Abstract
Although numerous studies indicate that inhibition of USP7 suppresses tumor growth by activating p53, the precise mechanism by which USP7 contributes to tumor growth through the p53-independent manner is not well understood. p53 is frequently mutated in most triple-negative breast cancers (TNBC), characterized as the very aggressive form of breast cancers with limited treatment options and poor patient outcomes. Here, we found that the oncoprotein Forkhead Box M1 (FOXM1) acts as a potential driver for tumor growth in TNBC and, surprisingly, through a proteomic screen, we identified USP7 as a major regulator of FOXM1 in TNBC cells. USP7 interacts with FOXM1 both in vitro and in vivo. USP7 stabilizes FOXM1 through deubiquitination. Conversely, RNAi-mediated USP7 knockdown in TNBC cells, dramatically reduced the levels of FOXM1. Moreover, based upon the proteolysis targeting chimera (PROTAC) technology, we generated PU7-1 (protein degrader for USP7-1), as a USP7 specific degrader. PU7-1 induces rapid USP7 degradation at low nanomolar concentrations in cells but shows no obvious effect on other USP family proteins. Strikingly, the treatment of TNBC cells with PU7-1 significantly abrogates FOXM1 functions and effectively suppresses cell growth in vitro. By using xenograft mouse models, we found that PU7-1 markedly represses tumor growth in vivo. Notably, ectopic overexpression of FOXM1 can reverse the tumor growth suppressive effects induced by PU7-1, underscored the specific effect on FOXM1 induced by USP7 inactivation. Together, our findings indicate that FOXM1 is a major target of USP7 in modulating tumor growth in a p53-independent manner and reveals the USP7 degrader as a potential therapeutic tool for the treatment of triple-negative breast cancers.
Similar content being viewed by others
Introduction
It is well established that tumor suppressor p53 is a short-life protein and its protein levels are tightly controlled by MDM2-mediated polyubiquitination and Ubiquitin-specific-processing proteases (USPs) -mediated deubiquitination [1,2,3,4,5,6]. USP7, also known as Herpesvirus-associated ubiquitin-specific protease (HAUSP), was considered as a promising therapeutic target for cancer due to its critical regulation of several oncoproteins and tumor suppressors such as MDM2, p53, N-MYC, PTEN and DNMT1 [2, 3, 7,8,9,10]. Numerous studies indicate a dynamic role of USP7 in MDM2-p53 network [2, 3]. On one hand, USP7 can stabilize p53 by deubiquitination; on the other hand, MDM2, a critical E3 ligase of p53, is also a target of USP7. Structure-based study revealed that MDM2 and p53 competitively recognize the same surface groove in USP7, whereas USP7 has higher binding affinity for MDM2 than p53 [11]. Therefore, ablation of USP7 predominantly leads to MDM2 degradation and ultimately p53 activation. These observations indicated that USP7 inhibition is potentially beneficial for therapeutic purposes in human cancers harboring wild-type p53. Later, several studies reported that p53 inactivation was unable to completely rescue the embryonic lethality in USP7 knockout mice [12, 13], implicating the existence of p53-independent substrates of USP7. Increasing evidence from our group and others confirmed that USP7 regulated tumor growth through both p53-dependent and p53-independent networks and inhibition of USP7 led to significant tumor suppression in many types of human cancer [2, 3, 7,8,9,10]. In the past decades, intensive efforts result in the development of several small-molecule inhibitors of USP7 [3, 14, 15]. However, due to issues in selectivity, potency, solubility, and metabolism properties in vivo, none of current USP7 inhibitors have entered clinical trials.
FOXM1 belongs to the forkhead box transcription factor family, in which members share an evolutionary conserved fork-head or winged-helix DNA-binding domain [16]. FOXM1 is a master regulator of cell cycle [16]. FOXM1 mediates the transcription of SKP2 and CKS1, which are key subunits of the SKP1-Cullin1-F-Box (SCF) ubiquitin ligase complex that targets the cyclin-dependent kinase inhibitors p21 and p27 for degradation during G1-S transition [17]. In addition, FOXM1 is essential for the transcription of the mitotic regulatory genes CCNB1/2, Aurora A/B, CDC25A/B, centromere proteins CENPA/B/F, PLK1 etc., modulating G2-M transition [17, 18]. Unlike other FOX family proteins, FOXM1 predominantly expresses in proliferating cells and overexpression of FOXM1 is a hallmark of various human cancers [16, 19]. High expression of FOXM1 is associated with a poor clinical prognosis of many malignancies, suggesting that it has an important role in cancer progression. Numerous studies demonstrate that FOXM1 plays a critical role in tumorigenesis, angiogenesis, invasion, and metastasis, as well as drug resistance [16, 19,20,21,22,23,24].
Breast cancer is the second leading cause of cancer death in women. The American Cancer Society estimates that about 43700 women will die from breast cancer in the United States in 2023. Triple-negative breast cancer (TNBC) represents the most invasive subtype of breast cancers that are characterized by the lack of expression of the estrogen receptor (ER), progesterone receptor (PR) and absence of epidermal growth factor receptor 2 (ERBB2, also known as HER2) amplification [25]. TNBC accounts for about 10–15% of all breast cancers and has a high prevalence in women of African descent and women who carry mutant BRCA1/2 gene [26]. It differs from other subtypes of breast cancer in more aggressive and metastatic behavior, fewer treatment options and worse prognosis than other subtypes of breast cancer. Despite the advances of the estrogen therapy and HER2-targeted therapy for breast cancer, chemotherapy has long been the only available therapeutic option for TNBC due to lack of therapeutic targets [27]. Given the limited benefit of chemotherapy, persistent efforts were focused on the development of new therapies for TNBC. Recently, PARP inhibitors have been successfully used in the triple-negative breast cancer with BRCA1/2 mutation [28]. Because, the prevalence rates of BRCA1/2 mutation in triple-negative breast cancer is about 20% [29,30,31], the majority of TNBC patients still do not benefit from PARP inhibitors. The molecular heterogeneity of the disease makes the optimal treatment strategy for patients with TNBC still a major unmet need.
