Abstract
Glioblastoma (GBM) is the most aggressive malignant primary brain tumor characterized by a highly heterogeneous and immunosuppressive tumor microenvironment (TME). The symbiotic interactions between glioblastoma stem cells (GSCs) and tumor-associated macrophages (TAM) in the TME are critical for tumor progression. Here, we identified that IFI35, a transcriptional regulatory factor, plays both cell-intrinsic and cell-extrinsic roles in maintaining GSCs and the immunosuppressive TME. IFI35 induced non-canonical NF-kB signaling through proteasomal processing of p105 to the DNA-binding transcription factor p50, which heterodimerizes with RELB (RELB/p50), and activated cell chemotaxis in a cell-autonomous manner. Further, IFI35 induced recruitment and maintenance of M2-like TAMs in TME in a paracrine manner. Targeting IFI35 effectively suppressed in vivo tumor growth and prolonged survival of orthotopic xenograft-bearing mice. Collectively, these findings reveal the tumor-promoting functions of IFI35 and suggest that targeting IFI35 or its downstream effectors may provide effective approaches to improve GBM treatment.
Similar content being viewed by others
Introduction
Glioblastoma (GBM), classified as World Health Organization grade IV glioma, is incurable and the most aggressive primary intracranial tumor, with a median survival of 14.6 months and a 5-year survival of 7% [1, 2]. The outcome of GBM patients is poor, despite the standard treatments of surgery, radiotherapy, and chemotherapy [3, 4]. Multiple studies using single-cell RNA sequencing analyses among other approaches have identified high levels of intra- and inter-tumoral heterogeneity and distinct cellular hierarchy [5,6,7]. The Glioblastoma stem cells (GSCs), sit on the apex of cellular hierarchy in GBM tumors and are defined by their self-renewal and tumor initiation capability. The infiltrative and migratory nature of glioblastoma stem cells contribute to tumor initiation, recurrence, and resistance to conventional therapies pose challenges in effectively eliminating this lethal cancer [8, 9].
GSCs respond to the changes in nutrient state and oxygen levels in tumor microenvironment, and modify their epigenome and gene expression program to maximize their fitness and survival [10, 11]. Unlike their normal counterparts, GSCs are small subpopulation of tumor cells expressing stemness-related markers, are capable of initiating tumors, and recapitulate tumor heterogeneity in serial transplantation assays in vivo [12]. Given the critical role of GSCs in the initiation and maintenance of GBM tumors, selectively targeting and ablating these tumor-initiating and -propagating populations may provide a potential target for glioblastoma treatment.
Inter- and intra-tumoral heterogeneity poses a major challenge to the development of targeted and immunotherapy for GBM treatment. The intracranial microenvironment (TME), in which tumor cells proliferate, adds to the diversity of inter- and intra-tumor heterogeneity of GBM [13]. The glioblastoma TME contains non-neoplastic cells, including infiltrating and resident immune cells, vascular components (e.g., endothelial cells and pericytes), and other brain cells (e.g., astrocytes, oligodendrocytes, and neurons) [14]. The understanding of the interactions between GBM cells and their TME is critical and is currently lacking. Emphasis has been placed on the immune system component of TME, particularly tumor-associated macrophages (TAMs). GSCs interact with surrounding TAMs to remodel the microenvironment, and in turn receive critical maintenance cues from their respective niches. Clinical progress remains restricted because of the heterogeneity and plasticity of the TME. Disrupting GSCs and their interactions with the intricate constituents of the TME may provide potentially useful therapeutic targets in GBM.
Interferon-induced protein 35 (IFI35) was originally identified as an interferon-inducible protein functioning in transcriptional regulation and inflammatory immune responses [15,16,17], but its biological and pathological mechanism in GBM remains elusive. Here, we provide evidence that IFI35 contributes to GSC proliferation and self-renewal and explore the mechanisms by which IFI35 supports GSCs and recruits TAMs. This study reveals new therapeutically available targets at the cell-autonomous and cell-extrinsic levels relying on IFI35.
Results
Combined multiomics analyses reveal a preferential expression of IFI35 in GSCs and GBM
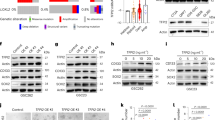
To identify genes specifically expressed in GBM and GSCs, we performed an integrative multiomics analysis of the Cancer Genome Atlas (TCGA) dataset of 528 GBMs and 10 non-tumor samples, proteomics data of 8 GSCs and 2 NSCs, and RNA sequencing data from 41 GSCs and 5 NSCs (Fig. 1A–D) [18]. We identified IFI35 as the solitary gene specifically enriched both in GBM specimens, and GSCs. Furthermore, we interrogated single-cell RNA sequencing data and observed the high enrichment of IFI35 in GBM tumor cells (Fig. 1E and Fig. S1A–C). We observed an 82.34% overlap of IFI35 expression with the stem cell marker SOX2 positive cells in single-cell RNA sequencing data, suggesting specific expression in stem-like populations (Fig. 1F, G). We thus hypothesized that IFI35 is preferentially expressed in GSCs. Quantitative reverse transcription (RT)-PCR and immunoblot analysis confirmed that the expression level of IFI35 was significantly elevated in 8 GSC compared to NSC lines (Fig. 1H, I). Further, we compared a cohort of nine glioblastomas with three non-tumor brain tissues. IFI35 mRNA and protein levels were elevated in glioblastomas compared to non-tumor brain tissue (Fig. 1H, I). Immunofluorescent staining with IFI35 and the GSC marker SOX2, further confirmed that IFI35 was enriched in patient-derived GSCs (839 and 2907) compared with NSCs (ENSA and HNP1; Fig. 1J). Moreover, there was a strong overlap between IFI35 and SOX2 positive tumor cells in GBM surgical specimens (Fig. 1K). Collectively, these data demonstrate that IFI35 is preferentially expressed in GBM and enriched in GSCs, suggesting that it could contribute to GBM pathology.
A Heatmap of differentially expressed genes (average change more than fourfold, adjusted P value < 0.05) in 528 GBMs and 10 non-tumors (TCGA_GBM HG-U133A). B Volcano plot of differentially expressed genes in eight GSCs and two NSCs using quantifies proteome sequence. As indicated by the dotted lines, cutoff was set as fold change >4, adjusted P value < 0.05. C Heatmap of differentially expressed genes (average change more than fourfold, adjusted P value < 0.05) in 41 GSCs and 5 NSCs using RNA sequencing [18]. D Venn diagram shows the overlap of TCGA GBM dataset, proteomics data of GSCs and NSCs, and RNA sequencing for GSCs and NSCs analysis for the genes. E Annotated cell clusters were visualized in a uniform manifold approximation and projection (UMAP) representation derived from GSE141383. F The overlap of IFI35 and SOX2 positive cells with analysis of single-cell RNA-seq of GEO database (GSE141383). G Quantitation of IFI35 and SOX2 positive cells with analysis of single-cell RNA-seq of GEO database (GSE141383). H qRT-PCR quantification of IFI35 mRNA levels in GSCs and NSCs (left), glioblastomas and normal brain tissues (right). Data were determined by unpaired t test and shown as mean ± SD. *P < 0.05, **P < 0.01. I Protein levels of IFI35 were assessed by immunoblotting in NSCs (HNP1 and ENSA) and GSCs (4121, 3028, 839, 2907, 387, CW738, MGG8, and RKI) (top), glioblastoma and normal brain tissue (bottom). J Immunofluorescent staining of IFI35 (green) and SOX2 (red) in 839 and 2907 GSCs and matched ENSA and HNP1 NSCs. Scale bar, 10 μm. K Immunofluorescent staining shows the fraction of IFI35+ cells in SOX2+ cells in GBM patients from glioblastoma specimens. Scale bar, 50 μm.
