Abstract
Escherichia coli and Klebsiella pneumoniae are important members of the Enterobacteriaceae family, involved in many infections. The increased resistance rate towards β-lactams and fluoroquinolones -which are the main therapeutic options- limits their treatment options. This study aimed to assess the local resistance patterns against different antimicrobials and to determine the extended-spectrum β-lactamase (ESBLs) producers. The study revealed that 36% of clinical isolates were ESBL producers, showing high resistance rates towards β-lactams and non-β-lactams, especially sulphamethoxazole-trimethoprim and fluoroquinolones. However, they were susceptible to chloramphenicol and doxycycline (33% and 20%; respectively). Also, the investigation aimed to screen the plasmid profile of quinolone-resistant ESBLs-producers and to detect the plasmid-mediated quinolone resistance genes including qnrA, qnrS, qnrB, qnrC, qnrD, and qnrVC. Moreover, the conjugative plasmid among the quinolone-resistant isolates was elucidated. The results showed that extracted plasmids of sizes ranging from ≈0.9 to 21.23 Kb, divided into 7 plasmid patterns were detected. A plasmid of approximately 21.23 Kb was found in all isolates and the QnrS gene was the most predominant gene. Moreover, the frequency of transconjugation within the same genus was higher than that recorded between different genera; where 68% of E. coli isolates transferred the resistance genes compared to Klebsiella isolates (36.6%). Plasmid profiles of transconjugants demonstrated great similarity, where 21.23 Kb plasmid was detected in all transconjugants. Since these transconjugants were quinolone-resistant ESBL producers, it has been suggested that quinolone resistance determinants might be carried on that plasmid.
Similar content being viewed by others
Introduction
Escherichia coli and Klebsiella pneumoniae are among the most medically important members of the family Enterobacteriaceae [1]. These bacteria cause a great number of infections including urinary tract infections, gastroenteritis, pneumonia, septicemia, and meningitis. Some of these diseases are associated with high mortality rates if not treated properly, so it is important to combat them with highly effective antibiotics [2]. The most commonly used antimicrobial agents for treating such infections are β-lactams including penicillins, cephalosporins, and other non-β-lactams such as aminoglycosides and quinolones [3]. Extensive use of antimicrobials and disinfectants has promoted the rapid development of bacterial resistance that become a global health problem, especially in developing countries [4].
Production of the extended-spectrum β-lactamases (ESBLs) has emerged as an important mechanism for β-lactams resistance, as they are capable of hydrolyzing broad spectrum β-lactams including third- and fourth-generation cephalosporins. However, these ESBLs can be inhibited by β-lactamase inhibitors like clavulanic acid, sulbactam, and tazobactam [5]. On the other hand, quinolone resistance has been associated with point mutations which result in single amino acid substitutions of DNA gyrase. In addition, the resistance was related to decreased accumulation inside the bacteria because of the impermeability of the membrane and/or overexpression of efflux pump systems. Previously, those mechanisms were known to be chromosomally mediated [6].
Moreover, resistance to quinolones can be mediated by plasmids that produce qnr protein which protects the quinolone target from inhibition [7]. Plasmid-mediated qnr genes have been widely associated with other relevant determinants of resistance in multi-resistance plasmids. The first plasmid-mediated quinolone resistance (PMQR) identified gene was qnrA [8].
This study aims to evaluate the resistance rates towards different classes of antimicrobials and to determine the rate of ESBL production among Escherichia coli and Klebsiella pneumoniae clinical isolates. Also, the study was designed to detect the six qnr families (including qnrA, qnrS, qnrB, qnrC, qnrD, and qnrVC) described among ESBLs producing K. pneumoniae and E. coli clinical isolates. A conjugation experiment was performed to test the possibility of transferring the plasmid-carrying quinolone resistance determinants in ESBLs-producing isolates as it contributes to the increased spread of antibiotic resistance in hospitalized patients.
