Abstract
The triggering receptor expressed on myeloid cells-1 (TREM-1) is a receptor expressed on innate immune cells. By promoting the amplification of inflammatory signals that are initially triggered by Toll-like receptors (TLRs), TREM-1 has been characterized as a major player in the pathophysiology of acute and chronic inflammatory diseases, such as septic shock, myocardial infarction, atherosclerosis, and inflammatory bowel diseases. However, the molecular events leading to the activation of TREM-1 in innate immune cells remain unknown. Here, we show that TREM-1 is activated by multimerization and that the levels of intracellular Ca2+ release, reactive oxygen species, and cytokine production correlate with the degree of TREM-1 aggregation. TREM-1 activation on primary human monocytes by LPS required a two-step process consisting of upregulation followed by clustering of TREM-1 at the cell surface, in contrast to primary human neutrophils, where LPS induced a rapid cell membrane reorganization of TREM-1, which confirmed that TREM-1 is regulated differently in primary human neutrophils and monocytes. In addition, we show that the ectodomain of TREM-1 is able to homooligomerize in a concentration-dependent manner, which suggests that the clustering of TREM-1 on the membrane promotes its oligomerization. We further show that the adapter protein DAP12 stabilizes TREM-1 surface expression and multimerization. TREM-1 multimerization at the cell surface is also mediated by its endogenous ligand, a conclusion supported by the ability of the TREM-1 inhibitor LR12 to limit TREM-1 multimerization. These results provide evidence for ligand-induced, receptor-mediated dimerization of TREM-1. Collectively, our findings uncover the mechanisms necessary for TREM-1 activation in monocytes and neutrophils.
Similar content being viewed by others
Introduction
The triggering receptor expressed on myeloid cells-1 (TREM-1) is an immune receptor expressed on innate immune cells, such as neutrophils, mature monocytes, macrophages,1 and on platelets.2 By promoting the amplification of inflammatory signals that are initially triggered by Toll-like receptor (TLR) and NOD-like receptor engagement, TREM-1 has been characterized as a major player in the pathophysiology of acute and chronic inflammatory diseases of different etiologies3,4 including septic shock,5 acute myocardial infarction,6 and atherosclerosis.7,8
TREM-1 is a single-pass 30-kDa glycoprotein receptor composed of an immunoglobulin (Ig)-like domain and associated with the adapter molecule DNAX activation protein of 12 kDa (DAP12).9 DAP12 contains an immunoreceptor tyrosine-based activation motif (ITAM) domain that serves as a kinase-docking site, allowing transduction of intracellular signals. The association of TREM-1 with DAP12 allows, subsequent to TREM-1 activation, the recruitment and tyrosine phosphorylation of spleen tyrosine kinase (SYK) and adaptor molecules, such as growth factor receptor-binding protein 2, activation of phosphatidylinositol 3-kinase, Bruton’s tyrosine kinase,10 and phospholipase C-γ (PLC-γ).11 Activation of these pathways leads to intracellular Ca2+ mobilization; actin cytoskeleton rearrangement; activation of transcription factors such as nuclear factor-kappa B (NF-κB); and subsequent cytokine production, oxidative burst, or phagocytosis.12
The connection between the TREM-1 and TLR pathways occurs at multiple levels.13 Indeed, the priming of monocytes and neutrophils through TLR2 and TLR4 activation is known to induce upregulation of TREM-1 expression in a myeloid differentiation factor 88 (MyD88)-dependent manner and involves the transcription factors NF-κB, PU.1, and activator protein 1.1,14,15 Following lipopolysaccharide (LPS) stimulation of neutrophils, TREM-1 is recruited to ganglioside M1-lipid rafts and colocalizes with TLR4.16 TREM-1 silencing induces down-expression of several key players of TLR pathways, such as MyD88, cluster of differentiation 14, NF-κB inhibitor alpha, inflammatory mediators interleukin 1 beta (IL-1β), monocyte chemotactic protein 1, and IL-10.17 Similar results were obtained with pharmacological inhibition of TREM-1 by the therapeutic peptide LR12, both in vitro and in vivo.18,19,20 TREM-1 ligation alone does not seem to induce sustained inflammation in resting primary human leukocytes, and it has been suggested that an initial priming through TLRs may be required to allow TREM-1 signaling.5,18 However, the molecular and mechanistic events leading to TREM-1-mediated amplification of the TLR pathway remain unknown.
Here, we show that activation of TREM-1 does not depend on its level of expression at the membrane but rather on the valency of its cross-linking. The activation of primary human neutrophils and monocytes through TLR4 induces, respectively, a one-step and two-step mobilization of TREM-1 at the membrane, ultimately leading to the clustering and multimerization of TREM-1. This multimerization controls signal transduction and is further stabilized by the adapter molecule DAP12 and the endogenous ligand of TREM-1.
Materials and methods
Peptide
LR12 peptide (LQEEDAGEYGCM) is a specific functional inhibitor of TREM-1.18 LR12-scr is a scrambled control peptide composed of the same amino acids from LR12 but in a random sequence (YQDVELCETGED). LR12 was chemically synthesized (Pepscan Presto BV, the Netherlands) as a COOH-terminally amidated peptide with >95% purity as confirmed by mass spectrometry (MS) coupled to high-performance liquid chromatography. The peptide was free of endotoxin according to the Limulus amebocyte lysate (LAL) assay (<0.02 UI/mg).
TREM-1 agonists αTREM-1 and Fab
Anti-human TREM-1 antibody fragment (Fab) was obtained by enzymatic digestion of the agonistic anti-TREM-1 antibody (αTREM-1) clone 193015 (MAB1278, Biotechne, R&D Systems, USA). Papain immobilized on an agarose resin (0.5 ml for 10 mg of IgG, Thermo Fisher Scientific, USA) was washed in digestion buffer before incubation at 37 °C for 16 h with αTREM-1. The resulting supernatant was removed and pooled with the supernatant of the resin after washing with a Tris pH 7.5 buffer. The remaining nondigested αTREM-1 fragments were removed by protein A affinity chromatography (Protein A Ceramic HyperD F, PALL Life Sciences, USA). Endotoxin levels were measured by the LAL assay, and endotoxin levels at working concentrations were below 2 UI/ml. The αTREM-1-Fab size and purity were confirmed on 13.5% polyacrylamide gel electrophoresis under reducing conditions and stained with Coomassie blue, and its ability to bind TREM-1 was assessed by fluorescence-activated cell sorting (FACS). αTREM-1-Fab and αTREM-1 were used at 10 µg/ml for monovalent and divalent stimulation of TREM-1.
Cell isolation, culture, and stimulation
The human myelomonocytic cell lines U937 (Culture Collections, Public Health England No. 85011440) and U937-TD (U937 stably transfected with hTREM-1 and human DAP1221) were cultured in RPMI 1640 Glutamax supplemented with 10% fetal calf serum (FCS), 25 mM Hepes, and 100 U/ml penicillin and streptomycin (all from Thermo Fisher Scientific). For some experiments, untransfected U937 cells were cultured under the same conditions supplemented with 100 nM of 1,25-dihydroxyvitamin D3 (Sigma-Aldrich, USA) to induce upregulation of TREM-1.22 For stimulation, cells were seeded at 0.5 × 106 cells/ml. Depending on the experiments, U937 and U937-TD cells were incubated under resting conditions or with TREM-1 agonists with and without a goat anti-mouse (H + L) antibody (GαM; Biotechne, R&D Systems) all at 10 µg/ml at indicated times.
Primary human neutrophils and monocytes were isolated from healthy blood donors by immunomagnetic negative cell sorting with EasySep™ Human Monocyte/Neutrophil Isolation Kits (StemCell, Canada). Purity was assessed by flow cytometry. Cells were suspended in RPMI 1640 Glutamax supplemented with 10% FCS, 25 mM Hepes, and 100 U/ml penicillin and streptomycin (all from Thermo Fisher Scientific) before stimulation. Human monocytes or neutrophils were incubated under resting conditions or with LPS from Escherichia coli 0127:B8 (100 ng/ml; Sigma-Aldrich) with or without TREM-1 agonists at the indicated times. When indicated, cells were incubated with the clinical-stage TREM-1 inhibitory peptide LR12 at 25 µg/ml18 or cytochalasin D (Sigma-Aldrich) at 5 µg/ml.23
TREM-1 expression was assessed by staining with anti-hTREM-1-allophycocyanin (APC) or the corresponding isotype-APC antibodies (Miltenyi Biotec, Germany), and data were collected by flow cytometry (C6 Accuri, BD, USA).