IFI35 supports GSC proliferation and self-renewal
To investigate the role of IFI35 in GSC proliferation and self-renewal, we performed loss-of-function and overexpression studies in our GSC models. Silencing IFI35 using two nonoverlapping short hairpin RNAs (shRNA) in two GSC models (839 and 2907 GSCs) reduced the mRNA and protein levels of IFI35 in GSCs (Fig. 2A, B). Knockdown of IFI35 decreased GSC proliferation, cell viability, and self-renewal, as measured by decreased neurosphere formation capability and in vitro limiting dilution assays (Fig. 2C–H). In addition, overexpression of IFI35 promoted GSC proliferation (Fig. 2I and Fig. S2A). To establish the specificity of the effects of IFI35 knockdown, we reconstituted the IFI35 expression in IFI35-depleted cells, restoring the decreased cell viability of GSCs (Fig. 2J and Fig. S2B). IFI35 targeting using shRNA transduction decreased the mRNA expression and protein levels of the GSC markers, SOX2, OLIG2, and MYC (Fig. S2C, D). Collectively, these data support a cell-intrinsic function of IFI35 to support GSC maintenance.
A IFI35 mRNA levels were assayed by qRT-PCR in GSCs with IFI35 knockdown using two independent shRNAs. A shCONT was used as the control. Data were determined by ANOVA and shown as mean ± SD from three independent experiments. **P < 0.01. B The protein levels of IFI35 after transduction in 839 and 2907 GSCs with two independent shRNAs. A shCONT was used as the control. Two independent shRNAs targeting IFI35 decreased the growth and self-renewal of GSCs compared with shCONT, as measured by cell number count (C), CellTiter-Glo assay (D), and sphere number quantification (E). Data are presented as mean ± SD from six independent experiments. **P < 0.01, one-way ANOVA with Dunnett’s multiple-comparison test. F The extreme limiting dilution assays revealed that knockdown of IFI35 in 839 (top) and 2907 (bottom) decreased the sphere formation. **P < 0.01. G, H Representative images of neurospheres of 839 and 2907 expressing shCONT, shIFI35-1, or shIFI35-2. Scale bar, 100 μm. Each image is representative of at least six independent experiments. I Overexpression of IFI35 increased the growth of 839 and 2907 GSCs compared with vector. Results were measured by a CellTiter-Glo assay. Data were determined. Data are presented as mean ± SD from six independent experiments. *P < 0.05, **P < 0.01, one-way ANOVA with Dunnett’s multiple-comparison test. J Overexpression of IFI35 rescued the decreased GSC growth caused by shRNA-mediated knockdown. Results were measured by a CellTiter-Glo assay. Data are presented as mean ± SD from six independent experiments. **P < 0.01, one-way ANOVA with Dunnett’s multiple-comparison test.
IFI35 interacts with TRIM21 to induce proteasomal processing of NFKB1 p105 to p50 in GSCs
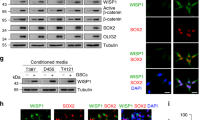
To understand the mechanism of IFI35 in maintaining GSCs, we performed immunoprecipitation followed by mass spectrometry using an anti-IFI35 antibody from cell lysates derived from 839 GSCs. By combining mass spectrometry data with IFI35-correlated genes in TCGA GBM dataset, TRIM21 was identified as top ranked IFI35-correlated gene which also showed a physical interaction with IFI35 at protein level. These observations suggest that TRIM21 might be a crucial factor in downstream IFI35 signaling (Fig. 3A–C). We further determined the correlations with IFI35-regulated KEGG pathway enrichment analysis in TCGA GBM database, demonstrating a high positive correlation between IFI35 and the NF-κB signaling pathway (Fig. 3D). Simultaneously, IFI35 showed an expression correlation with NFKB1 in TCGA dataset and the mass spectrometry data demonstrated that IFI35 directly interacted with NFKB1 (Fig. 3E, F). TRIM21 is an E3 ubiquitin ligase, involved in ubiquitination and proteasomal degradation of its targets [19]. We speculated that IFI35 may regulate the ubiquitination of NFKB1 through interaction with TRIM21, resulting in activation of the NF-κB signaling pathway. Based on Gene set enrichment analysis (GSEA) of genes correlated with IFI35 expression in the TCGA GBM mRNA expression database, IFI35 and TRIM21 positively correlated with NF-κB signaling pathway and ubiquitin dependent proteasomal protein catabolic process (Fig. 3G, H). Furthermore, we confirmed the interaction of IFI35, NFKB1, and TRIM21 in two patient-derived GSCs by immunoprecipitation with anti-IFI35, anti-NFKB1, or anti-TRIM21 antibodies (Fig. 3I). IFI35 regulates GSC biological function through TRIM21. Silencing TRIM21 reduced GSC proliferation as measured by cell number count, CellTiter-Glo assay, neurosphere formation capability, and in vitro limiting dilution assays (Fig. S3A–F). Overexpression of TRIM21 increased GSC proliferation (Fig. S3G, H). Moreover, silencing IFI35 reduced the interaction of TRIM21 with NFKB1 p105/p50 (Fig. S3I). And knocking down of TRIM21 affected the binding of IFI35 with NFKB1 p105/p50 (Fig. S3J). Immunofluorescence assays demonstrated that TRIM21 co-localized with IFI35 in the cytoplasm of GSCs, and NFKB1 co-localized with IFI35 both in the cytoplasm and nucleus of GSCs (Fig. 3J).
A Venn diagram shows the overlap of proteins identified by immunoprecipitation followed by mass spectrometry using anti-IFI35 antibody from cell lysates of 839 GSC and correlation with IFI35 in TCGA GBM dataset. B The correlation analysis of IFI35 with TRIM21 in TCGA GBM dataset. C Representative peptide of TRIM21 identified by immunoprecipitation followed by mass spectrometry using anti-IFI35 antibody in 839 GSC cell lysates. D Top 10 of KEGG signaling pathway enrichment analysis related with IFI35 expression in TCGA GBM dataset. E The correlation analysis of IFI35 with NFKB1 in TCGA GBM dataset. F Representative peptide of NFKB1 identified by immunoprecipitation followed by mass spectrometry using anti-IFI35 antibody in 839 GSC cell lysates. Gene Set Enrichment Analysis (GSEA) using Gene Ontology (GO) gene sets showing IFI35 (G) and TRIM21 (H) expression correlates with a transcriptional signature of positive regulation of NF-κB signaling and dependent proteasomal ubiquitin dependent protein catabolic process signaling in TCGA GBM dataset. I Immunoblot of immunoprecipitation with anti-IFI35, anti-NFKB1, anti-TRIM21 antibody in 839 and 2907 GSCs. The nonspecific IgG is the control. J The confocal image of IFI35, NFKB1, and TRIM21 distribution in 839 and 2907 GSCs. Scale bar, 10 μm.