Materials and methods
Isolation and identification of clinical isolates
A total of one hundred and fifty-two isolates were recovered from patients admitted to Mansoura University Hospitals from wounds, blood, urine, ventilator tubes, pus, and sputum. They were identified as 93 K. pneumoniae isolates (61%) and 59 E. coli isolates (39%) by standard microbiological methods.
Standard E. coli JM105 strain was obtained from Pharmacia (Rockville, USA) to be used in plasmid transconjugation experiments. For long-term storage, all isolates were preserved in tryptone soya broth (Oxoid, Hampshire, UK) with 20% glycerol and stored at −20 °C until use.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the standard disk diffusion method according to Clinical and Laboratory Standards Institute guidelines (CLSI, 2017). The tested antimicrobials belonged to different classes like β-lactams, aminoglycosides, fluoroquinolones, phenols, folate pathway inhibitors, and tetracyclines.
Antimicrobial disks were obtained from Oxoid (Hampshire, UK) including amoxicillin/clavulanate (AMC, 20/10 µg), cefotaxime (CTX, 30 µg), ceftriaxone (CRO, 30 µg), ceftazidime (CAZ, 30 µg), aztreonam (ATM, 30 µg), cefoxitin (FOX, 30 µg), cefuroxime (CXM, 30 µg), cefepime (FEB, 30 µg), gentamycin (CN, 10 µg), ciprofloxacin (CIPRO, 5 µg), levofloxacin (LEVO, 5 µg), norfloxacin (NOR, 5 µg), ofloxacin (OFX, 5 µg), chloramphenicol (C, 30 µg), sulphamethoxazole-trimethoprim (SXT, 23.27/1.25 µg), and doxycycline (DO, 30 µg).
Phenotypic detection of ESBL production
Clinical isolates that showed an inhibition zone less than 25 mm for ceftriaxone, 17 mm for cefpodoxime, and/or 27 mm for aztreonam were considered as potential ESBLs-producing isolates (CLSI, 2017).
ESBL production was confirmed by a double disk synergy test (DDST20) as described by Garrec et al. [9]. Briefly, disks representing third-generation cephalosporins including ceftriaxone (CRO, 30 μg), ceftazidime (CAZ, 30 μg), and cefotaxime (CTX, 30 μg) along with the fourth-generation cefepime (FEB, 30 μg) were placed at a distance of 20 mm from amoxicillin-clavulanate disk (AMC, 20/10 μg) which was placed in the center of MHA plate inoculated with the tested isolate. Plates were incubated at 37 °C for 16–18 h. Any enhancement or distortion of the inhibition zone of any antibiotic disk towards amoxicillin-clavulanic acid indicated ESBL production.
Plasmid profiling of ESBLs-producing K. pneumoniae and E. coli clinical isolates
ESBLs producing K. pneumoniae and E. coli isolates, selected by antimicrobial sensitivity test were subjected to plasmid extraction using Zyppy™ Plasmid Miniprep kit according to the manufacturer’s instructions and then analyzed by agarose gel electrophoresis according to Sambrook et al. [10]. The approximate sizes of the large plasmids were estimated by comparison to the plasmid marker (GeneRuler 1 kb Plus DNA ladder, ThermoFisher, USA).
Detection of plasmid-mediated qnr gene families in ESBLs-producing K. pneumoniae and E. coli clinical isolates
Genes encoding for quinolone resistance were detected by PCR using oligonucleotide primers listed in Table 1. According to White and colleagues, [11], each PCR mixture contained 15 μl of PCR master mix (New England Biolabs, USA), 2 μl of forward primer, 2 μl of reverse primer, 6 μl of template DNA, and nuclease-free water to 30 μl. PCR started by heating at 95 °C for 10 minutes, then running 35 cycles of 95 °C for 30 s, and 56 °C for 45 s for all genes except for qnrC and qnrVc, the annealing temperature was 56.5 °C and 58 °C; respectively. Finally, the temperature was raised to 72 °C for 45 s. The PCR products were separated on 1.5% agarose gel, stained with ethidium bromide, and visualized by a UV trans-illuminator.
