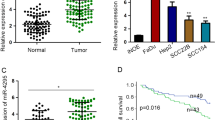
Studies manifested that miR-29c-3p functions in cancers like esophagus cancer and melanoma [24, 25]. Nevertheless, whether miR-29c-3p affects HNSCC progression remains unknown. We therefore used miR-29c-3p as our study object. Firstly, we discovered a notably lower miR-29c-3p expression in HNSCC tissue relative to normal tissue, utilizing differential analysis on the basis of TCGA-HNSC database (Fig. 1A). In addition, the survival analysis demonstrated that the prognosis of patients with lowly expressed miR-29c-3p was poorer than those with the high expression (Fig. 1B). Next, miR-29c-3p level was examined by qRT-PCR in human immortalized keratinocytes HaCaT and four HNSCC cell lines. The result exhibited that it was significantly downregulated HNSCC cell lines, compared with that in HaCaT cell line (Fig. 1C). Collectively, miR-29c-3p expressed lowly in HNSCC and its expression was the lowest in Detroit 562 cell line. Hence, Detroit 562 cell line was picked for subsequent assays.
Low miR-29c-3p level in HNSCC cells. A Box plot about miR-29c-3p expression in normal (blue) and tumor (red) tissue. B Survival curve about C1QTNF6 affecting patients’ prognosis. Abscissa represents time (unit: year) while ordinate represents survival rate. Expression is high in red and low in blue. C miR-29c-3p level in human immortalized keratinocytes HaCaT and HNSCC cell lines SCC-4, SCC-9, SCC-25 and Detroit 562. *Indicates P < 0.05
Overexpressed miR-29c-3p Inhibits the Progression of HNSCC Cells
To figure out how miR-29c-3p affected HNSCC cells, we constructed Detroit 562 cells with mimic NC and miR-mimic groups. Firstly, qRT-PCR result indicated that miR-29c-3p was notably upregulated in overexpressing group (Fig. 2A). The transfected cells therefore were used for the follow-up assays. Afterwards, through MTT assay, overexpressed miR-29c-3p markedly hampered the proliferation of Detroit 562 cells (Fig. 2B). Next, based on transwell assay, overexpressed miR-29c-3p caused a decrease in invasive ability of the cells (Fig. 2C). Furthermore, wound healing assay suggested that overexpressed miR-29c-3p strikingly weakened migratory ability of the cells (Fig. 2D). Finally, the tube formation of HUVEC cells was markedly decreased after the treatment of CM with Detroit562 cells transfected with miR-mimic (Fig. 2E). All in all, overexpressed miR-29c-3p repressed the proliferation, migration, invasion and angiogenesis of HNSCC cells.
Overexpressing miR-29c-3p restrains progression of HNSCC cells. A MiR-29c-3p expression in HNSCC cells Detroit 562. B The proliferative capability of Detroit 562 cells in different groups (miR-NC and miR-mimic). C The invasion of Detroit 562 cells (×100). D The migratory ability of Detroit 562 cells (×40). E HUVEC cells were cultured in CM from miR-mimic transfected Detroit562 and tube formation ability was assayed. *Suggests P < 0.05
MiR-29c-3p Inhibits C1QTNF6 in HNSCC
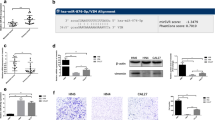
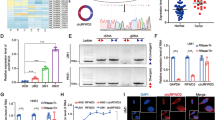
To investigate the downstream gene of miR-29c-3p in HNSCC, we carried out differential expression analysis on mRNA data and obtained 1,567 differentially expressed mRNAs, including 569 upregulated genes (Fig. 3A). Thereafter, miRTarBase, starbase and TargetScan databases were utilized to predict downstream genes of miR-29c-3p, which were then overlapped with upregulated mRNAs to acquire 11 target mRNAs (Fig. 3B). The correlation analysis revealed the strongest negative association between C1QTNF6 and miR-29c-3p (Fig. 3C). Besides, expression analysis indicated a remarkably high expression of C1QTNF6 in HNSCC tissue (Fig. 3D) and in patients with poor prognosis (Fig. 3E). To validate that miR-29c-3p regulated C1QTNF6, the binding site of miR-29c-3p and C1QTNF6 3′UTR was predicted through starBase database (Fig. 3F). Next, dual-luciferase assay verified this targeted relationship, manifesting that upregulating miR-29c-3p suppressed the luciferase activity of wt-C1QTNF6 3′UTR but had not impact on that of mut-C1QTNF6 3′UTR (Fig. 3G). Afterwards, qRT-PCR checked the C1QTNF6 mRNA level in Detroit 562 cell line. The outcome indicated a significant reduction in C1QTNF6 mRNA expression in miR-mimic group (Fig. 3H). Western blot assay also showed that overexpressed miR-29c-3p caused a remarkable restraint in C1QTNF6 protein level (Fig. 3I). These facts proved that miR-29c-3p hindered C1QTNF6 level in HNSCC cells.