To further investigate how IFI35 regulates NFKB1 p105/p50 through TRIM21, we silenced IFI35 or TRIM21 and examined the effect on NFKB1 in GSCs. The mRNA level of NFKB1 was not affected by knocking down of IFI35 in GSCs (Fig. 4A); however, the protein level of the NFKB1 precursor p105 was increased while the NF-κB active subunit p50 protein level was reduced (Fig. 4B). Activation of NF-κB is regulated via ubiquitin- and proteasome-mediated processes, one of which is the processing of the NFKB1 precursor p105 to the p50 active subunit [20]. We hypothesized that IFI35 may regulate the processing of p105 to p50 through ubiquitination by interacting with TRIM21. To assess this, we treated GSCs with cycloheximide (CHX), an inhibitor of de novo protein synthesis, under control conditions and following knockdown of IFI35 or TRIM21. Silencing of either TRIM21 or IFI35 decreased the proteasomal processing of p105 to p50 compared to control GSCs (Fig. 4C). Next, we co-expressed HA-tagged IFI35 or HA-tagged TRIM21 with His-ubiquitin in GSCs. After the immunoprecipitation of NFKB1 from cells treated with MG132, a proteasome inhibitor, we observed that overexpression of IFI35 or TRIM21 increased the ubiquitination level of p105. Blocking proteasomal degradation with MG132 inhibited the processing of p105 to p50 to a certain extent caused by IFI35 or TRIM21 (Fig. 4D, E and Fig. S4A, B). To examine the mechanistic basis of proteasomal degradation, we co-expressed IFI35 or TRIM21 with His-ubiquitin of wild-type ubiquitin, K48-linked, or K63-linked polyubiquitylation. Immunoprecipitation assays showed that IFI35 and TRIM21 effectively disassembled the K48-linked polyubiquitylation of NFKB1 but had no significant effect on the nondegradative K63-linked polyubiquitylation of NFKB1 (Fig. 4F). Furthermore, we expressed K48-resistant (K48R) and K63R form of ubiquitin in GSCs and found that K48R ubiquitin expression blocked the processing of p105 to p50 (Fig. 4G). Collectively, these data establish that IFI35 interacts with TRIM21 to regulate NFKB1 p105 processing through K48-linked ubiquitination and degradation leading to the formation of active and oncogenic p50 form of NFKB1.
A qRT-PCR analysis of NFKB1 mRNA expression in 839 and 2907 GSCs after transduction with shCONT, shIFI35 (left), or shTRIM21 (right). Data are presented as mean ± SD from three independent experiments. B Immunoblot analysis of IFI35, TRIM21, and NFKB1 levels in 839 and 2907 GSCs transduced with control shRNA, shIFI35 (left), or shTRIM21 (right). C 839 and 2907 GSCs expressing shCONT, shIFI35, or shTRIM21 were treated with 100 μg/ml CHX, harvested at the indicated times, and then subjected to immunoblot with antibodies against IFI35, TRIM21, and NFKB1. D, E 839 and 2907 GSCs were transduced with HA-IFI35 or HA-TRIM21, His-Ub and treated with 20 μM MG132 for 8 h. Extracts were immunoprecipitated with anti-NFKB1 antibody, followed by immunoblotting. F 839 and 2907 GSCs were co-transduced with Flag-NFKB1, HA-IFI35 or HA-TRIM21, and His-Ub, K48 or K63 plasmids for 72 h, then immunoprecipitated with anti-Flag antibody, followed by immunoblotting. G 839 and 2907 GSCs were co-transduced with Flag-NFKB1, HA-IFI35 or HA-TRIM21, and His-Ub, K48R or K63R plasmids for 72 h, then immunoprecipitated with anti-Flag antibody, followed by immunoblotting.
IFI35 signals via the RELB/p50 complex to transcriptionally regulate chemokines
To further delineate the molecular mechanisms through which IFI35 mediates its downstream effects in GSCs, we used GSEA enriched Gene Ontology (GO) analysis in TCGA GBM dataset and RNA-seq data after IFI35 knockout, and observed the overlap of IFI35-correlated pathways were highly enriched for leukocyte migration, humoral immune response, leukocyte chemotaxis, antigen processing and presentation, cell chemotaxis, and granulocyte migration (Fig. 5A–C and Fig. S5A, B). IFI35 correlates with a transcriptional signature of positive regulation of NF-κB transcription factor activity and DNA-binding transcription factor activity in TCGA GBM dataset and RNA-seq data (Fig. 5D, E and Fig. S5C). With analysis of the immunoprecipitation and mass spectrometry, we observed that IFI35 also interacted with transcription factor RELB, an important NF-κB signaling factor (Fig. 5F). IFI35 and RELB mRNA expression correlated in the TCGA GBM dataset (Fig. S5D). Knockdown of RELB in GSCs altered expression of gene sets related to chemotaxis or migration-associated genes and pathways (Fig. S5E–G). Since RELB/p50 is involved in the non-canonical pathway in NF-κB signaling [21], and our data showed that IFI35 regulates the ubiquitination of p105 to p50, we hypothesized that IFI35 may serve as a transcription co-regulatory factor and interact with RELB/p50 complex to transcriptionally regulate the target genes in GSCs. Immunoprecipitation and confocal imaging confirmed the interaction of IFI35 with RELB, IFI35 with p50, and RELB with p50 (Fig. 5G, H). We performed RNA-seq to evaluate downstream transcriptional effects of IFI35 knockout and assay for transposase-accessible chromatin with sequencing (ATAC-seq) to investigate genome wide changes in chromatin accessibility after decreasing IFI35 expression. Genes associated with cell chemotaxis displayed diminished chromatin accessibility at their transcription start sites when IFI35 was knocked out compared to control cells (Fig. 5I–K and Fig. S5H). qPCR assay confirmed the mRNA levels of cell chemotaxis-associated genes (CCL2, LOX, ANXA1, CX3CL1, WNT5A, SLIT2) were reduced upon IFI35 silencing (Fig. S5I). Furthermore, we performed co-silencing experiments, silencing IFI35 in combination with RELB, IFI35 in combination with NFKB1, and RELB with NFKB1, and found that the combined depletion of the two genes markedly reduced the mRNA levels of the downstream chemokines compared with shCONT (Fig. S5J–P). To directly confirm the binding of IFI35 with RELB/p50 complex on the promoter regions of chemokines in GSCs, we performed ChIP with IFI35, NFKB1, or RELB antibody, followed by qPCR with primer sets designed for the promoter region of chemokines (CCL2, LOX, ANXA1, CX3CL1, WNT5A, SLIT2). IFI35 along with RELB/p50 complex were enriched within the promoter regions of CCL2, LOX, ANXA1, CX3CL1, WNT5A, and SLIT2 compared with control IgG (Fig. 5L). Collectively, these results suggest that IFI35 functions in association with the RELB/P50 complex to activate the transcription of downstream chemokines in GSCs.