Conjugative transfer of plasmids harboring quinolone resistance determinants in ESBL-producers by liquid mating technique
Liquid mating technique was used for studying inter- and intra-generic spread of quinolone resistance determinants in ESBL-producers among the Enterobacteriaceae family according to Ozgumus et al. [12].
E. coli and K. pneumoniae ESBL-producing isolates were considered donor cells. While, E. coli JM105 standard strain was used as a recipient, after adapting to rifampicin antibiotic by growing on MHA plates containing serial concentrations of rifampicin.
A single colony of each isolate was separately inoculated in brain heart infusion (BHI) broth for 24 h at 37 °C. Then, one milliliter of each overnight culture was added to 100 mL of fresh BHI broth, and the flasks were incubated at 37 °C with shaking until reaching cell densities of 1 × 108 cells/ml for donors (ESBLs-producers) and 0.5 × 108 cells/ml for the recipient (E. coli JM105). Equal volumes from each donor isolate were mixed with the recipient standard strain (1 ml each) and incubated for 16 h at 37 °C without agitation in a 25-ml Erlenmeyer flask to provide a large surface area for better aeration.
Aliquots of 0.1 ml of the mating mixtures were plated into MH agar containing 100 μg ml−1 of rifampicin and 5 μg ml−1 of ciprofloxacin. The plates were incubated overnight, and the resulting colonies of trans-conjugants were subjected to plasmid extraction and the frequency of transfer was expressed relative to the number of donor cells.
Detection of quinolone resistance genes in transconjugants
Plasmid extraction for transconjugants was performed according to the manufacturer’s instructions supplied with the Zyppy™ Plasmid Miniprep kit, and then plasmid-coded quinolone resistance genes were amplified and detected by PCR as mentioned before.
Results
Antimicrobial susceptibility testing
K. pneumoniae and E. coli isolates exhibited different susceptibility rates towards the tested antimicrobials. As shown in Table 2, most K. pneumoniae isolates were highly resistant to β-lactams ranging from 93.5% to 65.6%, also Escherichia coli isolates exhibited high resistance rates ranging from 94.9% to 44%.
In comparison to β-lactams, K. pneumoniae and E. coli isolates exhibited a lesser resistance rate to quinolones Table 2. The lowest resistance rate was recorded with chloramphenicol (33.3%, and 34%) and doxycycline (17.2%, and 23.7%) for K. pneumoniae and E. coli isolates; respectively.
Double disk synergy test (DDST20)
From the initial antimicrobial susceptibility testing Table 2, seventy-one isolates (31 E. coli and 40 K. pneumoniae) exhibited reduced susceptibility to β-lactams including extended-spectrum cephalosporins, and were considered to be potential ESBL-producers according to CLSI guidelines.
All suspected isolates were subjected to a confirmation by DDST20 as shown in Fig. 1. The results revealed that only 55 isolates exhibited synergy between the amoxicillin/ clavulanic acid disk and the other disks representing extended-spectrum cephalosporins with an overall rate of 36% of the total number of isolates.
Seventy-five percent of K. pneumoniae isolates (30 isolates out of 40) were confirmed to be ESBLs-producers with a percentage of 32% of total K. pneumoniae isolates. Eighty percent of E. coli isolates (25 isolates out of 31) were confirmed to be ESBLs-producers with a percentage of 42% of total E. coli isolates.
Plasmid profiles of ESBLs-producing K. pneumoniae and E. coli isolates
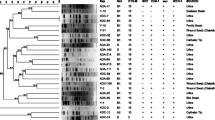
K. pneumoniae and E. coli isolates showed 7 different plasmid patterns. As shown in Table 3, the approximate sizes of plasmid DNA of ESBLs-producing K. pneumoniae isolates ranged from ≈1.3 to 21.23 kb, while for E. coli isolates the sizes ranged from ≈0.9 to 21.23 kb. A plasmid of ≈21.23 Kb was harbored by all K. pneumoniae and E. coli isolates.