MiR-29c-3p suppresses C1QTNF6 in HNSCC cells. A Volcano plot about results of differential expression analysis on mRNA-seq data in TCGA-HNSCC. High level in red and low level in green. B Venn plot of the predicted targets of miR-29c-3p and upregulated mRNAs. C Heat plot about the correlation between C1QTNF6 and miR-29c-3p. D Box plot about C1QTNF6 expression in normal group (blue) and tumor group (red). E Survival curve about effect of C1QTNF6 expression on patients’ prognosis. Abscissa represents time (unit: year) and ordinate represents survival curve. Red curve stands for high expression while blue curve stands for low expression. F Binding site of miR-29c-3p and C1QTNF6. G Luciferase activity of Detroit 562 cells in different transfection groups. H, I C1QTNF6 mRNA and protein levels in different transfection groups. *Denotes P < 0.05
MiR-29c-3p Targets C1QTNF6 to Suppress Progression of HNSCC
To prove how miR-29c-3p and C1QTNF6 interact in HNSCC, we established groups including miR-NC, oe-C1QTNF6 and miR-mimic + oe-C1QTNF6. Next, qRT-PCR and western blot assays ensured that mRNA and protein expression of C1QTNF6 was dramatically increased in oe-C1QTNF6 group, but suppressed in miR-mimic + oe-C1QTNF6 group (Fig. 4A and B). In addition, MTT assay showed that overexpressed C1QTNF6 strengthened the proliferative ability of HNSCC cells while simultaneously overexpressing C1QTNF6 and miR-29c-3p diminished such effect (Fig. 4C). Moreover, transwell and wound healing assays suggested that overexpressed C1QTNF6 also enhanced the migration invasion of HNSCC cells, whereas simultaneous overexpression of miR-29c-3p and C1QTNF6 weakened such effect (Fig. 4D and E). Finally, Detroit562 cells transfected with oe-C1QTNF6 promoted angiogenesis of HUVEC, but overexpressed miR-29c-3p and C1QTNF6 reversed angiogenesis ability of HUVEC (Fig. 4F). Hence, it could be concluded that miR-29c-3p downregulated C1QTNF6 expression to block the motor ability and angiogenesis of HNSCC cells.
MiR-29c-3p suppress HNSCC cell progression by targeting C1QTNF6. A C1QTNF6 mRNA expression in Detroit 562 cells in different groups (miR-NC + oe-NC, miR-NC + C1QTNF6 and miR-mimic + oe-C1QTNF6). B C1QTNF6 protein level in Detroit 526 cells. C Proliferative ability of Detroit 562 cells. D Invasive ability of Detroit 526 cells (×100). E Migratory ability of Detroit 562 cells (×40). F The tube formation ability of HUVEC in CM of Detroit562 cells. *Represents P < 0.05
Discussion
Sustained angiogenesis plays an imperative role in tumor growth and metastasis [26]. Hence, determining promising anti-angiogenic targets is an approach to the cancer treatment. Many studies have identified that many miRNAs were dysregulated in HNSCC cells and influenced the progression of this cancer [10, 27]. Finding aberrantly expressed miRNAs in HNSCC cells therefore was of great significance to investigate the angiogenesis mechanism of HNSCC.
To find out miRNA that plays significant role in HNSCC, we found through bioinformatics analysis that miR-29c-3p level was prominently low in cancer tissue, which was also related to patients’ poor prognosis. Previous studies clarified marked downregulation of miR-29c-3p in laryngeal squamous cell carcinoma and its low expression correlates with poor prognosis [28]. Furthermore, miR-29c-3p also obstructs angiogenesis of colorectal cancer cell by targeting VEGFA [18]. Similarly, we conducted cellular function assays and revealed that miR-29c-3p contained motor ability and angiogenesis of HNSCC cells.