Herein, we found that the oncogenic transcription factor FOXM1 is highly expressed in TNBC cell lines and tumors. Mass spectrometry analysis of FOXM1 protein complex identified USP7 as a novel interacting protein of FOXM1. Of note, further investigation demonstrates that USP7 directly binds, deubiquitinates and stabilizes FOXM1 protein. Moreover, several studies reported that the overexpression of USP7 in breast cancers was associated with poor prognosis and drug resistance [32,33,34]. It suggests that targeting USP7 to downregulate FOXM1 can be an effective strategy for TNBC treatment. Therefore, we proceeded to synthesize and screened a set of PROteolysis Targeting Chimeras (PROTAC) compounds to target USP7 for degradation and identified PU7-1 as a selective and potent USP7 degrader. As expected, PU7-1 significantly decreases FOXM1 protein levels and leads to inhibition of proliferation in TNBC cells with the IC50 of 1.8 μM in MDA-MB-468 and 2.8 μM in BT549. Notably, PU7-1 also markedly represses the growth of MDA-MB-468 xenografts in mouse model. Exogenous expression of FOXM1 rescued PU7-1-mediated growth inhibition in MDA-MB-468 cells and xenografts, implicating that targeting USP7 by PU7-1 in TNBC elicits tumor suppression by antagonizing FOXM1 network. Taken together, our data provide a novel insight and strategy to treat triple negative breast cancer.
Results
FOXM1 is highly expressed in TNBC
Overall survival analysis in The Cancer Genome Atlas (TCGA) pan-cancers revealed that higher expression of oncogenic transcription factor FOXM1 positively correlates with the shorter survival time of patients (Fig. S1A). Previous studies reported that FOXM1 is negatively regulated by wild-type p53 [35, 36]. Indeed, we downloaded the TCGA cancer dataset and compared the expression of FOXM1 between the wild-type and mutant p53 cohort. A significantly elevated FOXM1 expression was observed in cancers harboring mutant p53 (Fig. S1B). As the most aggressive subtype of breast cancer, TNBCs harbor mutant TP53 gene at much higher frequency than the other subtypes. According to the Catalogue Of Somatic Mutations In Cancer (COSMIC) database, 65% of TNBCs express mutant p53, in contrast, only 36% of all breast cancers have mutant p53 (Fig. S1C). Therefore, we speculate that TNBC is likely to express high levels of FOXM1. The FOXM1 expression profiles in TNBC versus normal tissue and TNBC versus other subtypes of BRCA were explored. As shown in Fig. 1A, the expression of FOXM1 in TNBC is about 3-fold higher than in normal tissue. Moreover, TNBC exhibits the highest FOXM1 expression levels in all four BRCA subtypes (Fig. S1D). To further validate it, we randomly screened several breast cancer cell lines and measured the FOXM1 protein levels in these cells by Western blot analysis. Three of them (MDA-MB-468, MDA-MB-231 and BT549 harboring p53 R273H, R280K and R249S respectively) are triple-negative breast cancer cells. As shown in Fig. 1B, the expression levels of FOXM1 are indeed higher in the breast cancer cell lines with p53 mutants than the one in the MCF7 cell line expressing wild type p53. Interestingly, the levels of FOXM1 are especially higher in three TNBC cell lines (lanes 1-3) than other breast cell lines with mutated p53 (lanes 4–6), suggesting that additional undefined mechanisms may be involved in upregulation of FOXM1 in TNBC cells. Taken together, these results demonstrate that oncogenic transcription factor FOXM1 is highly expressed in TNBC cells, and it is a potential therapeutic target for TNBC.
A FOXM1 expression in TCGA TNBC tumors versus normal tissues. B Western blot analysis was used to detect the expression of FOXM1 in human breast cancer cells MDA-MB-468 (p53 R273H), MDA-MB-231 (p53 R280K), BT549 (p53 R249S), BT474 (p53 E285K), T47D (p53 L194F), SKBR3(p53 R175H), MCF7(p53 wildtype). C Schematic representation of the SFB-FOXM1 protein used in the purification of SFB-FOXM1 complex. D Number of unique peptides for USP7 and known FOXM1-binding partners, identified by Mass spectrometry. E Immunoblots for USP7 after immunoprecipitation of F-FOXM1, with anti-Flag beads, from H1299 cells transfected with the constructs as indicated. F Immunoblots for FOXM1 after immunoprecipitation of HA-USP7, with anti-HA beads, from H1299 cells transfected with the constructs as indicated. G Immunoblots for endogenous USP7 after immunoprecipitation of endogenous FOXM1 from MDA-MB-468 and MDA-MB-231 cells. H Immunoblots for pulldown of purified F-FOXM1 with purified GST or GST-USP7. These data represent two independent experiments.