Protein phosphorylation analysis
Total proteins were extracted from stimulated cells using Phosphosafe extraction buffer (Novagen, USA), and the total protein concentration was measured by a Coomassie protein assay kit (Thermo Fisher Scientific) and used for normalization. Lysates were then analyzed by western blot (Criterion XT Bis-Tris Gel, 4–12%, Bio-Rad, USA, and polyvinylidene difluoride membrane, Millipore, France) and revealed with anti-phospho-SYK and anti-phospho-PLCγ (Cell Signaling, USA) with the corresponding secondary antibody conjugated to horseradish peroxidase (GE Healthcare, USA) and Super-Signal West Femto Substrate (Thermo Fisher Scientific). Anti-SYK and anti-PLCγ (Cell Signaling) were used for normalization on the same membranes after stripping for 20 min at 65 °C in stripping buffer (2% SDS, 62.5 mM Tris (pH 6.8), and 100 mM β-mercaptoethanol). Acquisition and normalization were performed with an LAS-4000 imager (Fujifilm, Japan) and Multi-Gauge software (LifeScience Fujifilm, Japan).
Cytokine measurements
Supernatants from stimulated cells were recovered after a 24-h stimulation, and IL-8 was measured using Human IL-8/CXCL8 and TNFα Quantikine ELISA Kit (Biotechne, R&D Systems) according to the manufacturer’s instructions.
Intracellular Ca2+ measurement
Ca2+ analysis was performed using flow cytometry (Fortessa and C6 Accuri, BD). U937 and U937-TD cells were washed twice in phosphate-buffered saline and loaded with Rhod-3-acetoxymethyl ester (10 µM) for 45 min and 2.5 mM probenecid (Thermo Fisher Scientific) in Ringer’s solution (in mM: 145 NaCl; 5.4 KCl; 2 CaCl2; 1 MgCl2; 10 glucose; 10 Hepes; and 0.1% bovine serum albumin (BSA), pH adjusted to 7.5). Primary human neutrophils and monocytes were washed and incubated with pluronic acid and fluo-3-acetoxymethyl ester (10 µM; Sigma-Aldrich) for 30 min in Ringer’s solution. After baseline acquisition for 1 min, intracellular Ca2+ mobilization was triggered by stimulation with TREM-1 agonists and monitored for 3 min before ionomycin incubation. The results were normalized to the baseline and the maximal signal induced by ionomycin during the last minute of acquisition (1 µM; Sigma-Aldrich).
Reactive oxygen species production
Flow cytometry quantification of intracellular reactive oxygen species (ROS) production after a 2-h stimulation was assessed using DCFDA, a fluorogenic substrate (Thermo Fisher Scientific), according to the manufacturer’s instructions. The results are expressed as the mean fluorescence intensity (MFI).
Confocal microscopy
Cells were seeded and stimulated on LabTek chambers (Thermo Fisher Scientific) for 24 h (monocytes) and 3 h (neutrophils). After stimulation, cells were washed and fixed with paraformaldehyde (4%) for 20 min and permeabilized with Triton 0.1% for 30 min prior to incubation with the indicated primary antibodies at 4 °C overnight (anti-hTREM-1-AF488 and anti-hDAP12-AF555 Bioss, USA). Nuclei were stained with TO-PRO-3 (1 μg/ml, Invitrogen, USA) for 1 h at 37 °C. After washing, coverslips were mounted using a solution of Vectashield (Vector Laboratories, USA). Confocal images were obtained using a Leica SP5 confocal laser-scanning microscope system (Leica, Germany) fitted with appropriated filter sets and acquired in sequential scan mode.
In situ proximity ligation assay
Primary human monocytes were seeded at 3 × 105 cells/well on LabTek chambers in complete medium and incubated overnight under resting conditions or with LPS (100 ng/ml) with or without LR12 (25 μg/ml), rhPGLYRP1 (1 µg/ml, Biotechne, R&D Systems), and cytochalasin D (5 µg/ml). After stimulation, cells were fixed in 4% paraformaldehyde for 20 min, washed, and blocked with Duolink blocking solution (Sigma-Aldrich) for 1 h at 37 °C. Cells were then incubated with anti-TREM-1 primary antibodies (proximity ligation assay (PLA) antibody 1, mouse anti-human TREM-1 (1:200, Bio-Rad) and PLA antibody 2, and rabbit anti-human TREM-1 (1:200, Abcam, UK)) and secondary antibodies conjugated with PLUS and MINUS oligonucleotide probes (anti‐rabbit PLA probe PLUS and anti‐mouse PLA probe MINUS) prior to incubation with Duolink Detection Reagents Red (Sigma-Aldrich). All images were taken using a ×63 oil immersion lens. Images were acquired by confocal microscopy (Leica SP2). Quantifications of the in situ PLA signal were performed by BlobFinder freeware (Center for Image Analysis, Sweden) and expressed as the ratio Blobs/cell. Alternatively, for competition assays, PLA antibodies 1 and 2 were incubated at various concentrations on U937-TD cells and revealed with APC-conjugated goat anti-mouse IgG (H + L).
Primary human neutrophils were incubated for 3 h at 3 × 105 cells/tube in resting conditions or with LPS (100 ng/ml) with or without LR12 (25 μg/ml). Cells were then washed and stained according to a protocol adapted for nonadherent cells and using the same PLA antibodies and reagents.24 Acquisitions were performed by flow cytometry and expressed as the MFI.
DAP12 silencing
Cells were transfected overnight at 37 °C and 5% CO2 with human DAP12 siRNA or a control siRNA (MISSION esiRNA human TYROBP and siRNA Universal Negative Control #1, Sigma-Aldrich) at a final concentration of 50 nM in the presence of an siRNA transfection reagent (MISSION siRNA Transfection Reagent, Sigma-Aldrich) according to the manufacturer’s instructions. After transfection, cells were stimulated as indicated for 24 h. Cells were then harvested for TREM-1 membrane expression, in situ PLA, protein extraction for western blotting with anti-hDAP12 and h-αTubulin antibodies (Cell Signaling), and RNA extraction.
Quantitative reverse transcriptase-polymerase chain reaction
Total RNAs were extracted from monocytes using the Direct-zol RNA MicroPrep kit (Zymo Research, USA) and quantified with a NanoDrop (Thermo Fisher Scientific) before being retrotranscribed using the iScript cDNA synthesis kit (Bio-Rad) and quantified by quantitative polymerase chain reaction (PCR) using the available probes (Qiagen, Quantitect Primers, the Netherlands) for human Dap12 and human β-actin (housekeeping gene). All PCRs were performed in a MyiQ Thermal Cycler and quantified by iQ5 software (Qiagen).
Proteins
Two types of recombinant hTREM-1 proteins were used. The first form, named hTREM-1-ECD(21–200), is a commercially available recombinant human TREM-1 (residues Ala21–Arg200) produced in a mammalian expression system with a purity >95% (reference C506, Novoprotein, USA).