As shown in Figs. 2 and 3, all isolates shared one common plasmid with a molecular size of approximately 21.23 Kb. Regarding K. pneumoniae, 20 out of 40 isolates (50%) harbored a single plasmid only (≈21.23 Kb). Eighteen isolates 18/40 (45%) harbored two plasmids, while two isolates harbored 3 plasmids with different molecular sizes, including the ≈21.23 Kb plasmid. E. coli plasmid extracts showed that 19 out of 31 isolates (61.3%) harbored the plasmid of ≈21.23 Kb only. Eight isolates 8/31 (25.8%) harbored 2 plasmids, with the common plasmid of ≈21.23 Kb, and four isolates harbored 4 different plasmids (12.9%).
Gel electrophoresis of plasmid DNA extracted from some representative ESBLs-producing K. pneumoniae isolates using Zyppy™ Plasmid Miniprep kit. The Plasmid DNA marker (GeneRuler 1 kb Plus DNA ladder) was run in lane 1. Bp: Base pair M: Marker 15 K: [15] represents the number of isolate, and (K) represents the type of isolate (K. pneumoniae)
Gel electrophoresis of plasmid DNA extracted from some representative ESBLs-producing E. coli isolates using Zyppy™ Plasmid Miniprep kit. The Plasmid DNA marker (GeneRuler 1 kb Plus DNA ladder) was run in lane 1. Bp: Base pair M: Marker 1E: [1] represents the number of isolate, and (E) represents the type of isolate (E. coli)
Detection of plasmid-mediated qnr gene families in ESBL-producing K. pneumoniae and E. coli clinical isolates
The data represented in Fig. 4 revealed that the qnrS gene (428 bp) was the most prominent as it was detected in the PCR products of 38/40 quinolone-resistant K. pneumoniae isolates (95%) and 27/31 quinolone-resistant E. coli isolates (87%). While the qnrC gene (135 bp) was determined in 35 K. pneumoniae isolates (87.5%) and the qnrD gene (691 bp) was determined in 34 isolates (85%). Both qnrC and qnrD genes were found in 24/31 quinolone-resistant E. coli isolates (77.4%). On the other hand, the least detected genes were qnrA (580 bp) and qnrB (264 bp). The qnrB gene was determined in 70% and 64.5% of K. pneumoniae and E. coli isolates; respectively, Fig. 5.
Conjugative transfer of plasmids harboring quinolone resistance genes in ESBL-producers
The results revealed that transconjugation within the same genus (intrageneric) was more likely to happen than in different genera (intergeneric). As among the tested ESBL-producing E. coli isolates, conjugation was performed successfully in 17 (68%) out of 25 isolates. While the trans-conjugation transfer occurred successfully in 11 (36.6%) out of 30 K. pneumoniae isolates.
The results of mating experiments demonstrated in Table 4 revealed that the frequency of plasmids transfer in ESBLs-producing E. coli ranged from 9.9 × 10−8 to 1.6 × 10−5 /donor, while the frequency of plasmids transfer from ESBLs-producing K. pneumoniae isolates ranged from 3.3 × 10−8 to 1.7 × 10−6/donor.
Plasmid profiles of transconjugants from either K. pneumoniae or E. coli isolates demonstrated great similarity. These transconjugants were found to show plasmids ranging from 2.9 Kb to 21.23 Kb as shown in Fig. 6. The plasmid of approximate size 21.23 Kb was detected in all E. coli and K. pneumoniae transconjugants. Since all transconjugants were quinolone-resistant and extended-spectrum β-lactamase producers, it had been suggested that quinolone resistance determinants might be carried on this plasmid.