To identify the downstream gene of miR-29c-3p, we found via bioinformatics methods that C1QTNF6 was targeted by miR-29c-3p. C1QTNF6 level was notably high in HNSCC tissue, which was related to poor prognosis. Thereafter, dual-luciferase, qRT-PCR and western blot assays proved that miR-29c-3p regulated C1QTNF6 expression. Besides, C1QTNF6 is prominently highly expressed in lung adenocarcinoma, which resulted in patients’ poor prognosis [29]. In hepatocellular cancer tissue, C1QTNF6 is significantly highly expressed [30, 31]. Knockdown of C1QTNF6 suppresses the progression but stimulates apoptosis of hepatocellular cancer cells by blocking AKT signaling pathway [30, 31]. These findings showed that C1QTNF6 plays pivotal roles in the progression of cancers. In our study, MTT, transwell, wound healing, angiogenesis and western blot assays verified the function of C1QTNF6 on HNSCC. These assays revealed that overexpressing C1QTNF6 remarkably promoted the proliferation, motor ability and angiogenesis of HNSCC cells. Most importantly, simultaneously overexpressing C1QTNF6 and miR-29c-3p reversed that promoted effect. Taken together, miR-29c-3p targeted C1QTNF6 expression to hinder progression of HNSCC cells. Additionally, miR-29c-3p/C1QTNF6 axis possibly regulated the cell progression as well as angiogenesis in HNSCC.
In conclusion, miR-29c-3p was less expressed in HNSCC and acted as an inhibitor. A negative association was presented between C1QTNF6 and miR-29c-3p. Besides, miR-29c-3p suppressed motor ability and angiogenesis of HNSCC cells via targeting C1QTNF6. Nevertheless, we did not conduct in vivo assays. It will be a focus in our future studies. In sum, miR-29c-3p had the potential to function in HNSCC targeted therapy. MiR-29c-3p/C1QTNF6 axis may also modulate the metastasis of HNSCC.
References
McDermott, J. D., & Bowles, D. W. (2019). Epidemiology of head and neck squamous cell carcinomas: Impact on staging and prevention strategies. Current Treatment Options in Oncology, 20, 43. https://doi.org/10.1007/s11864-019-0650-5
Gillison, M. L., et al. (2008). Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. Journal of the National Cancer Institute, 100, 407–420. https://doi.org/10.1093/jnci/djn025
Rettig, E. M., & D’Souza, G. (2015). Epidemiology of head and neck cancer. Surgical Oncology Clinics of North America, 24, 379–396. https://doi.org/10.1016/j.soc.2015.03.001
Takes, R. P., et al. (2012). Distant metastases from head and neck squamous cell carcinoma Part I. Basic aspects. Oral Oncology, 48, 775–779. https://doi.org/10.1016/j.oraloncology.2012.03.013
Sinha, P., et al. (2014). Distant metastasis in p16-positive oropharyngeal squamous cell carcinoma: A critical analysis of patterns and outcomes. Oral Oncology, 50, 45–51. https://doi.org/10.1016/j.oraloncology.2013.10.007
Bauer, E., et al. (2020). Extranodal extension is a strong prognosticator in HPV-positive oropharyngeal squamous cell carcinoma. The Laryngoscope, 130, 939–945. https://doi.org/10.1002/lary.28059
Sato, S., et al. (2019). EPHB2 carried on small extracellular vesicles induces tumor angiogenesis via activation of ephrin reverse signaling. JCI Insight. https://doi.org/10.1172/jci.insight.132447
Carmeliet, P., & Jain, R. K. (2000). Angiogenesis in cancer and other diseases. Nature, 407, 249–257. https://doi.org/10.1038/35025220
Bagnasco, L., et al. (2012). Role of angiogenesis inhibitors in colorectal cancer: Sensitive and insensitive tumors. Current Cancer Drug Targets, 12, 303–315. https://doi.org/10.2174/156800912800190929
Irimie-Aghiorghiesei, A. I., et al. (2019). Prognostic value of MiR-21: An updated meta-analysis in head and neck squamous cell carcinoma (HNSCC). Journal of Clinical Medicine. https://doi.org/10.3390/jcm8122041
Wang, Y., et al. (2017). MiR-34a suppresses HNSCC growth through modulating cell cycle arrest and senescence. Neoplasma, 64, 543–553. https://doi.org/10.4149/neo_2017_408
Vahabi, M., et al. (2019). miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. Journal of Experimental & Clinical Cancer Research, 38, 141. https://doi.org/10.1186/s13046-019-1119-x
Kolenda, T., Guglas, K., Teresiak, A., Blizniak, R., & Lamperska, K. (2019). Low let-7d and high miR-205 expression levels positively influence HNSCC patient outcome. Journal of Biomedical Science, 26, 17. https://doi.org/10.1186/s12929-019-0511-3