The second form of TREM-1, named hTREM-1-ECD(21–192), was produced in a prokaryotic expression system.25 hTREM-1-ECD(residues 21–192) synthetic gene was subcloned into a pCARGHO-1 vector (Patent WO2017194888) using NdeI and XhoI restriction sites. The overexpressed recombinant protein was produced, unlike previous studies,26 as a soluble fusion protein CRD-TREM-1(21–192) using a new CRD tag (patent pending) in E. coli C41(DE3) strain. Transfected cells were grown at 37 °C in Luria-Bertani broth until reaching OD600 nm = 0.8 and subsequently overnight at 30 °C in a medium supplemented with 1 mM isopropyl β-d-1-thiogalactopyranoside. Cells were harvested and resuspended in lysis buffer (Tris 20 mM, NaCl 300 mM, EDTA 3 mM, and dithiothreitol 1 mM, pH 8). Released recombinant protein was obtained by sonication. After centrifugation, the supernatant was loaded onto a Lactose-Sepharose column (Galab Laboratories, Germany). Elution of CRD-hTREM-1-ECD(21–192) (38 455 Da) was achieved using buffer A + lactose at 100 mM. The fusion protein was digested overnight at 4 °C using tobacco etch virus (TEV) protease. The CRD tag (18 815 Da), TEV protease, and hTREM-1-ECD(21–192) were separated by hydrophobic interaction chromatography onto Phenyl-Sepharose (GE Healthcare) using a decreasing linear ammonium sulfate gradient. Pooled fractions of hTREM-1-ECD(21–192) (19 640 Da) were then concentrated onto an Amicon-YM 10-kDa concentrator (Merck Millipore, Germany) by centrifugation until reaching a final concentration of 7.5 mg/ml. The identity of the protein was confirmed by electrospray ionization-MS (ESI-MS).
Native MS
Native MS measurements were performed with a Q-TOF-2 (Waters, UK) mass spectrometer equipped with a standard static nano-ESI source.27,28 Samples of recombinant hTREM-1-ECD(21–200) with or without LR12 peptide were prepared by dissolving the lyophilized powder in a 150 mM ammonium acetate solution (pH adjusted to 7.2 with ammonium hydroxide) followed by five sequential concentration and dilution cycles at 4 °C using a centrifugal filter with a molecular-weight (MW) cutoff of 10 kDa (Merck Millipore). The molar concentrations of these samples were determined with a Nanodrop UV spectrometer (Thermo Fisher Scientific). Tandem MS was performed by mass selection of precursor ions using a quadrupole mass analyzer followed by their collision with xenon molecules. Data analysis was performed with MassLynx (Waters) and Origin Pro (Origin Lab, USA) software. The relative abundance of the protein dimer was determined using a previously described method.29,30
High-mass matrix-assisted laser desorption/ionization MS
hTREM-1-ECD(21–200) was submitted to cross-linking using a CovalX K200 matrix-assisted laser desorption/ionization (MALDI) MS analysis kit (CovalX AG, Switzerland). hTREM-1-ECD(21–200) (1 mg/ml) was diluted at a ratio of 1:4 in K200 cross-linking buffer. The sample was mixed with 1 µl of K200 stabilizer reagent (2 mg/ml) and incubated at room temperature for 3 h. After mixing, 1 µl of each sample was spotted on the MALDI plate (SCOUT 384; AnchorChip). The analysis was repeated in triplicate. After crystallization at room temperature, the plate was introduced in the MALDI mass spectrometer. For control experiments, hTREM-1-ECD (0.250 mg/ml) was mixed with a solution of sinapinic acid matrix (10 mg/ml) in acetonitrile/water (1:1, v/v), trifluoroacetic acid 0.1%, and prepared for MALDI analysis as for cross-linking experiments.
Data were acquired by high-mass MALDI-time of flight immediately after crystallization. Analysis was performed using the standard nitrogen laser and focusing on different mass ranges from 0 to 1500 kDa. High-mass MALDI mass spectra were obtained using an Ultraflex III MALDI ToF/ToF instrument (Bruker, Germany) equipped with a HM4 high-mass retrofit system (CovalX AG). The instrument was operated in linear and positive modes by applying an accelerating voltage of 20 kV. The HM4 system was operated in high-mass mode with the acceleration voltage set to 20 kV and gain voltage set to 2.95 kV. The instrument was calibrated using clusters of insulin, BSA, and IgG. For each sample, three spots were analyzed (1000 laser shots/spot). The presented spectra correspond to the average of 1000 laser shots. The MS data were analyzed using Complex Tracker analysis software (CovalX).
Gel filtration
A Superdex 75 10/300 GL column linked to an ÄKTA Purifier system (GE Healthcare) was equilibrated in 20 mM Tris buffer, 150 mM NaCl, pH 8 or 20 mM phosphate buffer, and 150 mM NaCl, pH 8, and then calibrated with Bio-Rad Gel Filtration Standard (Bio-Rad). Injection of 100 or 5 µM hTREM-1-ECD(21–192) protein with or without LR12 peptide (50 µM) was applied to the column. The elution of the protein was monitored at 280 or 215 nm, respectively (this last wavelength is only compatible with phosphate buffers). The size of the proteins was calculated using the following equation: Ve = log (MW), where Ve is the elution volume.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) was performed with a VP-iTC (Malvern, UK). The reference power was set to 15 µcal/s, and the temperature was set to 25 °C. After an initial 60-s delay, 200 µM hTREM-1-ECD(21–192) was repeatedly injected into the mixing cell with a 6-min time interval to allow a return to the equilibrium state between each injection. Data were analyzed using Origin software provided by the manufacturer. Each heat measurement peak was integrated and converted to yield molar enthalpies.
Statistical analyses
All data, unless indicated, were normally distributed and are presented as the mean ± standard deviation. The statistical significance of differences between two groups was analyzed using two-tailed Student’s t-test using GraphPad Prism 7.01 (GraphPad Software, USA). A p-value < 0.05 was considered significant.
Results
Multimerization of TREM-1 is required for signal transduction
It was recently suggested that cross-linking of a potential TREM-1-ligand was necessary to activate TREM-1.31 To determine if the valency of TREM-1 cross-linking affects its downstream signaling, we generated a monovalent Fab fragment from the agonistic anti-TREM-1 antibody (clone 193015, “αTREM-1”; Supplementary Figure 1a, left panel). Its ability to bind to membrane TREM-1 was assessed by FACS (Supplementary Figure 1a, right panel). TREM-1 was then targeted either by Fab alone, Fab plus a goat anti-mouse secondary antibody (Fab + GαM), anti-TREM-1 (αTREM-1) alone, or anti-TREM-1 plus a goat anti-mouse antibody (αTREM-1 + GαM; Supplementary Figure 1b).
By using the U937 human monocytic cell line, we first measured early signaling events, such as intracellular Ca2+ mobilization, in response to gradual aggregation of TREM-1. No difference in Ca2+ influx was observed in U937 cells within the first few minutes of activation using these different conditions (Fig. 1a). However, in U937-TD cells, which overexpressed TREM-1 and DAP12 (Fig. 1b, left panel), intracellular Ca2+ mobilization was increased using Fab + GαM compared to the triggering of TREM-1 with Fab alone (Fig. 1b, right panel). This Ca2+ mobilization was further increased when TREM-1 was activated with αTREM-1 and αTREM-1 + GαM. The same profile of a graduated response was also observed in U937 cells pretreated with vitamin D3 for 48 h, which induced an upregulation of TREM-1 (Supplementary Figure 1c). To definitively show that TREM-1-mediated cell activation was dependent on receptor multimerization, we measured the level of intracellular Ca2+ mobilization at different concentrations of αTREM-1 and GαM. Ca2+ mobilization increased in a dose-dependent manner depending on the concentration of αTREM-1 and GαM (Fig. 1c, d). Therefore, Ca2+ mobilization subsequent to TREM-1 activation correlates with its level of multimerization.
a–d U937 and U937-TD cells were stimulated with TREM-1 agonists αTREM-1 and Fab (10 µg/ml) with or without GαM (10 µg/ml) when indicated. a Kinetics of intracellular calcium release in U937 cells. b Left panel: TREM-1 expression on U937 (gray) and U937-TD (black) cells. Right panel: kinetics of intracellular calcium release in U937-TD cells. c, d Kinetics of intracellular calcium release in U937-TD cells incubated c with increasing concentrations of αTREM-1 (0.1–20 µg/ml) and d with αTREM-1 (5 µg/ml) and increasing concentrations of GαM (2.5–10 µg/ml). e Western blot analysis of lysates of U937 and U937-TD cells at indicated times. f IL-8 concentrations in supernatants after a 24-h stimulation. Data information: data are representative of at least three independent experiments. MFI mean fluorescence intensity, ns nonsignificant. *p < 0.05, **p < 0.01, ***p < 0.001 vs. resting or as indicated, as determined by the two-tailed Student’s t-test
Next, we assessed the phosphorylation of well-characterized signaling molecules involved in Ca2+ mobilization in U937-TD cells using a longer time frame ranging from 2 min to 1 h. The kinetics of SYK and PLCγ phosphorylation were analyzed by western blot under conditions in which TREM-1 was stimulated following the same strategy as described above. Although a rapid phosphorylation of SYK and PLCγ was observed under conditions in which TREM-1 crosslinking was induced by Fab + GαM, αTREM-1, and αTREM-1 + GαM, no phosphorylation was induced by Fab alone (Fig. 1e). As for Ca2+ mobilization, no difference in the phosphorylation of these two proteins was observed in U937 cells (Fig. 1e).