FOXM1 directly interacts with USP7 deubiquitinase
Except nuclear factors, transcription factors are “undruggable” by small-molecule ligands due to significantly disordered structures and lack of small-molecule binding pockets. Hence, we seek to identify novel interaction partner of FOXM1, to achieve the repression of FOXM1 by modulating its binding partner. To this end, we established a cell line stably expressing SFB-tagged FOXM1 in human lung cancer cell H1299 and then isolated SFB-FOXM1-associated protein complex from the whole cell extracts using streptavidin beads (Figs. 1C and S2A). Analysis of this complex by liquid chromatography mass spectrometry/mass spectrometry (LC–MS/MS) identified several known FOXM1-binding partners such as Sirt1 [37], PLK1 [38], CDK1/2 [39], CREBBP [40] and CCNB1 [39], as well as a novel interacting protein USP7 (Supplementary Table S1). Of note, as much as 44 unique USP7 peptides obtained in FOXM1-associated protein complex (Figs. 1D and S2B). To validate the interaction between FOXM1 and USP7, we transfected H1299 cells with a vector expressing FLAG-tagged FOXM1 in the presence or absence of a vector expressing untagged USP7. As shown in Fig. 1E, USP7 was readily detected in the immunoprecipitated complex of FOXM1 by flag agarose beads. Conversely, FOXM1 was co-immunoprecipitated with HA-tagged USP7 using HA agarose beads (Fig. 1F). To evaluate this interaction under physiological conditions, we performed co-immunoprecipitation assays with endogenous proteins from human TNBC cells MDA-MB-468 and MDA-MB-231. As shown in Fig. 1G, endogenous USP7 protein was co-precipitated by a FOXM1-specific antibody in both TNBC cells. The interaction of endogenous FOXM1 and USP7 was also validated in H1299 and human head and neck squamous cell cancer CAL33 cells (Fig. S2C, D). To ascertain whether FOXM1 and USP7 interact directly, we performed in vitro GST pull-down assays by incubating purified Flag-tagged FOXM1 with a GST-fusion protein containing full-length USP7. As shown in Fig. 1H, FOXM1 strongly bound the GST-USP7 fusion protein but not GST alone. Thus, USP7 is a bona fide binding partner of FOXM1 both in vivo and in vitro.
USP7 deubiquitinates and stabilizes the FOXM1 protein
To explore the consequence of the interaction between FOXM1 and USP7, we tested if the function of USP7 as a deubiquitinase affects the FOXM1 protein stability. H1299 cells were transfected with Flag-tagged FOXM1 and increasing amounts of USP7 expression vectors. Western blot showed that FOXM1 protein levels were significantly increased upon USP7 expression in a dose-dependent manner (Fig. 2A). Conversely, USP7 depletion by specific siRNA dramatically reduced FOXM1 protein levels in several human cancer cells including triple-negative breast cancer MDA-MB-468, MDA-MB-231, BT549; head and neck squamous cell cancer CAL33 and lung cancer cell H460 (Figs. 2B and S2E). These results indicate that the USP7 expression upregulates FOXM1 protein levels.
A Immunoblots of the extracts from F-FOXM1 in CAL33 cells that were transfected with indicated constructs for 24 h. B MDA-MB-468, MDA-MB-231 and BT549 cells were transfected with non-targeting control siRNA or USP7 specific siRNA for 72 h. the expression of FOXM1 protein was detected by immunoblotting. C Immunoblots for FOXM1 of H1299 cells after transfection with F-FOXM1 alone, or together with wildtype USP7 (WT) or mutant USP7 C223S (CS) for 24 h. D H1299 cells were transfected with indicated constructs for 36 h and were treated with10 μM of MG132 for 4 h before harvest. After anti-Ni-NTA immunoprecipitation under the denaturing condition, the ubiquitination of FOXM1 was detected by immunoblots. E Immunoblots of FOXM1 in H1299 cells transfected with the constructs as indicated for 24 h, followed by incubation with 25 μg/ml of cycloheximide for additional 0, 2.5, 5 and 10 h. F Quantification of FOXM1 protein levels in (E) by ImageJ. These data represent two independent experiments.
To see if the deubiquitinase activity of USP7 is required, we utilized a USP7 mutation C223S that lacks the deubiquitinase activity. H1299 cells were co-transfected Flag-tagged FOXM1-expressing vector with wild-type USP7 or USP7 C223S mutant expressing vector. As expected, wild-type USP7, but not USP7 C223S mutant, was able to increase the FOXM1 protein levels (Fig. 2C). These results suggest that USP7 regulates FOXM1 through its deubiquitinase activity. We next investigated the effect of wild-type USP7 and USP7 C223S mutant on the FOXM1 polyubiquitination. As shown in Fig. 2D, co-transfection of wild-type USP7, but not USP7 C223S mutant, dramatically reduced the polyubiquitination levels of FOXM1, further indicating that USP7 regulates FOXM1 protein stability through deubiquitination. Consistently, we also determined the FOXM1 protein half-life in H1299 cells and found that co-expression of USP7 markedly extended the FOXM1 protein half-life from ~5 h to over 10 h (Fig. 2E, F). In addition, Pearson correlation analysis between the expression of FOXM1 signature and USP7 in BRCA revealed a positive correlation, albeit the correlation coefficient is small (Fig. S3A, B). Taken together, these results demonstrate that FOXM1 is a bona fide substrate of USP7 deubiquitinase.
PU7-1 is a potent and selective PROTAC degrader for USP7 protein
Recently, PROTAC technology is emerging as a breakthrough in drug discovery and development [41]. A PROTAC degrader contains three moieties: ligand for protein of target (POI), linker and ligand for cellular E3 ligase. Mechanistically, it has at least three major advantages over traditional small-molecule inhibitors. First, several studies have proved that PROTACs can achieve higher selectivity due to the need of recruiting both POI and E3 ligase to form a productive ternary complex [42, 43]. Second, PROTACs can achieve high potency at lower doses. Since PROTAC functions like a catalytic enzyme, it can be recycled after POI is degraded. The off-target toxicity caused by high dosage is often avoided. Third, all functions associated with the POI can be effectively blocked. Small-molecule inhibitors usually can abolish the activity of POI but are unable to block protein-protein interaction. PROTACs completely degrade POI and eliminate all corresponding cellular functions. Therefore, to repress FOXM1 in triple-negative breast cancer, we aimed to develop a novel USP7 PROTAC degrader.