A Venn diagram shows the overlap of TOP 50 of GSEA enriched Gene Ontology (GO) pathways using TCGA GBM dataset of IFI35 expression and TOP 50 of GSEA enriched GO pathways after IFI35 knockout. B The bar plot shows GSEA enriched GO pathways after IFI35 knockout. C GSEA scores calculated for RNA sequencing data using GO gene sets showing IFI35 expression correlates with a transcriptional signature of cell chemotaxis signaling (left) and leukocyte chemotaxis signaling (right) in 839 GSC. D Gene Ontology (GO) pathway enrichment analysis of transcription signaling pathways related with IFI35 expression in TCGA GBM dataset. E GSEA scores calculated for TCGA GBM dataset using GO gene set showing IFI35 expression correlates with a transcriptional signature of regulation of DNA-binding transcription factor activity in 839 GSC. F Representative peptide of RELB identified by immunoprecipitation followed by mass spectrometry using anti-IFI35 antibody in 839 GSC cell lysates. G Immunoblot of immunoprecipitation with anti-IFI35, anti-NFKB1, anti-RELB antibody in 839 and 2907 GSCs. The nonspecific IgG is the control. H The confocal image of IFI35, NFKB1, and RELB distribution in 839 and 2907 GSCs. Scale bar, 10 μm. I Heatmap shows down-regulated genes (average change more than 1.5-fold, adjusted P value < 0.05) after sgIFI35 knockout in cell chemotaxis signaling pathway. J IFI35 knockout-induced chromatin accessibility alteration of cell chemotaxis pathway from ATAC-seq. K CCL2, LOX, ANXA1, CX3CL1, WNT5A, and SLIT2 signals detected by ATAC-seq in 839 GSC transduced with sgCONT and sgIFI35. L Chip-PCR assays using nonspecific IgG, anti-IFI35, anti-RELB, or anti-NFKB1 antibody. Data were determined by ANOVA and shown as mean ± SD from three independent experiments. **P < 0.01.
IFI35 induces recruitment and maintenance of M2-like TAMs
Given the distinct role of IFI35 in immune regulation, we hypothesized that IFI35 modulate the TME in GBM. Since high immune signatures are associated with IFI35 (Fig. 6A, B), we speculated that IFI35 might be involved in immune regulation in GBM and its extracellular signaling in GSCs. To identify the specific immune compartment affected by IFI35 expression, we examined the TCGA GBM dataset for different types of immune cells using validated gene set signatures. The GSEA analysis demonstrated that IFI35 expression correlated most highly with a macrophage expression signature, compared to other immune fractions (Fig. 6C). We then treated THP1-derived M2-like macrophages with supernatants of IFI35-silenced GSCs and found the prevention of macrophage recruitment and predominant reduction of M2-like markers (CCL22, VEGF, CD206, ARG1) (Fig. 6D, E). As measured in xenograft specimens by Iba-1, the pan-macrophage marker, and CD206, a M2-like TAMs marker, we found that IFI35 disruption reduced the frequency of M2-like TAMs (Fig. 6F, G). We also examined the M2-like TAMs distribution in xenografts generated by implantation of GSCs expressing shRELB, and found that RELB disruption reduced the frequency of M2-like TAMs (Fig. S6A). Therefore, IFI35 signaling in GSCs activates cell chemotaxis to regulate recruitment and maintenance of M2-like TAMs.
A The immune scores showing IFI35 is associated with immune cell infiltration in TCGA GBM dataset. B The immune scores showing IFI35 is associated with immune cell infiltration with RNA sequencing analysis for GSCs and NSCs dataset. C GSEA for immune cells with correlation to IFI35 in TCGA GBM dataset. D THP1-derived M2 macrophages were treated with supernatants from GSCs transduced with either shIFI35 or shCONT for 24 h, transwell assays were performed to measure macrophages migration. E The mRNA level of M2-like markers (CCL22, VEGF, CD206, and ARG1) decreased in THP1-derived M2-like TAMs by stimulating with the supernatant of 839 or 2907 GSCs with loss of IFI35. Data are presented as mean ± SD from three independent experiments. F Immunofluorescent staining of IFI35 (green) and the pan-macrophage marker Iba1 (red) in GBM xenografts derived from 839 and 2907 GSCs expressing shCONT or shIFI35. Scale bar, 20 μm. G Immunofluorescent staining of the M2-like TAM marker CD206 (green) and the pan-macrophage marker Iba1 (red) in GBM xenografts derived from 839 and 2907 GSCs expressing shCONT or shIFI35. Scale bar, 20 μm.
IFI35 promotes in vivo tumor growth and has therapeutic potential in GSCs
To establish the in vivo effect of IFI35 in GBM growth, we silenced IFI35 in GSCs and implanted these GSCs in the brains of immunocompromised mice. Loss of IFI35 prolonged tumor latency and reduced tumor volumes compared with mice bearing GSCs transduced with a nontargeting control shRNA in two patient-derived GSCs (Fig. 7A–C). To determine the potential clinical significance of IFI35 in glioma patients, we investigated glioma tumor databases to examine the expression of IFI35. Analysis of the TCGA glioblastoma dataset revealed that IFI35 mRNA expression was upregulated in glioblastoma patient tumors compared with normal brain (Fig. S7A). IFI35 expression was positively correlated with tumor grade in the TCGA GBM LGG (low-grade glioma), CGGA GBM LGG, Gravendeel and Rembrandt databases (Fig. S7B, C). Moreover, we observed that TRIM21, NFKB1, and RELB were highly expressed in GBMs compared with non-tumor brain tissues in the TCGA GBM database (Fig. S7D). High expression of IFI35 was associated with poor prognosis in glioma and GBM patients in multiple databases including in TCGA GBM, TCGA GBM LGG, CGGA all, Rembrandt GBM, Rembrandt all, Gravendeel all, and Gravendeel GBM databases (Fig. 7D and Fig. S7E). Based on our findings that IFI35 interacts with TRIM21 to regulate NFKB1 ubiquitination and IFI35 with RELB/p50 complex regulates the downstream cell chemotaxis, we integrated the expression of IFI35 and TRIM21, IFI35 and NFKB1, IFI35 and RELB, which collectively informed prognosis to a greater degree (Fig. 7E, F). Taken together, our data elucidate that IFI35 and its downstream effectors in GSCs may provide potential therapeutic targets in GBM treatment.