Finally, we tested whether the level of TREM-1 aggregation also impacted late cellular function. To do so, we analyzed IL-8 secretion after an overnight stimulation with both U937 and U937-TD. In accordance with the previous findings, no IL-8 release by U937 and U937-TD cells was detected when cells were stimulated with Fab, whereas increased secretion was observed according to the valence of TREM-1 multimerization; higher concentrations of IL-8 were measured after tetravalent crosslinking of TREM-1 (αTREM-1 + GαM) compared to divalent stimulation (αTREM-1 or Fab + GαM; Fig. 1f). Although neither significant Ca2+ mobilization nor phosphorylation of SYK/PLCγ were observed after stimulation with untransfected U937 cells with αTREM-1, a weak release of IL-8 was induced under these conditions that could have been due to the low expression of TREM-1 on U937 cells and duration of stimulation. Globally, in both cell types, a similar response was observed when TREM-1 was crosslinked in a divalent manner either with Fab and a secondary antibody or with αTREM-1 alone.
Altogether, these data suggest that the TREM-1-mediated cellular response is correlated with its level of aggregation.
LPS priming of human primary neutrophils and monocytes is required to allow TREM-1 activation
Similar to U937-TD cells, primary human neutrophils express high levels of TREM-1 at the membrane under resting physiological conditions (Fig. 2a). To evaluate whether monovalent or multivalent stimulation of TREM-1 induced a graduated response, we stimulated freshly isolated primary human neutrophils with Fab or αTREM-1. Unexpectedly, the level of Ca2+ mobilization was quite low, and no difference was observed after monovalent triggering of TREM-1 with the Fab fragment compared to divalent triggering with the monoclonal antibody (Fig. 2b, left panel). Low production of ROS after a 2-h stimulation occurred only with αTREM-1 (Fig. 2c). Since TREM-1 is known to synergize with TLR4, we assessed the impact of LPS pre-treatment of neutrophils prior to TREM-1 crosslinking. After LPS stimulation, although no change in TREM-1 expression at the membrane was observed (Fig. 2a) even with an LPS concentration of up to 1 µg/ml (not shown), both Fab and αTREM-1 were able to induce intracellular Ca2+ release (Fig. 2b, right panel) and increased ROS production (Fig. 2c). A 3-h pre-treatment of neutrophils by LPS was sufficient to allow activation of TREM-1 (Fig. 2b, right panel). Under these pre-treated conditions, a graduated response was observed when comparing TREM-1 stimulation by Fab and αTREM-1 (Fig. 2b, c).
a–e Isolated primary human monocytes and neutrophils were incubated at indicated times in resting conditions or stimulated with LPS (100 ng/ml) and TREM-1 agonists (10 µg/ml) when indicated. a TREM-1 expression by FACS on neutrophils at indicated times. b Kinetics of intracellular calcium release in neutrophils in response to TREM-1 agonists with or without 3-h pre-treatment with LPS. c Neutrophil intracellular ROS production. d TREM-1 expression by FACS on monocytes at indicated times. e Kinetics of intracellular calcium release in monocytes in response to TREM-1 agonists with or without 24-h pre-treatment with LPS. f TNF-α concentrations in supernatants of 24-h-stimulated monocytes. Data information: data are representative of at least three independent experiments. MFI mean fluorescence intensity, DCF dichlorofluorescin, ns, nonsignificant. *p < 0.05, ***p < 0.001 vs. resting or as indicated, as determined by the two-tailed Student’s t-test
Primary human monocytes also express TREM-1 under resting physiological conditions, although at a lower level than primary neutrophils (Fig. 2d), and similar to neutrophils, no Ca2+ mobilization was observed after TREM-1 stimulation by Fab or αTREM-1 under resting conditions (Fig. 2e, left panel). We therefore assessed both TREM-1 expression at the membrane and TREM-1 activation after pre-treatment with LPS by using the same approach as with neutrophils. Monocytes required a 24-h stimulation by LPS to upregulate TREM-1 and increase its expression at the membrane (Fig. 2d). Similar to neutrophils, intracellular Ca2+ mobilization and release of tumor necrosis factor-α were induced by either Fab or αTREM-1 only under conditions in which monocytes were pre-stimulated by LPS (Fig. 2e, f). However, in monocytes, a 3-h pre-treatment with LPS was not sufficient to induce TREM-1 activation, and only a 24-h pre-treatment allowed activation of TREM-1 by stimulation with Fab and αTREM-1 (Fig. 2e, right panel).
Altogether, these results suggest that the activation of TREM-1 does not depend on its level of membrane expression in primary cells, but requires priming by LPS.
LPS priming of neutrophils induces a one-step clustering and multimerization of TREM-1
LPS priming seems to be necessary and sufficient to allow TREM-1 activation. We therefore wanted to decipher the mechanisms allowing the activation of TREM-1 following LPS activation. Freshly isolated circulating human neutrophils were plated on Labtek chambers under resting and activated conditions for further labeling of TREM-1 and observed by confocal microscopy. We observed diffuse labeling of TREM-1 at the surface of neutrophils under resting conditions, whereas 3-h LPS stimulation induced a clustering of TREM-1 at the membrane (Fig. 3a, upper panels, white arrows). The addition of cytochalasin D partly prevented this LPS-induced clustering of TREM-1, which therefore appears to be an active molecular mechanism driven by cytoskeleton rearrangements (Fig. 3a, lower panels). In addition, treatment of resting or LPS-activated neutrophils with cytochalasin D induced a slight decrease in membrane TREM-1 expression (Fig. 3b), suggesting that there is a dynamic turnover of TREM-1 at the membrane that is dependent on actin polymerization.
a–c Isolated human neutrophils were incubated at indicated times in resting conditions or stimulated with LPS (100 ng/ml) and/or cytochalasin D (5 µg/ml) when indicated. a TREM-1 (green) and nucleus (TO-PRO-3, blue) staining by confocal microscopy after 3 h stimulation (scale bar: 10 µm). b TREM-1 expression on isolated human neutrophils after 24 h. c TREM-1/TREM-1 interactions quantified by flow cytometry after 3 h. Data information: data are representative of at least three independent experiments. MFI mean fluorescence intensity, ns nonsignificant. **p < 0.01, ***p < 0.001 vs. resting or as indicated, as determined by the two-tailed Student’s t-test
Following LPS stimulation, an increase in TREM-1/TREM-1 interaction was measured by in situ PLA (Fig. 3c). In situ PLA allows precise detection of protein interactions with a maximum distance of 40 nm between two targets. As shown in supplementary Figure 2b, the two primary antibodies used for the in situ PLA assays competed against each other for binding to TREM-1, showing that our signal was specific for two distinct TREM-1 molecules. These data suggest that the clustering of TREM-1 at the membrane following LPS stimulation promotes its homo-oligomerization.
Overall, these results demonstrate that LPS priming of human neutrophils induces a rapid and active rearrangement of TREM-1 at the cell membrane, leading to TREM-1 clustering and homo-oligomerization.