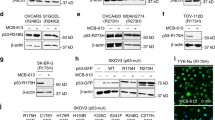
To specifically degrade USP7, we designed and synthesized a set of 66 PROTAC compounds that contain an usp7 specific binding moiety XL177A and an E3 ubiquitin ligase (CRBN or VHL) binding moiety connected by different linkers. All putative USP7 PROTACs were used to treat CAL33 cells that express high levels of USP7 protein, after which the USP7 protein levels were determined by western blot (Fig. S4A, B). Through initial screening, PU7-1 was identified as a potential USP7 degrader (Figs. 3A and S4). As shown in Fig. 3B, C, USP7 protein levels were dramatically reduced in CAL33 cells upon PU7-1 treatment in a dose- and time-dependent manner. Consistent with this effect, MDM2, a well-studied target of USP7 deubiquitinase, was decreased significantly in a similar manner. We also tested the effect of PU7-1 in several human cancer cells harboring wild-type p53. The data showed that PU7-1 treatment effectively reduced USP7 protein levels and activated p53 pathway (Fig. S5A). In addition, to exclude the possibility that PU7-1 affects the USP7 protein levels by regulating its transcription, we performed quantitative real-time PCR in CAL33 cells treated with PU7-1 for 72 h. Our data suggested that the expression levels of USP7 mRNA were not altered by PU7-1 treatment (Fig. 3D). In addition, we examined the half-life of USP7 protein in CAL33 cells in the presence or absence of PU7-1 treatment. As shown in Fig. 3E, F, the PU7-1 treatment remarkably decreased the USP7 half-life from over 24 h to ~15 h. We also treated CAL33 cells with different dosages of PU7-1 ranging from 0.0001 to 10 μM for 24 h. Western blot and densitometry analysis of USP7 protein bands using IMAGE J software revealed that PU7-1 effectively decreased USP7 protein at a DC50 of 4.3 nM (Figs. S5B, 3G and Supplementary Table S2). These findings indicated that PU7-1 can effectively decrease the protein levels of USP7 through targeting its protein stability.
A Chemical structure of USP7 PROTAC degrader PU7-1. B CAL33 cells were treated with indicated dosages of PU7-1 for 72 h. USP7 protein levels were detected by immunoblots. C CAL33 cells were treated with 10 μM of PU7-1 for indicated times. USP7 protein levels were measured by immunoblots. D Quantitative real-time PCR were used to determine the USP7 mRNA levels in CAL33 cells treated with or without PU7-1 for 72 h. ns, p > 0.05. E Immunoblots of USP7 in CAL33 cells treated with or without 10 μM of PU7-1 48 h, then incubated with 25 μg/ml of cycloheximide for additional 0, 8, 12 and 24 h. F Quantification of USP7 protein levels in (E) by ImageJ. G CAL33 cells were treated with indicated concentrations of PU7-1 for 24 h. USP7 protein levels were detected by immunoblots, then quantified by ImageJ after normalized to internal control β-actin. Half maximal degradation concentration (DC50) was calculated by Graphpad Prism. H Immunoblots of CRBN and USP7 in control and CRBN crispr knockout (KO) CAL33 cells that were treated with or without 2.5 μM of PU7-1 for 72 h. I Immunoblots of USP7 in CAL33 cells that were treated with DMSO or 5 μM of PU7-1 for 48 h, followed by the treatment with 5 μM of MLN4924, or 25 μM of Chloroquine (CLQ) or 5 μM of MG132 for 24 h before harvest. J, Heatmap of the deubiquitinases in quantitative proteomics analysis of A549 cells that were treated with 2.5 μM PU7-1 versus DMSO for 72 h. These data represent two independent experiments.
To investigate the mechanism of the effect of PU7-1, CRBN knockout (KO) CAL33 cell line was generated using CRISPR-cas9 technology. We treated control and CRBN KO cells with 2.5 μM of PU7-1 for 72 h and then detected the expression levels of USP7 and CRBN proteins by western blot analysis. As expected, CRBN protein was undetectable in the CRBN KO cells (Fig. 3H). Notably, CRBN depletion completely abrogated the USP7 degradation mediated by PU7-1 treatment. In addition, PU7-1N, in which a methyl group was installed to impair the binding of CRBN, also exhibited reduced capability to degrade USP7 protein (Fig. S5C). Furthermore, Cal33 cells were first treated with 5 μM of PU7-1 for 48 h to degrade the USP7 proteins, and then treated with 5 μM of MLN4924, 25 μM of Chloroquine (CLQ) or 5 μM of MG132 for additional 24 h. Western blot analysis revealed that both Cullin E3 ligase inhibitor MLN4924 and proteosome inhibitor MG132 significantly rescued the USP7 degradation mediated by PU7-1, whereas lysosome inhibitor Chloroquine (CLQ) had no effect on USP7 abundance (Fig. 3I). Taken together, these data demonstrated that the USP7 degradation by PU7-1 requires cellular CRBN/Cullin E3 ligase via ubiquitination-proteosome pathway.
Lastly, because of the high homology in USP protein family, we explored the selectivity of PU7-1 against potential off-target effects. Western blot analysis of several USPs including USP47, the closest homolog of USP7, were performed in CAL33 cells treated with 10 μM of PU7-1 for 72 h. The result indicated that USP7 was the only protein degraded upon the treatment, while the protein levels of the other USPs had no change (Fig. S5D). To further investigate the selectivity of PU7-1 proteome-wide, we performed mass spectrometry-based quantitative proteomic analysis in human lung cancer A549 cells treated with or without 2.5 μM of PU7-1 for 72 h. As shown in Fig. S5E and Supplementary Table S2, of ~5500 proteins quantified, USP7 was the most significantly decreased protein upon the treatment with PU7-1. In contrast, there was no measurable decrease in the protein levels of the other 29 deubiquitinases (Fig. 3J). Our findings indicated that PU7-1 is an effective and selective PROTAC degrader for USP7 protein.