a Gel electrophoresis of plasmid DNA extracted from some representative E.coli transconjugants. b Gel electrophoresis of plasmid DNA extracted from some representative K. pneumoniae transconjugants. The Plasmid DNA marker (GeneRuler 1 kb Plus DNA ladder) was run in lane 1 and plasmid DNA extracted from the Standard E. coli JM105 strain was run in lane 2. Bp Base pair, M Marker, T 10 Transconjugant nu. 10
Screening for plasmid-mediated quinolone resistance (PMQR) determinants in transconjugants of K. pneumoniae and E. coli clinical isolates
The results revealed that qnrS and qnrD genes were transferred in all transconjugants. QnrB gene was transferred by 81.8% as it was detected in 18 transconjugants out of 22 donors which were originally confirmed to harbor the qnrB gene. In addition, the qnrC gene was transferred by 76.9% (20/26 donors). On the other hand, qnrVc and qnrA genes were the least detected in the plasmid extract of transconjugants where qnrVc was transferred by 56% and qnrA by 40% only.
Discussion
Escherichia coli and K. pneumoniae are the most frequent bacteria involved in human infections. Several antibiotic classes such as β-lactams, sulphamethoxazole-trimethoprim, and fluoroquinolones are considered appropriate choices for the treatment of such infections. Unfortunately, the increased local resistance rate towards these antibiotics especially β-lactams and fluoro-quinolones limits their role in the treatment [13].
Our result showed a high resistance rate towards β-lactams including extended-spectrum cephalosporins ranging from (64.4% to 94.9%) for E. coli and K. pneumoniae clinical isolates. These results came in agreement with a previous study performed in Egypt, Kadry et al. [14] reported considerable resistance rates toward third-generation cephalosporins like cefoperazone (52.9%) and ceftazidime (43.5%), followed by the fourth generation cefepime (32.1%). In the current study, 36% of isolates were confirmed to be ESBL-producers by DDST20, which was parallel to the percentage reported by Gad [15]. Another study showed that the percentage of ESBL-producing isolates reached 50% of the total isolates [16], while Morsi and Tash [17] stated that ESBL production was detected in 54.6% of isolates confirming the increased rate of ESBL producers in Egypt.
On the other hand, our research revealed that ESBL-producing isolates were susceptible to other antibiotics than β-lactams, so they could offer a good treatment option for such MDR isolates, where less resistance rate was recorded with chloramphenicol (33.3%, 34%), and doxycycline (17.2%, 23.7%) for K. pneumonia and E. coli isolates; respectively. This was confirmed by Cunha et al. [18], who reported a case of persistent ESBL-producing Escherichia coli chronic prostatitis refractory to antibiotic therapy but with treatment by a combination of fosfomycin and doxycycline patient was rapidly cured. Another study demonstrated that the combination of polymyxin B and chloramphenicol managed to enhance bacterial killing and suppressed the emergence of resistance when used against MDR K. pneumoniae [19].
Plasmid profiling is a useful epidemiologic and typing tool that correlates with antimicrobial resistance patterns. Our results revealed that quinolone-resistant ESBL producers of K. pneumoniae showed 7 plasmid patterns ranging from ≈1.3 to 21.23 Kb and the plasmid of ≈21.23 Kb was harbored by all isolates. A comparable result by Aladag and Durak [20], found that the molecular sizes of plasmids extracted from ESBLs of Klebsiella isolates ranged between 1.6 and 30.1 Kb. Some of the isolates had more than one plasmid and revealed 8 plasmid profiles. The plasmid with 19.3 Kb size was the most common in the tested strains and was capable of transferring by conjugation [20].
The prevalence of 21.23 Kb plasmid in the tested strains from different infections indicated a worldwide problem. In a study performed by Daini and Adesemowo [21], they found that plasmids’ patterns of E. coli isolates ranged between 0.12 kb to 23.13 kb and grouped into 7 plasmid profiles. The resistant strains carried a common R–plasmid of 23.13 kb [12, 22]. Many researchers detected plasmids of 21 Kb in ESBL producers Klebsiella pneumoniae strains that were resistant to a wide range of antibiotics and can be transferred among members of the Enterobacteriaceae family by conjugation [20, 23].