LPS priming of monocytes induces a two-step clustering and multimerization of TREM-1
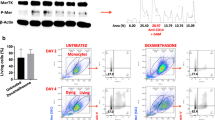
Consistent with the above results of primary monocytes and contrary to those of primary neutrophils, a 3-h stimulation of monocytes did not induce any change in the TREM-1 expression levels at the membrane, and no clustering or dimerization was detectable by confocal microscopy and in situ PLA, respectively (Fig. 4a, b). Since primary monocytes express TREM-1 at low levels under resting conditions, we assessed the effect of a longer LPS stimulation. After a 24-h LPS stimulation of monocytes, we observed upregulation of TREM-1 by confocal microscopy and, similar to neutrophils, clustering (Fig. 4a, left panel, white arrows) and dimerization (Fig. 4b) of TREM-1 at the membrane. This TREM-1 mobilization was also prevented by cytochalasin D treatment. Indeed, even if the LPS-induced TREM-1 upregulation was decreased in the presence of cytochalasin D (Fig. 4a, right panel), diffuse TREM-1 labeling was observed (Fig. 4a, left panel), which provides evidence of an absence of TREM-1 clustering. A decrease in the average number of TREM-1/TREM-1 interactions, quantified as blobs/cell, was consistently observed with cytochalasin D (Fig. 4b).
a–b Isolated human monocytes were incubated at indicated times in resting conditions or stimulated with LPS (100 ng/ml) and/or cytochalasin D (5 µg/ml) when indicated. a Left panel: TREM-1 (green) and nucleus (TO-PRO-3, blue) staining by confocal microscopy after 3 and 24 h stimulation. Right panel: TREM-1 expression on isolated human monocytes after 24 h stimulation (scale bar: 10 µm). b Left panel: TREM-1 in situ PLA (red blobs) and nucleus (blue) by confocal microscopy after 3 and 24 h stimulation (scale bar: 10 µm). Right panel: quantification of average blobs/cell. Data information: data are representative of at least three independent experiments. MFI mean fluorescence intensity, ns nonsignificant. *p < 0.05, **p < 0.01, ***p < 0.001 vs. resting or as indicated, as determined by the two-tailed Student’s t-test
These results show that in primary monocytes, LPS induces a two-step mobilization of TREM-1 at the membrane, consisting of upregulation of membrane expression, followed by TREM-1 multimerization.
TREM-1 extracellular domain dimerizes in vitro
To characterize the homo-oligomerization of TREM-1, we used several approaches with recombinant proteins. Kelker et al. have shown by analytical ultracentrifugation that recombinant hTREM-1-ECD(21–194) produced in E. coli, consisting of the full-length ECD domain, was present as a monomer in solution. The authors observed an elution at lower retention volumes during gel filtration experiments compared to the theoretical size of the protein.25 We produced a fragment of TREM-1, TREM-1-ECD(21–192) (Supplementary Figure 3), and performed gel filtration experiments. The theoretical size of this recombinant non-glycosylated TREM-1-ECD(21–192) is 19.6 kDa. At 100 µM, the protein eluted as a single symmetrical peak with a similar retention volume as in the results of Kelker et al. and corresponding to a MW of ~40.4 kDa (Fig. 5a, dashed line). This MW was compatible with the theoretical size of a globular dimeric state of non-glycosylated TREM-1-ECD(21–192) (Fig. 5a). We then used another biophysical method to validate the existence of TREM-1 as a dimer in solution (Fig. 5b). ITC is a powerful approach for investigating self-association processes of protein complexes32,33,34 and yields the equilibrium constant or the oligomerization midpoint, which is also known as the critical transition concentration (CTC). The CTC defines the transition point at which half of the injected oligomers dissociate. The ITC curve obtained after injection of hTREM-1-ECD(21–192) at a concentration of 200 µM, at which we would expect the protein to be mainly present as a dimer according to our results, was indeed found to be typical of the dissociation of the dimer. We measured a CTC of 7 µM for hTREM-1-ECD(21–192), corresponding to the inflection point of the sigmoid curve or the x-intercept of the second derivative (Fig. 5b, right panel). Above the CTC, the injected protein does not dissociate and the peaks are equivalent to the level of heat dilution, compatible with a protein that is completely monomeric in solution. By injecting the protein at a concentration of 5 µM on the gel filtration column, we obtained a broader peak at a higher elution volume. This peak was compatible with an equilibrium between a globular dimeric form of the protein (measured value of 32.4 kDa) and globular monomer appearing as a shoulder on the chromatogram (size of ~20.4 kDa and fully compatible with the theoretical monomer size of 19.6 kDa; Fig. 5a, in red). Therefore, we demonstrated that the TREM-1 extracellular domain is in equilibrium between its monomeric and dimeric forms in solution.
a Characterization of hTREM-1-ECD(21–192) (100 μM, dashed line and 5 µM, red line) by gel filtration chromatography. Elution was monitored by absorbance at 280 nm except for hTREM-1-ECD(21–192) at low concentration (5 µM), which was monitored at 215 nm. b Left panel: titration isotherms of hTREM-1-ECD(21–192) determined by isothermal titration calorimetry (ITC). Right panel: the critical transition concentration (CTC) corresponds to the x-intercept of the second derivative (C: TREM-1 monomer concentration; H: enthalpy). c Characterization of hTREM-1-ECD(21–192) (5 µM, red line) and hTREM-1-ECD(21–192) (5 µM) in competition with LR12 peptide at 50 µM (dashed line) by gel filtration chromatography. Data information: data are representative of at least three independent experiments
The TREM-1 inhibitor LR12 is known to act as a decoy receptor.18 To assess whether this peptide was able to affect the monomer/dimer equilibrium, we incubated TREM-1 and LR12 at a ratio of 5 µM recombinant TREM-1 and 50 µM LR12. Under these conditions, we did not observe any shift from the dimeric to the monomeric form of TREM-1-ECD(21–192) in solution (Fig. 5c), suggesting that LR12 is not able to compete with TREM-1 homo-oligomerization in solution.
With then used a recombinant hTREM-1-ECD(21–200) produced in mammalian cells to confirm the dimerization properties of TREM-1 in vitro. In native MS measurements, 2 µM hTREM-1-ECD(21–200) gave rise to MS signals exhibiting a unimodal profile (Fig. 6a). When increasing the concentration of TREM-1 above 4 µM, an additional distribution of signals emerged, indicating the formation of species with higher MWs. At higher protein concentrations, we observed a higher relative abundance of the larger species (Fig. 6a). Upon mass selection and subsequent gas-phase dissociation, the subpopulations of the protein ions represented by the first and second distributions of signals released fragments in the same manner, revealing the identical chemical nature of these species (Fig. 6b). Based on the empirical correlation between the MW and average charge state of the protein ions observed in native MS, the average MW of the larger species was estimated to be twofold that of the smaller species. Thus, we attributed the two signal distributions to TREM-1 monomer and dimer. The tailing feature of the second signal distribution implies the possible presence of larger oligomers (Fig. 6a, lower panels). Figure 6c illustrates the dependence of the level of dimerization on the TREM-1 concentration. The relative abundance of dimeric TREM-1 increased rapidly with the increase in protein concentration below 30 µM. At ~12 µM, 50% of TREM-1 molecules were in the dimeric state. Under these conditions, the addition of increasing concentrations, from 6 to 24 µM, of LR12 peptide did not impact the fractional mass of the dimer (Fig. 6d).