A In vivo bioluminescent imaging of tumor growth was performed in nude mice bearing glioblastoma xenografts derived from 5 × 104 GSCs (839 and 2907) transduced with shCONT, shIFI35-1, or shIFI35-2 on days 3, 8, 15, and 50. The control groups (mice bearing 839 and 2907 GSCs) on day 50 were dead. The groups of shIFI35-1 and shIFI35-2 formed a tumor in mice brains. B Kaplan–Meier survival curves of immunodeficiency mice with intracranial 839 or 2907 GSCs expressing shCONT, shIFI35-1, or shIFI35-2. n = 6 mice per group. C Representative images of hematoxylin and eosin staining of mouse brains collected on days 15 and 50 after transplantation of 839 or 2907 GSCs expressing shCONT, shIFI35-1, or shIFI35-2. Scale bar, 2 mm. Each image is representative of at least three similar experiments. D Kaplan–Meier survival analysis of IFI35 by the median of different glioma datasets: TCGA GBM, TCGA GBM LGG, CGGA all, and Rembrandt GBM datasets. E Kaplan–Meier survival analysis of IFI35 combined with TRIM21, NFKB1, and RELB by the median of TCGA GBM dataset. F Kaplan–Meier survival analysis of IFI35 combined with TRIM21, NFKB1, and RELB by the median of TCGA GBM LGG dataset.
Discussion
The intricate mechanisms that support interactions between GSCs and their TME contribute to tumor initiation, propagation, therapy resistance, invasion, and immune escape [22,23,24]. IFI35 has been implicated in regulating cell homeostasis and innate immunity in inflammation-related diseases such as liver injury, rheumatoid arthritis, and systemic lupus erythematosus [25,26,27], but the function of IFI35 in GBM tumors has not been elucidated well. In this study, we identified that IFI35 is overexpressed in GSCs and GBM tumors where it acts as a crucial regulator of GSC proliferation and non-canonical NF-κB signaling by TRIM21 induced ubiquitination of p105 leading to the formation of transcriptionally active p50. IFI35 associates with RELB/p50 in GSCs to activate cell chemotaxis leading to the recruitment of M2-like macrophages in TME (Fig. S7F). Targeting IFI35 and its downstream NF-κB signaling could provide additional strategies for glioblastoma therapeutic improvement. IFI35 is a leucine zipper protein regulated by interferon. Previous studies have demonstrated IFI35 as an immunoregulatory factor that was detected in a variety of cell types, including monocytes/macrophages, epithelial cells, fibroblasts, and tumor infiltrating immune cells [28, 29]. IFI35 is induced by toll-like receptors (TLR) signaling that triggers innate immunity and exacerbates immune responses upon detection of damage associated molecular patterns (DAMPs) [25]. IFI35 can translocate from the cytoplasm to the nucleus upon specific viral infection [15]. IFI35, N-myc and STAT interacting protein (NMI) were previously reported to interact [25, 30]. However, the function of IFI35 in cancers especially GBM remains unknown. Our study demonstrated that IFI35 supports GSC proliferation and self-renewal through interacting with TRIM21 complex. We identified TRIM21, a E3 ubiquitin ligase, as an interacting partner of IFI35 which mediated the ubiquitination and proteasomal processing of NFKB1 p105 to p50. TRIM21 was previously shown to activate the NF-κB, AP1, IRF3, IRF5, and IRF7 mediated antiviral pathways by conjugating K63-linked ubiquitin chains. Moreover, TRIM21 is also reported to downregulate the DNA sensor DDX41 by K48-linked ubiquitination and subsequent proteasomal degradation [31,32,33,34]. Our results revealed that mutation of K48-linked ubiquitin chain decreased the ubiquitination of NFKB1 p105 and increased the protein level of p50. IFI35 interacts with TRIM21 complex to regulate NFKB1 p105 processing to p50 through K48-linked ubiquitin chain in GSCs.
The NF-κB family proteins are critical transcriptional regulators involved in cell proliferation, survival, transformation, and angiogenesis in the pathogenesis of GBM [35,36,37]. Several important steps in NF-κB activation are mediated by the ubiquitin system. The majority of substrates of the ubiquitin proteasome system are completely degraded in to amino acids [38]. With the exception of the precursors-NF-kB1 (p105) and NF-kB2 (p100), which can undergo limited proteolytic processing by the proteasome to yield the active subunits of p50 and p52 to generate NF-κB transcriptional dimeric complexes [39,40,41]. The canonical NF-κB pathway relies on inducible degradation of the IκB-kinase (IKK) complex, particularly IκBα, leading to nuclear translocation of various NF-κB complexes, predominantly the RELA/p50 dimer. As opposed to the canonical pathway, the non-canonical or alternative pathway of NF-kB is dependent on the translocation of RELB subunits p50 or p52 into the nucleus and activation of transcription of downstream target genes [40, 42, 43]. Our findings demonstrated the chemokines CCL2, LOX, ANXA1, CX3CL1, WNT5A, and SLIT2 are the downstream target genes of IFI35 induced non-canonical NF-κB signaling involving RELB/p50 activation to induce the recruitment of surrounding immune cells especially macrophages.
TAMs and microglia are the most abundant immune cells within the TME, accounting for up to 50% of total cells in the GBM tumor mass, which play a critical role in promoting tumor progression and inducing immunosuppressive TME [44, 45]. TAMs are polarized as classically activated or M1-like phenotype and alternatively activated macrophages or M2-like phenotype. Emerging evidence demonstrates that the M1/M2 dichotomy is not sufficient to characterize the complexity and plasticity of TAMs in response to different environmental conditions in TME [46, 47]. TAM subpopulations infiltrate into GBM tumors, resulting in a context-dependent symbiotic interaction between TAMs and tumor cells. Targeting TAMs infiltration and immunosuppressive reprogramming may provide therapeutic strategies for clinical GBM patients. In this study, we identified that IFI35 regulates the maintenance of GSCs and recruitment of TAMs. GSCs recruit TAMs through secretion of a series of chemokines and promote an M2-like phenotype. IFI35 was reported to be associated with M1-like macrophages to function as an anti-tumor effect in triple negative breast cancer [48]. However, our data provide, for the first time, definitive evidence that IFI35 induced non-canonical NF-kB signaling and acted to transcriptionally regulate chemokines in TME.
In conclusion, our results identified IFI35 as both a transcriptional and translational regulatory factor in maintaining GSCs and recruitment of TAMs in GBM. Upregulated IFI35 levels in GSCs and plays both cell-intrinsic and cell-extrinsic roles in maintaining the immunosuppressive TME. IFI35 promoted TRIM21 induced proteasomal processing of p105 to p50 leading to the activation of non-canonical NF-kB signaling by the formation of RELB/p50 complex in a cell-autonomous manner, and recruitment and maintenance of M2-like TAMs in TME in a paracrine manner. Targeting IFI35 and its downstream signaling effectors, including NFKB1, TRIM21, and RELB may provide effective therapeutic strategies for GBM treatment.
Methods
Patient-derived and cell line culture
Glioblastoma tissues of GSCs were obtained from excess surgical resection samples from patients who underwent treatment at Case Western Reserve University (Cleveland, OH) after review by neuropathology with appropriate consent and in accordance with an IRB-approved protocol (090401). Patient-derived xenografts were produced and propagated as a renewable source of GSCs, in order to minimize in vitro artifacts from cell culture. STR analyses were performed on the tumor model used for authentication. GSCs were cultured as neurospheres in Neurobasal medium (Invitrogen) supplemented with B27 supplement without vitamin A (Life Technologies), Glutamax (Life Technologies), sodium pyruvate (Life Technologies), penicillin/streptomycin, 20 ng/mL EGF and basic fibroblast growth factor (bFGF, R&D Systems) at 37 °C with 20% oxygen and 5% carbon dioxide.