PU7-1 treatment leads to FOXM1 degradation and consequently suppresses its transcriptional targets
Since our results demonstrate that FOXM1 is a bona fide target of USP7 deubiquitinase, we next examined the effect of PU7-1 on FOXM1. As expected, FOXM1 protein levels were significantly decreased in a dose- and time-dependent manner in CAL33 cells upon PU7-1 treatment, well correlated with USP7 protein levels (Fig. 4A, B). Consistently, the FOXM1 half-life was significantly reduced from more than 6 h to 3 h upon PU7-1 treatment in CAL33 cells (Fig. 4C, D). These results demonstrate that the treatment with USP7 degrader PU7-1 results in the destabilization of FOXM1. This effect was readily blocked by Cullin inhibitor MLN4924 and proteosome inhibitor MG132, but not by lysosome inhibitor BafA1 (Fig. 4E), suggesting that PU7-1-induced destabilization of FOXM1 requires the ubiquitination-proteosome pathway.
A Immunoblots of FOXM1 protein in CAL33 cells that were treated with indicated concentrations of PU7-1 for 96 h. B Immunoblots of FOXM1 protein in CAL33 cells that were treated with 10 μM of PU7-1 for indicated times. C Immunoblots of FOXM1 in CAL33 cells treated with or without 10 μM of PU7-1 for 48 h, then incubated with 50 μg/ml of cycloheximide for additional 0, 2, 4 and 6 h. D Quantification of FOXM1 protein levels in (C) by ImageJ. E Immunoblots of FOXM1 in CAL33 cells that were treated with 10 μM of PU7-1 for 72 h and treated with 1 μM of MLN4924 and BafA1 for 24 h and 10 μM of MG132 for 6 h before harvest. F, G CAL33 cells were treated with or without with 10 μM of PU7-1 for 96 h. Total RNA was extracted and subjected to RNAseq analysis. Differentially expressed genes were further analyzed. Top10 downregulated KEGG enriched pathways were shown in (F). Heatmap in (G) represents the downregulation of known FOXM1 target genes. H qPCR analysis of FOXM1 target genes in CAL33 cells treated with 10 μM of PU7-1 for 96 h. These data represent two independent experiments.
Transcription factor FOXM1 is a master regulator of the cell cycle. It tightly controls the cell cycle progression and maintains sustained cell proliferation via its various downstream target genes crucial for cell cycle regulation [16]. To explore the functional consequences of PU7-1-mediated FOXM1 degradation, we performed a transcriptome analysis. CAL33 cells were treated with or without 10 μM of PU7-1 for 96 h and total RNA were extracted for RNAseq analysis. We identified ~5000 differentially expressed genes (DEGs) in DMSO-treated versus PU7-1-treated cells (Supplementary Table S3). Of note, gene ontology analysis of the DEGs revealed that FOXM1 pathway was one of the top hits repressed by PU7-1 (Fig. 4F). Gene expression profiles demonstrated that FOXM1 downstream target genes were significantly downregulated upon PU7-1 treatment (Fig. 4G). This effect was further validated in CAL33 cells by quantitative real-time PCR (Fig. 4H). Our data suggest that PU7-1 is highly effective in suppressing the transcriptional activities of FOXM1. Consistent with this observation, PU7-1-treated CAL33 cells exhibited a significantly lower percentage of BrdU incorporation, an indicator of DNA replication and cell proliferation, suggesting slowed cell cycle progression (Fig. S6A, B). Notably, colony formation assay revealed that PU7-1 induced marked growth inhibition in several human cancer cell lines including CAL33, BT474 (breast cancer), DU145 (prostate cancer) and SK-N-DZ (brain cancer) (Fig. S6C, D). Collectively, these data indicated that the USP7 degrader PU7-1 can effectively repress FOXM1 transcription network and induce dramatic inhibition of cell proliferation.
PU7-1 mediates FOXM1 degradation, cell proliferation inhibition and tumor suppression in TNBC
Given the highly expression of FOXM1 in triple-negative breast cancer, we next investigate the effects of the USP7 degrader PU7-1 in TNBC cells. First, three TNBC cell lines MDA-MB-468, MDA-MB-231 and BT549 were treated with 10 μM of PU7-1 for 96 h. The protein levels of FOXM1 were determined by Western blot. As shown in Fig. 5A, PU7-1 treatment dramatically decreased the FOXM1 protein levels in all TNBC cells. Next, we examined the effects of PU7-1 on TNBC cell proliferation. MDA-MB468 and BT549 were incubated in the complete growth medium with or without 10 μM of PU7-1 for two weeks. Colonies were stained by crystal violet dye that stains nucleic acids and proteins and is used to reflect the cell number. As shown in Fig. 5B, much less colonies were observed in PU7-1-treated wells. Furthermore, the dye bound to the cells was solubilized with 30% acetic acid and measured by the absorbance at 600 nm. The viability of PU7-1-treated cells is about 20% of the control cells in MDA-MB-468 or 30% of the control cells in BT549 (Fig. 5C). In addition, BrdU incorporation assay in BT549 exhibited similar effects (Fig. S7A, B). The IC50 of PU7-1 in TNBC cells were then determined using a luminescent cell viability assay. As shown in Fig. 5D, PU7-1 treatment led to the proliferation inhibition in TNBC cells with IC50 of 1.8 μM in MDA-MB-468 and 2.8 μM in BT549. These results demonstrate that the USP7 degrader PU7-1 induces significant cell proliferation inhibition in TNBC cells. Lastly, we utilized the xenograft mouse model to investigate the in vivo efficacy of PU7-1 in TNBC. Two million of MDA-MB-468 cells were subcutaneously inoculated into 6–8-week female nude mice. PU7-1 was intraperitonially administrated at the dosage of 37.5 mg/kg daily for 3 weeks, 6 days per week (Fig. 5E). At the endpoint, tumors were dissected, weighed, and photographed. The results showed that PU7-1 significantly reduced the size and weight of the formed tumors in xenograft mouse model using MDA-MB-468 cells (Fig. 5F, G), indicating a promising therapeutic potential of PU7-1 in treating triple-negative breast cancer. In addition, the expression of USP7, FOXM1 and the proliferation marker Ki67 in tumors was examined by immunohistochemistry. As shown in Fig. S7C, tumor sections from PU7-1-treated mice displayed dramatically lower levels of USP7, FOXM1 and Ki67. Collectively, our findings demonstrated that the USP7 degrader PU7-1 inhibits cellular proliferation and tumor growth in TNBC.