The role of plasmid in antibiotic resistance was examined through conjugation, and we found that among the tested ESBL-producing E. coli isolates, conjugation was performed successfully in 17 out of 25 isolates with percentage of 68%, while the conjugation transfer occurred only in 36.6% with frequencies between 9.9 × 10−8 to 1.65 × 10−5/donor for E. coli and 3.3 × 10−8 to 1.78 × 10−6/donor for K. pneumoniae isolates. So, we proved that transconjugation within the same genus (intrageneric) was more likely to happen than in different genera (intergeneric).
Conjugation results were comparable to that reported by Gangoué-Piéboji et al. [24] who found that 38.7% of ESBL-producing K. pneumoniae were able to transfer the ESBL genes to E. coli HK 225 and the transfer frequencies were between 9 × 10−8 and 6.7 × 10−4 per donor. In addition, Ozgumus et al. [12], reported that ESBL-producing strains including E. coli, K. pneumoniae, and Enterobacter aerogenes were able to transfer their ESBL genes to a recipient E. coli J53-2 by conjugation at a transfer frequency of 5 × 10−8 to 10−4/donor.
Our results revealed that a plasmid with an approximate molecular size of 21.23 Kb was detected in all transconjugants that were confirmed to exhibit co-resistance against extended-spectrum β-lactams as well as quinolones. As a consequence, we suggested that ESBL genes and qnr determinants are probably carried on this plasmid. Our suggestion agreed with Gangoué-Piéboji et al. [24], who found that the ESBL-producers usually carry a multi-resistant plasmid having genes conferring resistance to β-lactam and non-β-lactam antibiotics. Also, Yhiler et al. [25] stated that the PMQR gene was co-carried with the ESBL gene in the plasmid.
Our PCR data revealed that the qnrS gene was the most prominent, while the qnrA gene was detected in the PCR products of 72.5% and 74% of quinolone-resistant ESBL-producing K. pneumoniae and E. coli isolates; respectively. Moreover, qnrB was found in 70% and 64.5% of isolates; respectively, which agreed with Yhiler et al. [25], who confirmed that the qnrB gene was detected by a percentage of 62.4%. Yang et al. [26] found that the qnr genes were detected in 42% of E. coli and 65.5% of K. pneumoniae quinolone-resistant clinical isolates. QnrB gene was present in 25.4% of isolates, while qnrS and qnrA were found in 12.2% and 8.6% of isolates, respectively. Valadbeigi et al. [27] confirmed the presence of qnrS and qnrB in 47.5% and 2.5% of clinical isolates, respectively.
QnrVc gene was detected in 77.5% of isolates and this agreed with that reported by Poirel et al. [28], who confirmed the prevalence of qnrVc in 70% of isolates. This study confirmed the presence of the qnrC gene in 87.5% of K. pneumoniae isolates and 77.4% of E. coli, while the qnrD gene was determined in 85% of K. pneumoniae and 77.4% of E. coli isolates. These results were higher than those reported by Silva-Sanchez et al. [29] who stated that qnrB was the most predominant gene and was present in 71.4% of isolates followed by qnrS which was detected in 24.4% of isolates then qnrA with a percentage of 18.3%, while qnrC and qnrD genes were not identified [29].
In conclusion, ESBLs producing clinical isolates exhibited MDR resistance nature with a high resistance rate towards different antibiotic classes. These MDR infections are associated with high public health costs, therapeutic failures, limited antibacterial choice, increased duration of hospitalization, and rising mortality rates. Our study confirmed that plasmid-mediated quinolone resistance (PMQR) genes are often carried on the same plasmid as the ESBLs genes. Unfortunately, these plasmids can be transferred by conjugation. It is worth mentioning that transconjugation within the same genus (intrageneric) was more likely to happen than in different genera (intergeneric) leading to the dissemination of PMQR determinants and the spread of multidrug resistance in different Enterobacteriaceae species that may lead to serious health problems. Therefore, regular monitoring of antibacterial resistance is a must, also more effort is required to enhance the awareness of people about antibiotic misuse to avoid the emergence of MDR isolates.