a Native mass spectra of hTREM-1-ECD(21–200) at different concentrations. Signals representing monomeric and dimeric proteins are highlighted in orange and blue, respectively. (TREM-1-ECD(21–200) displays three predicted sites of N-glycosylation. Indeed, we observed that the high micro-heterogeneity of protein glycosylation resulted in overlapping signals from a wide distribution of glycoforms carrying different charges, making the charge states of TREM-1 ions unresolvable.) b MS signals of monomeric and dimeric hTREM-1-ECD(21–200): b, 1 native mass spectrum of 30 µM hTREM-1-ECD(21–200). Subpopulations of protein ions represented by the first and second peaks were mass-selected for subsequent collision induced dissociation (CID), releasing series of fragments. b, 2 and b, 3 show the resulting patterns of fragmentation derived from the corresponding mass selections. c Fractional mass of hTREM-1-ECD(21–200) dimer as a function of the protomer concentration of TREM-1. The dashed line indicates the level where half of TREM-1 molecules populates the dimeric state. d Fractional mass of hTREM-1-ECD(21–200) dimer as a function of the protomer concentration of TREM-1 (12 µM) in the presence of the LR12 peptide at different concentrations. The dashed line indicates the dimerization level in the absence of any peptide. The inset shows an exemplary mass spectrum of 12 µM TREM-1 in the presence of 18 µM LR12 peptide. e Analysis by CovalX HM4 system high-mass detector of hTREM-1-ECD(21–200) (0.250 mg/ml, 7.5 µM) without crosslinking (upper panel, in black) and after chemical crosslinking (lower panel, in red). Data information: data are representative of at least three independent experiments
We then used a crosslinking approach to further confirm the TREM-1 quaternary structure in solution. Using a combination of chemical crosslinking and high-mass MALDI MS analysis allows for an accurate characterization of non-covalent protein complexes.35,36 In a control experiment, hTREM-1-ECD(21–200) without crosslinking was detected only as a monomer of 32.54 kDa (Fig. 6e, upper panel). No other species were detected in the control experiment. After crosslinking (Fig. 6e, lower panel), two additional species were detected compared to the control experiment with m/z = 71.41 and 142.08 kDa. These species corresponded, respectively, to the non-covalent dimer and tetramer of TREM-1. The increase in TREM-1 MW after crosslinking is due to the mono-linking and intra-linking reactions that occur when crosslinking reagents are mixed with proteins. The addition of LR12 peptide at a ratio as high as 1:10 before chemical crosslinking did not induce any change in the formation of both dimeric and tetrameric oligomers of TREM-1. Indeed, for three different TREM-1 vs. LR12- or control LR12-scrambled peptide ratios (1:1; 1:5, and 1:10), we performed high-mass MALDI analysis of the TREM-1 non-covalent multimers. For each peptide and every ratio, non-covalent multimers of TREM-1 were observed (with tetramers as the maximum), and no significant difference was observed between LR12 and LR12-scr on multimerization of TREM-1 (Supplementary Figure 4a and b). The lower-mass spectrometric signal/noise ratio observed for these spectra was mainly due to the presence of high concentrations of peptides (to a maximum of 80 µM).
Altogether, these results strongly suggest that TREM-1-ECD is able to homo-oligomerize in a concentration-dependent manner. The TREM-1 inhibitory peptide LR12 does not directly prevent TREM-1 homo-oligomerization in solution.
The TREM-1 inhibitor LR12 is able to decrease TREM-1 dimerization at the cell surface
Peptidoglycan recognition protein 1 (PGLYRP1) has been recently suggested to be a TREM-1 ligand.31 We evaluated whether PGLYRP1 supplementation on resting and LPS-activated monocytes impacted TREM-1 dimerization. We did not observe any significant effects of PGLYRP1 when added either to resting or LPS-activated cells (Supplementary figure 5b).
We have previously shown that LPS-activated TREM-1-expressing myeloid cells were able to release an as-yet-unknown TREM-1 ligand, which in turn can activate TREM-1 in an autocrine manner.18 The addition of the TREM-1 antagonist peptide LR12 was able to decrease the activation of myeloid cells by specifically inhibiting TREM-1. The mechanism of action of this peptide has been attributed to competition with the TREM-1 ligand.18 We therefore used LR12 to evaluate the impact of the TREM-1-ligand on TREM-1 multimerization.
First, we assessed the impact of LR12 on both TREM-1 expression and clustering on neutrophils and monocytes by co-incubation during LPS stimulation. We observed that LR12 had no impact either on TREM-1 membrane expression (Fig. 7a) or on TREM-1 clustering (Fig. 7b) on both neutrophils and monocytes after a 3 or 24-h stimulation with LPS, respectively. Furthermore, we showed by surface plasmon resonance that LR12 had no impact on the binding of PLA antibodies on hTREM-1 (supplementary figure 5a). However, a significant decrease in TREM-1/TREM-1 interactions was observed in both neutrophils and monocytes when co-stimulated with LPS and LR12 compared to LPS alone (Fig. 7c).
a–c Isolated human primary neutrophils (upper panels) and monocytes (lower panels) were incubated, respectively, during 3 or 24 h in resting conditions or stimulated with LPS (100 ng/ml) and/or with LR12 peptide (25 µg/ml) when indicated. a TREM-1 expression by FACS on neutrophils and monocytes. b TREM-1 (green) and nucleus (TO-PRO-3, blue) staining by confocal microscopy (scale bar: 10 µm). c TREM-1/TREM-1 interactions (scale bar: 10 µm). Data information: data are representative of at least three independent experiments. MFI mean fluorescence intensity, ns, nonsignificant. **p < 0.01, ***p < 0.001 vs. resting or as indicated, as determined by the two-tailed Student’s t-test
These results show that LR12 has no effect on TREM-1 expression and clustering but is able to induce a decrease in TREM-1 homo-oligomerization at the cell surface of LPS-activated monocytes and neutrophils. LR12 competition with the interaction between TREM-1 and its ligand further suggests that the ligand could promote TREM-1 homo-oligomerization.
DAP12 stabilizes TREM-1 surface expression and multimerization
Considering that the adapter protein DAP12 is essential for TREM-1 activation, we assessed whether DAP12 was involved in TREM-1 mobilization and multimerization at the membrane. Indeed, TREM-1 and DAP12 colocalize in resting neutrophils and LPS-activated monocytes and neutrophils (Supplementary Figure 5, 6a and b).
The expression of DAP12 was knocked down using siRNA in both primary monocytes and neutrophils. A decrease of 60% in DAP12 mRNA and 30% in DAP12 protein levels was achieved with the DAP12-specific siRNA in both cell types compared to control siRNA (Fig. 8a). TREM-1 expression in resting and LPS-activated cells was then measured. Whereas in resting monocytes DAP12 knockdown induced no change in TREM-1 expression, a decrease in LPS-induced TREM-1 upregulation was found (Fig. 8b). TREM-1 expression was decreased in both resting and LPS-activated neutrophils when DAP12 was knocked down, suggesting that DAP12 is able to stabilize TREM-1 expression in these cells (Fig. 8c). Furthermore, even after upregulation of TREM-1 in LPS-treated monocytes, DAP12 knockdown was also associated with a decrease in TREM-1 dimerization (Fig. 8d).
a Normalized DAP12 mRNA and protein quantification in native and DAP12-silenced conditions. b–d Isolated human primary neutrophils and monocytes were incubated 12 h with siRNAs and incubated during 24 h in resting conditions or stimulated with LPS (100 ng/ml). b TREM-1 expression on monocytes. c TREM-1 expression on neutrophils. d TREM-1/TREM-1 interactions (red blobs) and nucleus (blue) on monocytes after 24 h by confocal microscopy (scale bar: 10 µm). Data information: data are representative of at least three different experiments. ns nonsignificant. *p < 0.05, **p < 0.01, ***p < 0.001 vs. resting or as indicated, as determined by the two-tailed Student’s t-test
Altogether, these data suggest that DAP12 is not only involved in the downstream signaling of TREM-1 but is also able to stabilize TREM-1 expression at the cell surface, favoring its multimerization.
DISCUSSION
TREM-1 associates with DAP12, an ITAM-bearing transmembrane signaling dimer, to initiate intracellular signaling cascades, resulting in amplification of the inflammatory reaction, especially through synergism with TLR signaling. However, the molecular mechanisms by which TREM-1 is activated are still elusive, including the nature and valence of its ligand and avidity of TREM-1 for its ligand. Fortin et al.16 observed that the engagement of TREM-1 by the agonistic αTREM-1 clone 193015 elicits similar intracellular pathways in monocytes and neutrophils. These pathways include phosphorylation of SYK and PLCγ and the release of inflammatory chemokines and cytokines.
We used these different early and late specific readouts of TREM-1 activation to show that TREM-1 needs to multimerize to be active in the U937-TD monocytic cell line. Indeed, a multivalent ligation of TREM-1 induced TREM-1 activation. By contrast, monovalent triggering of TREM-1 elicited a weak or no response, leading to the conclusion that TREM-1 crosslinking is critical for its activation. Moreover, TREM-1-mediated U937-TD activation increased in a dose-dependent manner with the concentration of αTREM-1 and GαM. These data are consistent with recent findings by Read et al., who identified PGLYRP1 as a potential ligand for TREM-1. They demonstrated that even if PGLYRP1 was able to bind to TREM-1, it was not able to activate it and required crosslinking by peptidoglycan to induce TREM-1 activation in a reporter cell system expressing a TREM-1:DAP12 fusion protein.31 These results could have been due to a lack of cell-induced TREM-1 multimerization by the reporter cells, but the same results were obtained with primary monocytes and neutrophils.