A Immunoblots of FOXM1 in TNBC cell lines MDA-MB-468, MDA-MB-231 and BT549 treated with or without 10 μM of PU7-1 for 96 h. B, C Representative images (B) and normalized absorbance at 600 nm (C) of colony-formation assays in MDA-MB468 and BT549 treated with or without 10 μM of PU7-1 for 2 weeks. D Representative dose-dependent curves of CellTiter-Glo cell viability assay in MDA-MB-468 and BT549. E Treatment schedule for xenograft mouse models. F, G Representative images of MDA-MB-468 tumors (F) and quantification of tumor weight (G) in xenograft mouse model. **p < 0.01. These data represent two independent experiments.
FOXM1 is required for the PU7-1-mediated growth inhibition and tumor suppression in TNBC
To investigate if PU7-1-mediated effects in TNBC depend on FOXM1, we first tried to establish a FOXM1 crispr knockout cell line. Unfortunately, we were unable to generate the FOXM1 knockout cell line, likely because FOXM1 is an essential gene for cell survival. Previous studies demonstrated that FOXM1 is critical for embryonic development, and FOXM1−/− mice exhibits embryonic lethal phenotype due to multiple abnormalities in the major organs [44]. Alternatively, we generated an inducible FOXM1 overexpression cell line in MDA-MB-468 cells (MDA-MB-468 FOXM1 cells). As expected, doxycycline induces high levels of FOXM1 expression in MDA-MB-468 FOXM1 overexpression cells (Fig. 6A). The effects of PU7-1 on cell proliferation were compared in control and doxycycline-treated MDA-MB-468 FOXM1 cells using BrdU incorporation assay. As shown in Fig. 6B–D, PU7-1 significantly reduced BrdU positive cells from 32% to 15% in control cells, whereas no reduction of BrdU positive cells was observed after PU7-1 treatment in FOXM1 overexpression cells induced by doxycycline. These results indicated that FOXM1 is required for PU7-1-mediated proliferation inhibition in TNBC cells. Next, we tested the role of FOXM1 in PU7-1-induced tumor suppression. MDA-MB-468 FOXM1 overexpression cells were subcutaneously inoculated into 6-8-week female nude mice fed with doxycycline diet at the dosage of 625 mg/kg to induce exogenous FOXM1 expression. On day 7 after inoculation, mice were randomly separated into two groups. Vehicle or 37.5 mg/kg of PU7-1 were intraperitoneally administrated at the timeline shown in Fig. 5E. At the endpoint, tumors were dissected, weighed, and photographed. As shown in Fig. 6E, F, there was no significant difference in tumor sizes and weights between control and PU7-1 group, suggesting that replenishing of FOXM1 completely abrogates the tumor suppression mediated by PU7-1. These findings demonstrated the critical role of FOXM1 in the PU7-1-mediated proliferation inhibition and tumor suppression in triple-negative breast cancer.
A Immunoblots of FOXM1 in MDA-MB-468 FOXM1 cells treated with or without 1 μg/ml of doxycycline for 72 h. B MDA-MB-468 FOXM1 inducible cells were induced by 1 μg/ml of doxycycline overnight, then DMSO or 10 μM of PU7-1 were added for 72 h. FOXM1 protein levels were determined by immunoblotting. C, D Representative images of BrdU staining (C) and quantification of BrdU positive (BrdU+) cells (D) in MDA-MB468 FOXM1 inducible cells that were induced with 1 μg/ml Doxycycline overnight followed by the treatment of 10 μM of PU7-1 for 72 h. **p < 0.01. E, F Representative images of MDA-MB-468 FOXM1 tumors (E) and quantification of tumor weight (F) in xenograft mouse model. **p < 0.01. These data represent two independent experiments.
Discussion
In this study, we demonstrated the overexpression of oncogenic transcription factor FOXM1 in triple negative breast cancer. Further investigation of its interaction partners identified USP7 as the bona fide deubiquitinase of FOXM1. Given the “undruggable” nature of transcription factors, we utilized PROTAC technology and discovered a novel USP7 degrader PU7-1 to downregulate FOXM1 in TNBC. Notably, PU7-1 significantly elicited cell proliferation inhibition and tumor suppression via a FOXM1-dependent mechanism. These findings confirmed that targeting FOXM1 by USP7 degrader PU7-1 is a feasible and promising strategy, suggesting that downregulation of FOXM1 through inactivation of USP7 is a potential therapeutic option for triple-negative breast cancer. Moreover, Since FOXM1 overexpression in various human malignancies including head and neck cancer, liver cancer, colon cancer, prostate cancer, lung cancer, glioma and ovarian cancer [45, 46], it will be very interesting to explore the effect of PU7-1 in other human cancers.