Data availability
Data will be available upon request.
References
Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, Shallom JM, et al. Phylogeny of gammaproteobacteria. J Bacteriol. 2010;192:2305–14.
Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012-2013. JAMA. 2015;314:1479–87.
Pitout JDD. Extraintestinal pathogenic Escherichia coli: A combination of virulence with antibiotic resistance. Front Microbiol. 2012;3:9.
D’Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–61.
Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: A clinical update. Clin Microbiol Rev. 2005;18:657–86.
Livermore DM. β-Lactams: mode of action and mechanisms of bacterial resistance. Antibiot Lab Med. 1996;2:502–78.
Guiral E, Gonçalves Quiles M, Muñoz L, Moreno-Morales J, Alejo-Cancho I, Salvador P, et al. Emergence of resistance to quinolones and β-lactam antibiotics in enteroaggregative and enterotoxigenic Escherichia coli causing traveler’s diarrhea. Antimicrobial Agents Chemother. 2019;63:e01745-18.
Alheib O, Al Kayali R, Abajy MY. Prevalence of plasmid-mediated quinolone resistance (PMQR) determinants among extended spectrum beta-lactamases (ESBL)-producing isolates of Escherichia coli and Klebsiella pneumoniae in Aleppo, Syria. Arch Clin Infect Dis. 2015;10:e20631.
Garrec H, Drieux-Rouzet L, Golmard JL, Jarlier V, Robert J. Comparison of nine phenotypic methods for detection of extended-spectrum β-lactamase production by enterobacteriaceae. J Clin Microbiol. 2011;49:1048–57.
Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual (No. Ed. 2, pp. xxxviii+−1546). USA: University of Texas South Western Medical Center; 1989.
White PA, McIver CJ, Deng YM, Rawlinson WD. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol Lett. 2000;182:265–9.
Ozgumus OB, Tosun I, Aydin F, Kilic AO. Horizontol dissemination of TEM-and SHV-typr beta-lactamase genes-carrying resistance plasmids amongst clonical isolates of Enterobacteriaceae. Braz J Microbiol. 2008;39:636–43.
Kadry AA, El-Antrawy MA, El-Ganiny AM. Management of clinical infections of Escherichia coli by new β-lactam/β-lactamase inhibitor combinations. Iranian. J Microbiol. 2022;14:466–74.
Kadry AA, El-Antrawy MA, El-Ganiny AM. Impact of short chain fatty acids (SCFAs) on antimicrobial activity of new β-lactam/β-lactamase inhibitor combinations and on virulence of Escherichia coli isolates. J Antibiotics. 2023;76:225–35.
Gad AH. The Occurrence of Multidrug Resistant E. Coli which Produce ESBL and Cause Urinary Tract Infections. J Appl Microbiol. 2017;1:8.
M Al-Kashef N, A Al-sayed M, A Kadry A, M El-Ganiny A. Extended spectrum β-Lactamase production and the Co-existed antibiotic resistance among Uropathogenic Escherichia coli isolates. Zagazig J Pharm Sci. 2018;27:64–72.
Morsi SS, Tash RME. Virulence Determinants among Extended-Spectrum B-Lactamases producers of Uropathogenic Escherichia coli isolates In Zagagig University Hospitals, Egypt. Egyptian J Med Microbiol. 2016;25:101–8.
Cunha BA, Gran A, Raza M. Persistent extended-spectrum β-lactamase-positive Escherichia coli chronic prostatitis successfully treated with a combination of fosfomycin and doxycycline. Int J Antimicrobial Agents. 2015;45:427–9.