The nonmalignant NSC models ENSA and HNP1 were used in this study. ENSA (ENStem-A) is a human embryonic stem-derived neural progenitor cell (Millipore Sigma, Cat# SCC003). HNP1 (STEMEZ HNP1) is a human neural progenitor cell (Neuro-mics, Cat# HN60001). NSCs were cultured in Neurobasal media supplemented with B27, 20 ng/ml EGF and bFGF. 293FT cells (ThermoFisher Scientific, Cat# R70007) were used to generate lentiviral particles as described in the “Methods” details section.
In vivo tumorigenesis and animal models
Intracranial xenograft models were established by implanting 5 × 104 human-derived GSCs (839 GSC and 2907 GSC) into the cerebral cortex of BALB/c-Nude (Strain No. D000521, GemPharmatech, Nanjing, China) mice at a depth of 3.5 mm. Female mice at 4–6 weeks of age were randomly assigned to each treatment group (n = 6 mice per group) for the animal experiments used in the study. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC-2006033-2) at Nanjing Medical University in accordance with NIH and institutional guidelines. All experiments used in this study were approved by the Institutional Ethics Committee of Nanjing Medical University. Pathogen-free environments were provided at the Animal Core Facility of Nanjing Medical University. Seven days after GSCs implantation, the mice were observed daily and euthanized when they presented neurological signs or became moribund. Brains were harvested and fixed in 4% formaldehyde, cryopreserved in 30% sucrose, and then cryosectioned. Sections were stained with hematoxylin and eosin for histologic analysis.
Proliferation and neurosphere formation assays
Cell proliferation experiments were conducted by plating cells of interest at a density of 2000 cells/well in 96-well plates with six replicates. CellTiter-Glo (Promega) was used to measure cell proliferation. All data were normalized to day 0 and presented as mean ± SD. Neurosphere formation was measured by in vitro limiting dilution, as previously described [49]. Briefly, decreasing numbers of cells per well (50, 20, 10, 5, and 1) were plated into 96-well plates. The presence and number of neurospheres in each well were recorded 7 days after plating. Extreme limiting dilution analysis was conducted with software available at http://bioinf.wehi.edu.au/software/elda, as previously described [50]. All neurosphere and proliferation experiments were performed six times.
Plasmids and lentiviral transduction
All shRNAs used were obtained from Sigma-Aldrich and are listed in Table S1. Nonoverlapping shRNAs were selected based on knockdown efficiency and were then used for all following experiments. Lentiviral particles were generated in 293FT cells in Neurobasal complete medium (Life Technologies) with co-transfection with the packaging vectors psPAX2 and pMD2.G (Addgene) using a standard calcium phosphate transfection method in neurobasal complete medium.
RNA isolation and quantitative RT-PCR
Total cellular RNA from cells were isolated by Trizol reagent (Takara). The PrimeScript cDNA Synthesis Kit (Takara) was used for reverse transcription into cDNA. Quantitative real-time PCR was performed with an Applied Biosystems Stepone Plus cycler using SYBR-Green PCR Master Mix (ThermoFisher Scientific). All the primers used are listed in Table S1.
Western blotting
For western blot analysis, cells and tissues were collected and lysed in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% NP-40, 0.1% SDS and supplemented with protease inhibitors), then incubated on ice for 30 min. Lysates were centrifuged at 4 °C for 10 min at 12,000 rpm, and supernatants were collected. Supernatants were subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were blocked with 5% non-fat milk for 1 h and then immunoblotted with primary antibodies against IFI35 (abcam, ab233415), NFKB1 (Cell Signaling, 13586), RELB (Cell Signaling, 10544), TRIM21 (Proteintech, 12108-1-AP), SOX2 (Cell Signaling, 3579), OLIG2 (Sigma-Aldrich, MABN50), MYC (Cell Signaling, 18583), His-Tag (Cell Signaling, 12698), Flag-Tag (Cell Signaling, 14793S), HA-Tag (Cell Signaling, 3724S) and β-Tubulin (Abcepta, AM1031A) overnight at 4 °C followed by the HRP-conjugated antibody at room temperature for 2 h. Antibody validation was performed per manufacturer’s instructions. The evaluation of immunoblot was imaged using ECL analysis.
Immunofluorescence
Immunofluorescence experiments were performed using GBM cells or samples from GBM surgical specimens. Tumor samples from GBM patients or xenografts grown in mice were fixed in 4% paraformaldehyde overnight at 4 °C, followed by overnight cryoprotection with 20% sucrose in PBS at 4 °C. Sections were stained with primary antibodies anti-IFI35 (abcam, ab233415), anti-SOX2 (R&D, AF2018), anti-CD206 (CST, 24595 S), anti-Iba-1 (Santa Cruz, sc-32725). For GSCs, cells were fixed using 4% paraformaldehyde for 30 min and washed with PBS, permeabilized with 0.45% Triton X-100 for 10 min and blocked in 3% bovine serum albumin (BSA) at room temperature for 2 h. Cells were incubated with appropriate antibodies anti-IFI35 (santa, sc-393513), anti-SOX2 (R&D, AF2018), anti-NFKB1 (Cell Signaling, 13586, Proteintech, 66992-1-lg), anti-RELB (Cell Signaling, 10544), anti-TRIM21 (Proteintech, 12108-1-AP). Images were captured with a Zeiss LSM700 microscope (Carl Zeiss, USA).
Co-immunoprecipitation (Co-IP)
GSCs were collected and lysed in IP buffer (20 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 1 mM DTT) containing PMSF and cocktail protease inhibitor at 4 °C for 30 min, and followed by centrifugation at 14,000 × g for 10 min at 4 °C. The antibody of anti-IFI35 (Santa Cruz, sc-393513), anti-NFKB1 (Cell Signaling, 13586), anti-RELB (Cell Signaling, 10544), anti-TRIM21 (Proteintech, 12108-1-AP), or nonspecific IgG was added to the supernatants and incubated at 4 °C overnight. The supernatants were immunoprecipitated by incubation with Protein A/G sepharose beads (Beyotime) at 4 °C for 2 h. Captured proteins were washed with IP buffer three times before immunoblotting.
Chromatin immunoprecipitation (ChIP)
In total, 4 × 106 cells per condition were collected, and ChIP assay was performed using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling, #9003) according to the manufacturer’s protocols. Briefly, 10 µl IFI35 antibody (abcam, ab233415), anti-NFKB1 (Cell Signaling, 13586), anti-RELB (Cell Signaling, 10544), or nonspecific IgG were used for the immunoprecipitation of the DNA-protein immunocomplexes. Crosslinking was reversed by heating for 6 h at 65 °C, followed by digestion with proteinase K. The collected DNA was subsequently analyzed by qPCR. All the primers used are listed in Table S1.