Besides being a monotherapy, inactivation of FOXM1 by PU7-1 also can synergize with current TNBC treatments. In the past few years, FDA have approved two PARP inhibitors (Olaparib and Talazoparib) as monotherapy to treat HER2-negative metastatic breast cancer with germline BRCA1/2 mutations. The mechanism of action of PARP inhibitors in cancer treatment is the synthetic lethality [47,48,49], in which the cells do not die when when either one of the two genes is inhibited individually, but die when both of the two genes are inhibited. Poly(ADP-ribose) polymerase (PARPs) are a family of related enzymes that contribute to the recognition of DNA single-strand breaks (SSB) and facilitate DNA repair to maintain genomic stability [50]. BRCA1/2 genes encode tumor suppressor proteins that are crucial for the accurate repair of DNA double-strand breaks by homologous recombination (HR). Breast cancer cells with BRCA1/2 deficiency are unable to perform HR. In the absence of HR, alternative DNA repair mechanisms such as PARP-dependent SSB repair are used for cell survival. Both preclinical and clinical studies have demonstrated that Inhibition of PARP has been synthetically lethal in cancer cells with BRCA deficiency [49, 51,52,53]. As mentioned above, about 10%-20% of TNBCs carry BRCA mutations. Therefore, it is not surprising that only a fraction of TNBC patients can benefit from the PARP inhibition. Of note, most patients that initially responded to PARP inhibitors develop resistance to these agents, resulting in disease relapse [54]. Thus, it is still urgent to make sustained effort on broadening the therapeutic options for triple negative breast cancer. Previous studies have reported that FOXM1-deficient cells exhibite increased DNA breaks and reduce expression of BRCA2, indicating an important role of FOXM1 in DNA repair [55, 56]. A study has reported that PARP inhibitor Olaparib induced the expression and nuclear localization of FOXM1 and targeting FOXM1 disrupted Olaparib-induced adaptive resistance in ovarian cancer [57]. Although future experiments are required, it is possible that the combination of FOXM1 degradation by PU7-1and PARP inhibition may enhance the sensitivity to PAPR inhibition in tumor suppression and overcame the adaptive resistance induced by PARP inhibitors in the treatment of TNBCs.
Immune checkpoint inhibitors such as anti-PD-1/PD-L1 antibodies are the major therapeutic advancements in oncology and have rapidly become a standard-of-care treatment for several solid tumor types [58]. Nevertheless, only a small portion (1–10%) of advanced-stage TNBC patients had long-lasting benefit beyond 24 months in the setting of single-agent therapy. A recent study reported that FOXM1 regulates PD-L1 expression by binding directly to its promoter and FOXM1 inhibition enhances the therapeutic outcome of lung cancer immunotherapy by repressing PD-L1 expression [59]. Interestingly, overexpression of PD-L1 was observed in TNBC cells [60]. Hence, targeting FOXM1 by PU7-1 likely suppresses PD-L1 expression, block PD-L1-mediated immune evasion and enhances the efficacy of immune checkpoint inhibitors in triple negative breast cancer. Moreover, USP7 was reported to modulate regulatory T (Treg) function through the rapid temporal control of Forkhead box protein P3(Foxp3) protein stability [61]. Transcription factor Foxp3 is crucial for the development and immunosuppression function of Treg cells. USP7 was found to deubiquitinate and stabilize Foxp3 in Treg cells. Treatment with USP7 inhibitor decreased Foxp3 protein levels and impaired Treg-mediated immunosuppression. These findings suggest that USP7 inactivation may promote anti-tumor immunity through both PD-L1 downregulation by FOXM1 repression in tumor cells and suppression of Tregs immune capacity in the tumor microenvironment. Thus, the effect of PU7-1 in immunoproficient condition, with both wild-type or mutant p53, is worth of further investigation.
Methods
Statistical analyses
Data are presented as the means ± SD or ±SEM. Statistics were performed using unpaired two-sided student’s t test for comparing two sets of data with the software GraphPad Prism. A P value of <0.05 was considered significant.
Mice
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Columbia University under the supervision of the Institute of Comparative Medicine. For xenograft mouse model, 2 × 106 MDA-MB-468 cells were mixed with Matrigel (Corning, #354248) at a 1:1 ratio (Volume) and injected subcutaneous into 6–8-week-old female nude mice (Charles River, #088). When tumors are palpable (~7 days after injection of tumor cells), mice were treated with either vehicle or PU7-1 at 37.5 mg/kg with daily intraperitoneal injections as indicated schedules. Once mice were euthanized, tumors were collected and weighed for analysis. The immunohistochemistry of tumor sections was performed by the Molecular Pathology core facility in Columbia University Herbert Irving Comprehensive Cancer Center.
Western blot
Cells were lysed in FLAG lysis buffer (50 mM Tris-HCl (pH 7.3), 137 mM NaCl, 10 mM NaF, 0.5 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 10% glycerol) freshly supplemented with protease inhibitor cocktail. Protein concentration was determined by the Bio-Rad protein assay. Whole cell extracts were then resolved by Novex Tris-Glycine gel (Thermo Scientific) and transferred onto a Nitrocellulose membrane (Millipore). Membranes were blocked with 5% (w/v) nonfat milk in Tris-buffered saline with Tween-20 (TBST), incubated with primary antibodies, then secondary HRP-conjugated antibodies (Jackson ImmunoResearch, 1:10000 dilution) and detected on autoradiographic films after incubating with ECL (Thermo Scientific, #32106). Primary antibodies used in this study include anti-USP7 (Cell Signaling, #4833), anti-CRBN (Cell Signaling, #71810); anti-P53 (SantaCruz Biotechnology, sc-126), anti-PUMA (Santa Cruz Biotechnology Cat# sc-28226); anti-p21 (Santa Cruz Biotechnology Cat# sc-53870); anti-FOXM1(Cell Signaling, #5436); anti-Actin (Sigma-Aldrich Cat# A5441); anti-vinculin (Sigma-Aldrich Cat# V9131).