Hayes, J., Peruzzi, P. P., & Lawler, S. (2014). MicroRNAs in cancer: Biomarkers, functions and therapy. Trends in Molecular Medicine, 20, 460–469. https://doi.org/10.1016/j.molmed.2014.06.005
Garzon, R., Marcucci, G., & Croce, C. M. (2010). Targeting microRNAs in cancer: Rationale, strategies and challenges. Nature Reviews. Drug Discovery, 9, 775–789. https://doi.org/10.1038/nrd3179
Kinoshita, T., et al. (2013). Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. British Journal of Cancer, 109, 2636–2645. https://doi.org/10.1038/bjc.2013.607
Li, Y., et al. (2020). TUG1 enhances high glucose-impaired endothelial progenitor cell function via miR-29c-3p/PDGF-BB/Wnt signaling. Stem Cell Research & Therapy, 11, 441. https://doi.org/10.1186/s13287-020-01958-3
Chen, C., et al. (2020). The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. Journal of Experimental & Clinical Cancer Research, 39, 91. https://doi.org/10.1186/s13046-020-01594-y
Lee, W., Kim, M. J., Park, E. J., Choi, Y. J., & Park, S. Y. (2010). C1qTNF-related protein-6 mediates fatty acid oxidation via the activation of the AMP-activated protein kinase. FEBS Letters, 584, 968–972. https://doi.org/10.1016/j.febslet.2010.01.040
Wong, G. W., et al. (2008). Molecular, biochemical and functional characterizations of C1q/TNF family members: Adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. The Biochemical Journal, 416, 161–177. https://doi.org/10.1042/BJ20081240
Murayama, M. A., et al. (2015). CTRP6 is an endogenous complement regulator that can effectively treat induced arthritis. Nature Communications, 6, 8483. https://doi.org/10.1038/ncomms9483
Qu, H. X., et al. (2019). C1QTNF6 is overexpressed in gastric carcinoma and contributes to the proliferation and migration of gastric carcinoma cells. International Journal of Molecular Medicine, 43, 621–629. https://doi.org/10.3892/ijmm.2018.3978
Wang, L., Liu, Z., Duan, L., Ma, B., & Sun, Z. (2015). C1q tumor necrosis factor-related protein 6 (CTRP6) inhibits the proliferation and migration of ovarian cancer cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi, 31, 1664–1668.
Wang, H., et al. (2020). MiR-29c-3p suppresses the migration, invasion and cell cycle in esophageal carcinoma via CCNA2/p53 axis. Frontiers in Bioengineering and Biotechnology, 8, 75. https://doi.org/10.3389/fbioe.2020.00075
Wang, Y., Liu, G., Ren, L., Wang, K., & Liu, A. (2019). Long non-coding RNA TUG1 recruits miR29c3p from its target gene RGS1 to promote proliferation and metastasis of melanoma cells. International Journal of Oncology, 54, 1317–1326. https://doi.org/10.3892/ijo.2019.4699
Carmeliet, P., & Jain, R. K. (2011). Molecular mechanisms and clinical applications of angiogenesis. Nature, 473, 298–307. https://doi.org/10.1038/nature10144
Guo, L. M., et al. (2018). MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biology & Therapy, 19, 973–983. https://doi.org/10.1080/15384047.2018.1450112
Fang, R., Huang, Y., Xie, J., Zhang, J., & Ji, X. (2019). Downregulation of miR-29c-3p is associated with a poor prognosis in patients with laryngeal squamous cell carcinoma. Diagnostic Pathology, 14, 109. https://doi.org/10.1186/s13000-019-0893-2
Han, M., Wang, B., Zhu, M., & Zhang, Y. (2019). C1QTNF6 as a novel biomarker regulates cellular behaviors in A549 cells and exacerbates the outcome of lung adenocarcinoma patients. In Vitro Cellular & Developmental Biology Animal, 55, 614–621. https://doi.org/10.1007/s11626-019-00377-w
Takeuchi, T., Adachi, Y., & Nagayama, T. (2011). Expression of a secretory protein C1qTNF6, a C1qTNF family member, in hepatocellular carcinoma. Analytical Cellular Pathology (Amsterdam), 34, 113–121. https://doi.org/10.3233/ACP-2011-009
Wan, X., Zheng, C., & Dong, L. (2019). Inhibition of CTRP6 prevented survival and migration in hepatocellular carcinoma through inactivating the AKT signaling pathway. Journal of Cellular Biochemistry, 120, 17059–17066. https://doi.org/10.1002/jcb.28967
Funding
This study is supported by Scientific Research Project of Hunan Provincial Health Commission (No. 20200216).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, F., Qi, H. MiR-29c-3p/C1QTNF6 Restrains the Angiogenesis and Cell Proliferation, Migration and Invasion in Head and Neck Squamous Cell Carcinoma. Mol Biotechnol 65, 913–921 (2023). https://doi.org/10.1007/s12033-022-00591-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-022-00591-0