THP1-derived macrophage culture and migration assay
THP1 cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere with 5% CO2. To obtain THP1-derived primary macrophages (M0-like cells), THP1 cells (1 × 106 cells/ml) were treated with 10 ng/ml phorbol 12-myristate 13-acetate (PMA) for 24 h. To obtain THP1-derived M2-like macrophages (M2-like cells), THP1 cells (2 × 105 cells/well) were treated with 10 ng/mL PMA for 24 h, and then cultured with 25 ng/mL IL4 and 25 ng/mL IL13 for another 48 h. For the coculture system, we stimulated M2-like cells with the supernatant of GSCs transduction with the corresponding shRNAs. Cell migration assay was performed using a transwell coculture system in 24-well plates (Corning). Primary macrophages were seeded at a density of 1 × 106 cells/well to top wells with an 8.0-mm pore size, then cocultured with the supernatant of GSCs transduction with the corresponding shRNAs in bottom wells for 48 h, and migrated cells were stained.
RNA-seq and data analysis
Total RNA was extracted with Trizol reagent (Takara) according to the manufacturer’s instructions. RNA sequencing was performed using BGISEQ-500 platforms. FASTQ sequencing reads were trimmed using Trim Galore (RRID:SCR_011847), and transcript quantification was conducted using Salmon with the quasi-mapping mode [51]. Salmon “quant” files were converted using Tximport [52], and differential expression analysis was performed using DESeq2 [53]. GSEA was performed by selecting differentially expressed genes (FDR-corrected P value < 0.05), generating a pre-ranked list, and importing the pre-ranked list into the GSEA desktop application (http://www.gsea-msigdb.org/gsea/index.jsp).
ATAC-seq and data analysis
ATAC-seq was performed by BGI-Shenzhen (https://en.genomics.cn). Cells were harvest and washed in ice-cold PBS followed by centrifugation at 500 × g for 5 min. Cells were resuspended in 50 µl cold lysis buffer (10 mM Tris-HCl pH 7.5, 3 mM MgCl2, 10 mM NaCl, 0.1% Igepal CA-630) and mixed end over end. After centrifugation at 500 × g for 10 min, cell pellet was resuspended in transposition reaction mix (25 μl TD-Buffer, 2.5 μl Tn5, 22.5 μl water) and incubated at 37 °C for 30 min with gentle mixing. Immediately following the transposition reaction, MinElute PCR Purification Kit (Qiagen) was used to purify transposed DNA. Finally, the PCR amplification was performed to amplify purified DNA and qPCR-based methods were used to quantify ATAC-seq libraries. The libraries were mixed in equimolar ratios and sequenced on MGI2000 platform. ATAC-Seq raw reads were filtered to remove low-quality or adapter sequences by SOAPnuke. Cleaned reads were mapped to the reference genome of hg38 with Bowtie2 and generated SAM files were converted to BAM files using Samtools using the view function. BAM files which were converted to BigWig files using Deeptools. MACS2 was used to call peaks for open chromatin regions. IDR was applied to measure the reproducibility of findings identified from replicate samples.
Statistical analysis
All numerical data are presented as the mean ± SD from at least three independent experiments. Statistical analyses were performed using GraphPad Prism with two-tailed Student’s t-test for two groups, or with one-way ANOVA with Dunnett’s multiple-comparison test for multiple groups.
For Kaplan–Meier survival curves, statistical differences between groups were determined by log-rank test. Blinding and randomization were performed in all experiments. The correlation between gene expression and patient survival was performed through analysis of different brain tumor datasets, downloaded from TCGA, CGGA, Gravendeel, and Rembrandt brain tumor datasets, downloaded from TCGA data portal or National Center for Biotechnology Information Gene Expression Omnibus (GEO) database.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
RNA-seq (GSE242225), ATAC-seq (GSE242570), and single-cell RNA-seq (GSE141383, GSE117891) data generated for this study are available in Gene Expression Omnibus Database (GEO). All data accessed from external sources and prior publications have been referenced in the text and corresponding figure legends. Additional data will be made available upon request.
References
Miller KD, Ostrom QT, Kruchko C, Patil N, Tihan T, Cioffi G, et al. Brain and other central nervous system tumor statistics, 2021. CA Cancer J Clin. 2021;71:381–406.
Ostrom QT, Price M, Neff C, Cioffi G, Waite KA, Kruchko C, et al. CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro Oncol. 2022;24:v1–v95.
Perry JR, Laperriere N, O’Callaghan CJ, Brandes AA, Menten J, Phillips C, et al. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N Engl J Med. 2017;376:1027–37.
Karachi A, Dastmalchi F, Mitchell DA, Rahman M. Temozolomide for immunomodulation in the treatment of glioblastoma. Neuro Oncol. 2018;20:1566–72.
Hernandez Martinez A, Madurga R, Garcia-Romero N, Ayuso-Sacido A. Unravelling glioblastoma heterogeneity by means of single-cell RNA sequencing. Cancer Lett. 2022;527:66–79.
Yeo AT, Rawal S, Delcuze B, Christofides A, Atayde A, Strauss L, et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat Immunol. 2022;23:971–84.
LeBlanc VG, Trinh DL, Aslanpour S, Hughes M, Livingstone D, Jin D, et al. Single-cell landscapes of primary glioblastomas and matched explants and cell lines show variable retention of inter- and intratumor heterogeneity. Cancer Cell. 2022;40:379–392.e379.
Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28:318–28.
Santangelo A, Rossato M, Lombardi G, Benfatto S, Lavezzari D, De Salvo GL, et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro Oncol. 2021;23:264–76.
Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–17.
Prager BC, Xie Q, Bao S, Rich JN. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019;24:41–53.
Biserova K, Jakovlevs A, Uljanovs R, Strumfa I. Cancer stem cells: significance in origin, pathogenesis and treatment of glioblastoma. Cells. 2021;10:621.
Dapash M, Hou D, Castro B, Lee-Chang C, Lesniak MS. The interplay between glioblastoma and its microenvironment. Cells. 2021;10:2257.
Hernandez A, Domenech M, Munoz-Marmol AM, Carrato C, Balana C. Glioblastoma: relationship between metabolism and immunosuppressive microenvironment. Cells. 2021;10:3529.
Yang H, Winkler W, Wu X. Interferon inducer IFI35 regulates RIG-I-mediated innate antiviral response through mutual antagonism with Influenza protein NS1. J Virol. 2021;95:e00283–00221.
Chai D, Shi SY, Sobhani N, Ding J, Zhang Z, Jiang N, et al. IFI35 promotes renal cancer progression by inhibiting pSTAT1/pSTAT6-dependent autophagy. Cancers. 2022;14:2861.
Jing X, Yao Y, Wu D, Hong H, Feng X, Xu N, et al. IFP35 family proteins promote neuroinflammation and multiple sclerosis. Proc Natl Acad Sci USA. 2021;118:e2102642118.