We then confirmed these results under more physiological conditions by using primary human monocytes and neutrophils. Under resting conditions, TREM-1 is expressed at low levels in monocytes and high levels in neutrophils. Only a limited in situ PLA signal was measured in both cell types, suggesting that TREM-1 is predominantly monomeric in non-activated conditions. Indeed, TREM-1 labeling in these resting cells was diffuse at the membrane, and monovalent stimulation of TREM-1 was not able to induce activation of TREM-1. Moreover, divalent ligation of TREM-1 was also not able to induce activation of TREM-1 without a pre-stimulation step with LPS. These results suggests that LPS-induced clustering and dimerization of TREM-1 at the membrane of both monocytes and neutrophils is required for TREM-1 activation. TREM-1 was previously found to be rapidly mobilized in lipid rafts after TLR4 activation in neutrophils, a phenomenon expected to facilitate the assembly of extracellular and intracellular TLR4 and TREM-1-pathway mediators.16 We indeed observed a colocalization of TREM-1 and DAP12 after LPS activation. Under these LPS-induced conditions, in which TREM-1 clustering and dimerization were observed in monocytes and neutrophils, the monovalent agonist Fab was able to induce activation of TREM-1, similar to multivalent ligation of the receptor. We thus postulate that LPS activation of primary human monocytes and neutrophils induces a clustering of TREM-1 that promotes TREM-1 multimerization required for its activation.
Our results suggest that TREM-1 is quickly activated in neutrophils, whereas a two-step activation is necessary for monocytes, requiring upregulation of TREM-1 expression at the membrane prior to clustering and multimerization. This differential regulation of TREM-1 expression and activation on monocytes and neutrophils is consistent with previous findings by Prüfer et al.,37 who found significant differences in the function of TREM-1 in human neutrophils and monocytes. Consistently, neutrophils are known to be rapidly mobilized and the first leukocytes recruited to inflammatory sites38 within the first 2–3 h following tissue injury.39 Compared to neutrophils, monocytes are recruited as a second wave of leukocytes to sites of injury within 1–7 days.40 Rapid TREM-1 mobilization at the cell surface of neutrophils could therefore be a key mechanism for the rapid recruitment and migration of neutrophils to injured tissues through ligand-sensing.
Radaev et al.26 showed by structural analysis that the recombinant hTREM-1-ECD(17-133), produced in E. coli as inclusion bodies and then reconstituted in vitro, displays a potential intrinsic ability to oligomerize and forms a “head-to-tail” dimer. However, the physiological relevance of such a head-to-tail dimer of TREM-1 in the context of our findings remains obscure. Controversially, Kelker et al.25 have found different fragments of hTREM-1-ECD to be in a monomeric state. In particular, they showed by analytical ultracentrifugation that recombinant hTREM-1-ECD(21–194), purified from E. coli inclusion bodies with a similar protocol, was only present as a monomer in solution. The authors eluted the in vitro refolded protein at a lower retention volume in gel filtration experiments by secondary structure constraints. We produced the same protein in an E. coli expression system, however in a soluble form, and observed the same apparent shift in retention time. We attributed this shift to the formation of a dimer of TREM-1 that could be dissociated both by dilution before gel filtration and using ITC. The appearance of oligomeric states of TREM-1 in solution was definitively confirmed by native MS and after chemical crosslinking of TREM-1 by using a recombinant full-length hTREM-1-ECD produced in a mammalian cell system. Even if the precise orientation of the TREM-1 ectodomains within homo-oligomers cannot be characterized by these approaches, we confirmed that TREM-1 was in a concentration-dependent dynamic equilibrium between monomeric and homo-oligomeric states in solution with a CTC between 7 and 12 µM by using two different recombinant forms of hTREM-1-ECD either produced as soluble proteins in E. coli or in mammalian cells.
Different studies suggest that 35% of cell proteins are oligomeric and mainly homo-oligomeric41 with the average oligomeric state being tetrameric.42 TREM-1 appears to have a dissociation constant within the micromolar range in vitro,which is considered to be a weak affinity. However, environmental conditions can impact oligomerization. The phenomena of TREM-1 upregulation and clustering are thus believed to allow a local increase in receptor concentration at the membrane and to promote its transient oligomerization. Other parameters seem to also influence TREM-1 oligomerization, such as DAP12 expression. DAP12 was indeed found to impact the expression and surface stability of Ig-like receptors.43 Similarly, DAP12 silencing was associated with a decrease in both TREM-1 upregulation and oligomerization at the membrane. We have previously shown that LPS stimulation of human neutrophils induces the release of an endogenous ligand of TREM-1 and that incubation with LR12 is able to dose-dependently decrease the binding of the ligand to the TREM-1 receptor,18 resulting in a decrease of LPS-mediated cell activation via functional inhibition of downstream signaling to TREM-1, which confirms that this endogenous ligand released after LPS activation is able to functionally activate TREM-1. The LR12 peptide had no effect either on TREM-1 clustering at the membrane or on its dimerization in solution. However, LR12 was able to strongly decrease the level of TREM-1 oligomerization at the membrane. This suggests that the binding of the natural endogenous ligand of TREM-1 could impact TREM-1 dimerization. The LR12 peptide is representative of a region on TREM-1 located in the F-G loop of the Ig-like domain,25 a region identified as a potential ligand-binding site by surfaces and electrostatic potential calculations and prone to conformational changes upon ligand binding. Conformational changes following ligand-binding could promote and stabilize TREM-1 multimerization. Thus, the decrease in TREM-1 oligomerization at the membrane of activated cells in the presence of LR12 provides evidence for a ligand-induced receptor-mediated dimerization of TREM-1. Supplementation of the newly identified TREM-1 ligand, PGLYRP1, had no impact on TREM-1 dimerization, which suggests that another ligand is released after LPS activation. This result is consistent with the literature, since PGLYRP1 has been identified to be released and to activate TREM-1 only after activation of primary human cells with PeptidoGlycan.31
This study sheds new light on the mechanism of TREM-1 receptor activation following stimulation of human monocytes and neutrophils through TLR4 by combining biochemical, biophysical, and cellular approaches. We have previously shown that LR12 is able to dampen LPS activation of leukocytes by specifically inhibiting TREM-1-ligand–TREM-1-mediated signals, which is associated with a decrease in the activation of downstream signaling to TREM-1.18 This study sheds new light on the underlying mechanism by which it can inhibit TREM-1. The work presented here is also a first step toward understanding the molecular mechanisms leading to the interaction of TREM-1 and its ligand, as this ligand is still elusive despite the well-characterized role of TREM-1 in inflammatory diseases. In addition to the paracrine effect of TREM-1 ligand release, the potential contribution of cytoskeleton-related kinase activation has not been evaluated and could be further studied.
References
Bouchon, A., Dietrich, J. & Colonna, M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J. Immunol. 164, 4991–4995 (2000).
Jolly, L. et al. Triggering receptor expressed on myeloid cells-1: a new player in platelet aggregation. Thromb. Haemost. 117, 1772–1781 (2017).
Derive, M., Massin, F. & Gibot, S. Triggering receptor expressed on myeloid cells-1 as a new therapeutic target during inflammatory diseases. Self Nonself 1, 225–230 (2010).
Tammaro, A. et al. TREM-1 and its potential ligands in non-infectious diseases: from biology to clinical perspectives. Pharmacol. Ther. 177, 81–95 (2017).
Bouchon, A., Facchetti, F., Weigand, M. A. & Colonna, M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 410, 1103–1107 (2001).
Boufenzer, A. et al. TREM-1 mediates inflammatory injury and cardiac remodeling following myocardial infarction. Circ. Res 116, 1772–1782 (2015).