Abdul Rahim N, Cheah SE, Johnson MD, Yu H, Sidjabat HE, Boyce J, et al. Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two ‘old’antibiotics—polymyxin B and chloramphenicol. J Antimicrobial Chemother. 2015;70:2589–97.
Aladag M, Durak Y. Investigation of some antibiotic susceptibilities, plasmid profiles and ESBL characteristics of K. pneumoniae isolated from urinary system infections. World Appl Sci J. 2009;6:630–6.
Daini O, Adesemowo A. Antimicrobial susceptibility patterns and R-plasmids of clinical strains of Escherichia coli. Aust J Basic Appl Sci. 2008;2:397–400.
Helfand MS, Bonomo RA. Current challenges in antimicrobial chemotherapy: the impact of extended-spectrum β-lactamases and metallo-β-lactamases on the treatment of resistant Gram-negative pathogens. Curr Opin Pharmacol. 2005;5:452–8.
de Souza Lopes AC, Rodrigues JF, de Morais Júnior MA. Molecular typing of Klebsiella pneumoniae isolates from public hospitals in Recife, Brazil. Microbiological Res. 2005;160:37–46.
Gangoué-Piéboji J, Bedenic B, Koulla-Shiro S, Randegger C, Adiogo D, Ngassam P, et al. Extended-spectrum-β-lactamase-producing Enterobacteriaceae in Yaounde, Cameroon. J Clin Microbiol. 2005;43:3273–7.
Yhiler NY, Bassey BE, Paul IE, Francis UM, Anne A, Okocha-Ejeko A. Extended spectrum beta-lactamase production and plasmid mediated quinolone resistance among lactose non-fermenting nterobacteriaceae isolated from poultry sources in Calabar, Nigeria. Afr J Microbiol Res. 2019;13:400–6.
Yang H, Chen H, Yang Q, Chen M, Wang H. High prevalence of plasmid-mediated quinolone resistance genes qnr and aac (6′)-Ib-cr in clinical isolates of Enterobacteriaceae from nine teaching hospitals in China. Antimicrobial Agents Chemother. 2008;52:4268–73.
Valadbeigi H, HatamiLak M, Maleki A, Kouhsari E, Sadeghifard N. Molecular characteristics, antimicrobial resistance profiles, and antibiotic resistance determinants in uropathogenic fluoroquinolone resistant-Escherichia coli isolates. Gene Rep. 2020;18:100584.
Poirel L, Leviandier C, Nordmann P. Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrobial Agents Chemother. 2006;50:3992–7.
Silva-Sanchez J, Barrios H, Reyna-Flores F, Bello-Diaz M, Sanchez-Perez A, Rojas T, et al. Prevalence and characterization of plasmid-mediated quinolone Resistance genes in extended-Spectrum β-lactamase–producing Enterobacteriaceae isolates in Mexico. Microb Drug Resistance. 2011;17:497–505.
Kraychete GB, Botelho LAB, Campana EH, Picão RC, Bonelli RR. Updated multiplex PCR for detection of all six plasmid-mediated qnr gene families. Antimicrob Agents Chemother 2016;60:7524–6.
Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother 2007;60:394–7.
Acknowledgements
The authors appreciate the assistance of the staff of Mansoura University Hospitals and their assistants in collecting specimens.
Author information
Authors and Affiliations
Contributions
AAK contributed to the study conception, data analysis, supervision, review, and editing. MAE-A contributed to methodology, resources, data collection, and analysis, writing draft, review, and editing. AME-G contributed to data analysis, writing draft, review, editing, and validation. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval and consent to participate
All experimental tests were carried out according to approval number (FPDU15/2022) from the Faculty of Pharmacy, Delta University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kadry, A.A., El-Antrawy, M.A. & El-Ganiny, A.M. Investigation of plasmid-mediated quinolone resistance among extended-spectrum β-lactamase isolates of E. coli and K. pneumoniae. J Antibiot (2024). https://doi.org/10.1038/s41429-024-00761-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41429-024-00761-z
- Springer Japan KK