Mack SC, Singh I, Wang X, Hirsch R, Wu Q, Villagomez R, et al. Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J Exp Med. 2019;216:1071–90.
Li JY, Zhao Y, Gong S, Wang MM, Liu X, He QM, et al. TRIM21 inhibits irradiation-induced mitochondrial DNA release and impairs antitumour immunity in nasopharyngeal carcinoma tumour models. Nat Commun. 2023;14:865.
Kravtsova-Ivantsiv Y, Cohen S, Ciechanover A. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-κB precursor. Adv Exp Med Biol. 2011;691:95–106.
Gasparini C, Celeghini C, Monasta L, Zauli G. NF-κB pathways in hematological malignancies. Cell Mol Life Sci. 2014;71:2083–102.
Xuan W, Hsu WH, Khan F, Dunterman M, Pang L, Wainwright DA, et al. Circadian regulator CLOCK drives immunosuppression in glioblastoma. Cancer Immunol Res. 2022;10:770–84.
Tao W, Chu C, Zhou W, Huang Z, Zhai K, Fang X, et al. Dual role of WISP1 in maintaining glioma stem cells and tumor-supportive macrophages in glioblastoma. Nat Commun. 2020;11:3015.
Yin J, Kim SS, Choi E, Oh YT, Lin W, Kim TH, et al. ARS2/MAGL signaling in glioblastoma stem cells promotes self-renewal and M2-like polarization of tumor-associated macrophages. Nat Commun. 2020;11:2978.
Xiahou Z, Wang X, Shen J, Zhu X, Xu F, Hu R, et al. NMI and IFP35 serve as proinflammatory DAMPs during cellular infection and injury. Nat Commun. 2017;8:950.
Zhang L, Zhu H, Li Y, Dai X, Zhou B, Li Q, et al. The role of IFI35 in lupus nephritis and related mechanisms. Mod Rheumatol. 2017;27:1010–8.
Weix J, Haupl T, Raio L, Villiger PM, Forger F. The physiologic increase in expression of some type I IFN-inducible genes during pregnancy is not associated with improved disease activity in pregnant patients with rheumatoid arthritis. Transl Res. 2013;161:505–12.
Bange FC, Vogel U, Flohr T, Kiekenbeck M, Denecke B, Bottger EC. IFP 35 is an interferon-induced leucine zipper protein that undergoes interferon-regulated cellular redistribution. J Biol Chem. 1994;269:1091–8.
Tan J, Qiao W, Wang J, Xu F, Li Y, Zhou J, et al. IFP35 is involved in the antiviral function of interferon by association with the viral tas transactivator of bovine foamy virus. J Virol. 2008;82:4275–83.
Zhang L, Tang Y, Tie Y, Tian C, Wang J, Dong Y, et al. The PH domain containing protein CKIP-1 binds to IFP35 and Nmi and is involved in cytokine signaling. Cell Signal. 2007;19:932–44.
Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14:172–8.
McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14:327–36.
Lin L, Wang X, Chen Z, Deng T, Yan Y, Dong W, et al. TRIM21 restricts influenza A virus replication by ubiquitination-dependent degradation of M1. PLoS Pathog. 2023;19:e1011472.
Yang K, Shi HX, Liu XY, Shan YF, Wei B, Chen S, et al. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol. 2009;182:3782–92.
Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–46.
Friedmann-Morvinski D, Narasimamurthy R, Xia Y, Myskiw C, Soda Y, Verma IM. Targeting NF-κB in glioblastoma: a therapeutic approach. Sci Adv. 2016;2:e1501292.
Cahill KE, Morshed RA, Yamini B. Nuclear factor-κB in glioblastoma: insights into regulators and targeted therapy. Neuro Oncol. 2016;18:329–39.
Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–62.
Kravtsova-Ivantsiv Y, Shomer I, Cohen-Kaplan V, Snijder B, Superti-Furga G, Gonen H, et al. KPC1-mediated ubiquitination and proteasomal processing of NF-κB1 p105 to p50 restricts tumor growth. Cell. 2015;161:333–47.
Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Signal Transduct Target Ther. 2020;5:209.
Savinova OV, Hoffmann A, Ghosh G. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high- molecular-weight heterogeneous complexes. Mol Cell. 2009;34:591–602.
Sun SC. The non-canonical NF-κB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17:545–58.
Roy P, Mukherjee T, Chatterjee B, Vijayaragavan B, Banoth B, Basak S. Non-canonical NFκB mutations reinforce pro-survival TNF response in multiple myeloma through an autoregulatory RelB:p50 NFκB pathway. Oncogene. 2017;36:1417–29.
Xiao Y, Wang Z, Zhao M, Deng Y, Yang M, Su G, et al. Single-cell transcriptomics revealed subtype-specific tumor immune microenvironments in human glioblastomas. Front Immunol. 2022;13:914236.
Bikfalvi A, da Costa CA, Avril T, Barnier JV, Bauchet L, Brisson L, et al. Challenges in glioblastoma research: focus on the tumor microenvironment. Trends Cancer. 2023;9:9–27.
Xuan W, Lesniak MS, James CD, Heimberger AB, Chen P. Context-dependent glioblastoma-macrophage/microglia symbiosis and associated mechanisms. Trends Immunol. 2021;42:280–92.
Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–72.
Xu B, Sun H, Song X, Liu Q, Jin W. Mapping the tumor microenvironment in TNBC and deep exploration for M1 macrophages-associated prognostic genes. Front Immunol. 2022;13:923481.
Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16:1373–82.
Xie Q, Wu Q, Horbinski CM, Flavahan WA, Yang K, Zhou W, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci. 2015;18:501–10.
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–9.
Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Funding
This work was supported by the National Natural Science Foundation of China (82072779 to XXW, 82172667 to XFW) and Jiangsu Provincial Key Research Development Program of China (BE2022770 to YC).
Author information
Authors and Affiliations
Contributions
XXW, QZ, JNR, and KY conceptualized and supervised the study. DL, XFW, KC, DS conducted most of the experiments, analyzed the data, prepared the figures and contributed to the original paper preparation, review, and editing. GC, WY, QL, CL performed the data analysis and bioinformatics analysis under the supervision of ZS, XF, QW, and WG; DG, HY, JG, YL, TK, JY, HY, and KS interpreted the data. ZZ, JM, QW, FL, WT, SCM, YC, JZ, CL, NZ, YY, and XQ commented on the study, RCG and DD revised the manuscript and supervised the work. All authors discussed the results and approved submission of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC-2006033-2) at Nanjing Medical University in accordance with NIH and institutional guidelines. Human fresh tumor samples were obtained with informed consent of patients, and all experimental procedures were performed in accordance with the recognized ethical guidelines of the Declaration of Helsinki, and Ethics Committee of the First Affiliated Hospital of Nanjing Medical University (2021-SR-076).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, D., Wang, X., Chen, K. et al. IFI35 regulates non-canonical NF-κB signaling to maintain glioblastoma stem cells and recruit tumor-associated macrophages. Cell Death Differ 31, 738–752 (2024). https://doi.org/10.1038/s41418-024-01292-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-024-01292-8
- Springer Nature Limited