Zysset, D. et al. TREM-1 links dyslipidemia to inflammation and lipid deposition in atherosclerosis. Nat. Commun. 7, 13151 (2016).
Joffre, J. et al. Genetic and pharmacological inhibition of TREM-1 limits the development of experimental atherosclerosis. J. Am. Coll. Cardiol. 68, 2776–2793 (2016).
Tessarz, A. S. & Cerwenka, A. The TREM-1/DAP12 pathway. Immunol. Lett. 116, 111–116 (2008).
Ormsby, T. et al. Btk is a positive regulator in the TREM-1/DAP12 signaling pathway. Blood 118, 936–945 (2011).
McVicar, D. W. et al. DAP12-mediated signal transduction in natural killer cells. A dominant role for the Syk protein-tyrosine kinase. J. Biol. Chem. 273, 32934–32942 (1998).
Radsak, M. P., Salih, H. R., Rammensee, H.-G. & Schild, H. Triggering receptor expressed on myeloid cells-1 in neutrophil inflammatory responses: differential regulation of activation and survival. J. Immunol. 172, 4956–4963 (2004).
Klesney-Tait, J. & Colonna, M. Uncovering the TREM-1-TLR connection. Am. J. Physiol. Lung Cell Mol. Physiol. 293, L1374–L1376 (2007).
Gibot, S. Clinical review: Role of triggering receptor expressed on myeloid cells-1 during sepsis. Crit. Care 9, 485 (2005).
Knapp, S. et al. Cutting edge: expression patterns of surface and soluble triggering receptor expressed on myeloid cells-1 in human endotoxemia. J. Immunol. 173, 7131–7134 (2004).
Fortin, C. F., Lesur, O. & Fulop, T. Effects of TREM-1 activation in human neutrophils: activation of signaling pathways, recruitment into lipid rafts and association with TLR4. Int. Immunol. 19, 41–50 (2007).
Ornatowska, M. et al. Functional genomics of silencing TREM-1 on TLR4 signaling in macrophages. Am. J. Physiol. Lung Cell Mol. Physiol. 293, L1377–L1384 (2007).
Derive, M. et al. Soluble TREM-like transcript-1 regulates leukocyte activation and controls microbial sepsis. J. Immunol. 188, 5585–5592 (2012).
Derive, M., Boufenzer, A. & Gibot, S. Attenuation of responses to endotoxin by the triggering receptor expressed on myeloid cells-1 inhibitor LR12 in nonhuman primate. J. Am. Soc. Anesthesiol. 120, 935–942 (2014).
Derive, M. et al. Effects of a TREM-like transcript 1-derived peptide during hypodynamic septic shock in pigs. Shock 39, 176–182 (2013).
Tessarz, A. S. et al. Non-T cell activation linker (NTAL) negatively regulates TREM-1/DAP12-induced inflammatory cytokine production in myeloid cells. J. Immunol. 178, 1991–1999 (2007).
Lee, B. et al. 1α,25-Dihydroxyvitamin D3 upregulates HIF-1 and TREM-1 via mTOR signaling. Immunol. Lett. 163, 14–21 (2015).
Wenzel, J. et al. Measurement of TLR-induced macrophage spreading by automated image analysis: differential role of Myd88 and MAPK in early and late responses. Front. Physiol. 2, 71 (2011).
Leuchowius, K.-J., & Weibrecht, I. & Söderberg, O. In situ proximity ligation assay for microscopy and flow cytometry. Curr. Protoc. Cytom. Chapter 9, Unit 9.36 (2011).
Kelker, M. S. et al. Crystal structure of human triggering receptor expressed on myeloid cells 1 (TREM-1) at 1.47 A. J. Mol. Biol. 342, 1237–1248 (2004).
Radaev, S., Kattah, M., Rostro, B., Colonna, M. & Sun, P. D. Crystal structure of the human myeloid cell activating receptor TREM-1. Structure 11, 1527–1535 (2003).
van den Heuvel, R. H. H. et al. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal. Chem. 78, 7473–7483 (2006).
Snijder, J. & Heck, A. J. R. Analytical approaches for size and mass analysis of large protein assemblies. Annu Rev. Anal. Chem. Palo Alto Calif. 7, 43–64 (2014).
Wang, G. et al. Molecular basis of assembly and activation of complement component C1 in complex with immunoglobulin G1 and antigen. Mol. Cell 63, 135–145 (2016).
Wang, G., Johnson, A. J. & Kaltashov, I. A. Evaluation of electrospray ionization mass spectrometry as a tool for characterization of small soluble protein aggregates. Anal. Chem. 84, 1718–1724 (2012).
Read, C. B. et al. Cutting Edge: identification of neutrophil PGLYRP1 as a ligand for TREM-1. J. Immunol. 194, 1417–1421 (2015).
Barranco-Medina, S., Kakorin, S., Lázaro, J. J. & Dietz, K.-J. Thermodynamics of the dimer-decamer transition of reduced human and plant 2-cys peroxiredoxin. Biochemistry 47, 7196–7204 (2008).
Burrows, S. D. et al. Determination of the monomer-dimer equilibrium of interleukin-8 reveals it is a monomer at physiological concentrations. Biochemistry 33, 12741–12745 (1994).
Luke, K., Apiyo, D. & Wittung-Stafshede, P. Dissecting homo-heptamer thermodynamics by isothermal titration calorimetry: entropy-driven assembly of co-chaperonin protein 10. Biophys. J. 89, 3332–3336 (2005).
Nazabal, A., Wenzel, R. J. & Zenobi, R. Immunoassays with direct mass spectrometric detection. Anal. Chem. 78, 3562–3570 (2006).
Bich, C. et al. Characterization of antibody-antigen interactions: comparison between surface plasmon resonance measurements and high-mass matrix-assisted laser desorption/ionization mass spectrometry. Anal. Biochem. 375, 35–45 (2008).
Prüfer, S. et al. Distinct signaling cascades of TREM-1, TLR and NLR in neutrophils and monocytic cells. J. Innate Immun. 6, 339–352 (2014).
Kolaczkowska, E. & Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013).
McDonald, B. et al. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science 330, 362–366 (2010).
Libby, P., Nahrendorf, M. & Swirski, F. K. Leukocytes link local and systemic inflammation in ischemic cardiovascular disease: an expanded ‘cardiovascular continuum’. J. Am. Coll. Cardiol. 67, 1091–1103 (2016).
Goodsell, D. S. & Olson, A. J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29, 105–153 (2000).
Goodsell, D. S. Inside a living cell. Trends Biochem. Sci. 16, 203–206 (1991).
Mulrooney, T. J., Posch, P. E. & Hurley, C. K. DAP12 impacts trafficking and surface stability of killer immunoglobulin-like receptors on natural killer cells. J. Leukoc. Biol. 94, 301–313 (2013).
Acknowledgements
We thank Sebastien Hupont and Dominique Dumas from the Cellular imaging plateform (PTIBC) at Lorraine University for providing us full access to confocal microscopy and for their technical support. We also thank Jean-Marc Alberto from Inserm U954 for his help with the in situ proximity ligation assay experiments and analysis with the Blobfinder software. We thank Dr. Hortense Mazon for electrospray ionization-mass spectrometry spectra carried out in the Mass Spectometry platform of Université de Lorraine (SCMS). Microcalorimetry and surface plasmon resonance were performed at the Molecular Interactions and Biophysics platform of Université de Lorraine (SCBIM). G.W. and A.J.R.H. are supported by the Roadmap Initiative Proteins@Work (project number 184.032.201), funded by the Netherlands Organization for Scientific Research (NWO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.D. and S.G. are co-founders of Inotrem SA, a Company developing TREM-1 inhibitors. P.L. is co-founder of Inatherys SA, a Company developing therapeutic monoclonal antibodies. A.N. is co-founder of CovalX GmbH, a Company providing services for the analysis of protein complexes. The remaining authors declare that they have no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Carrasco, K., Boufenzer, A., Jolly, L. et al. TREM-1 multimerization is essential for its activation on monocytes and neutrophils. Cell Mol Immunol 16, 460–472 (2019). https://doi.org/10.1038/s41423-018-0003-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-018-0003-5
- Springer Nature Limited