Co-immunoprecipitation
For the co-immunoprecipitation assay of ectopically expressed proteins, cells were lysed in BC100 buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.2% Triton X-100, and 10% glycerol) freshly supplemented with protease inhibitor cocktail. Whole cell extracts were incubated with M2(Flag) agarose beads (Sigma, A2220) or HA agarose beads (Sigma, A2095) at 4 °C overnight. After four washes with BC100 buffer, the immunopricipitates were eluted with Flag-peptide (Sigma, F3290) or HA-peptide (Sigma, I2149) for 2 h at 4 °C.
For endogenous co-immunoprecipitation assay, cells were lysed in BC100 buffer freshly supplemented with protease inhibitor cocktail. Whole cell extracts were incubated with IgG, anti-USP7 or anti-Foxm1 antibodies as indicated at 4 °C overnight. Next day protein A sepharose (GE healthcare, #17-0780-01) was then added for 4 h at 4 °C. After four washes with BC100 buffer, the bound proteins were eluted by boiling with 1X SDS loading buffer.
Deubiquitination assay
H1299 cells were transfected with FLAG-FOXM1, USP7, and His-ubiquitin as indicated. At 24 h post-transfection, 10 μM MG132 were added for additional 4 h. 5% of the cells were lysed in FLAG lysis buffer and saved as input. The rest of the cells were lysed in phosphate/guanidine buffer (6 M Guanidine-HCl, 0.1 M Na2HPO4, 0.1 M NaH2PO4, 10 mM Tris-HCl, pH 8.0, freshly supplemented with 10 mM β-mercaptoethanol and 5 mM imidazole) with mild sonication. Cell extracts were incubated with Ni-NTA agarose (Qiagen, # 30210) at 4 °C overnight. Ni-NTA beads were washed once with lysis buffer, and then four times with urea wash buffer (8 M Urea, 0.1 m Na2HPO4, 0.1 M NaH2PO4, 10 mM Tris-HCl, pH 6.8, freshly supplemented with 10 mM β-mercaptoethanol and 5 mM imidazole). The bound proteins were eluted with elution buffer (0.5 M Imidazole, 10 mM Tris-HCl, pH 6.8, freshly supplemented with 10 mM β-mercaptoethanol).
GST pulldown
GST and GST-USP7 proteins were expressed and purified from BL21 E.coli (Milliporesigma, Cat#70954-4). F-FOXM1 proteins were purified by M2 IP from H1299 cells transiently transfected with F-FOXM1constructs. Purified F-FOXM1 proteins were incubated with GST or GST-USP7-conjugated GST resin at 4 °C overnight. The beads were washed 5 times with BC100 buffer, and then boiled with 1X SDS loading buffer. Precipitates were subjected to western blot analysis and Ponceau S staining.
Colony formation assay
1000 cells (10000 cells for BT474) were seeded in 12-well plates. Next day cells were treated with DMSO or PU7-1 for 2 weeks. Drugs were refreshed every 48 h. After fixation with cold methanol, cells were stained with 0.5% crystal violet solution for 10 min, and then washed with distilled water. For quantification purpose, 1 ml of 30% acetic acid was added to de-stain for 15 min with gentle rotation. The absorbance at 600 nm was measured by using a plate reader in triplicates.
BrdU incorporation
Cells were incubated with 10 ug/ml BrdU (BD, 51-2420KC) for 3 h and fixed with cold 70% ethanol. DNA was denatured in 1.5 N HCL for 30 min. Cells were blocked with 1%BSA in PBS, and incubated with anti-BrdU antibody (Roche, clone#BMC9318) at 4 °C overnight. After three washes with 1%BSA in PBS, cells were incubated with AlexaFlour 488 conjugated anti-mouse IgG (Thermo scientific, #A32723). Counterstaining with DAPI was performed to visualize the nuclei.
Cell viability assay
Cells were seeded in 96-well white plates (Corning, #3610) and incubated with serially diluted compounds for 6 days. Cell viability was determined using Celltiter glo2.0 cell viability assay (Promega, #G9243) as manufacturer’s instruction. IC50 values were determined after fitting curves using Graphpad Prism software.
USP7 siRNA knockdown assay
Cells were reversely transfected with non-targeting control or USP7 specific siRNA oligos purchased from Dharmacon for 72 h. The transfection was performed using Lipofectamine3000 (Thermo Scientific, # L3000015) following the manufacturer’s instruction.
Generation of CRNB knockout cell line and inducible FOXM1 cell line
For CRBN knockout cell line, the plenti-px330-CRBN-T1-pGK-Pur construct was purchased from Addgene. HEK293 cells were transiently transfected with plenti-px330-CRBN-T1-pGK-Pur, VSVG and ∆8.9 constructs for lentivirus packaging. Viruses were harvested at 48 h post-transfection. CAL33 cells were transfected with CAS9 constructs and infected with lentiviral particles, and then selected with 1ug/ml puromycin. CRBN knockout clones were identified by western blot using CRBN specific antibody.
For inducible FOXM1 cell line, the pCW57.1-FOXM1b construct was purchased from Addgene. Lentiviruses were prepared as above. MDA-MB-468 cells were transduced with lentiviruses, and then selected with 1 ug/ml puromycin. The FOXM1 expression was induced with 1 ug/ml doxycycline (Sigma, D9891).
Data availability
All uncropped blots and chemistry details were provided in supplementary file. All other data supporting the finding of this study are available from the corresponding author upon reasonable request.
References
Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–96.
Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–53.
Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–86.
Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–15.
Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–5.
Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22.
Harakandi C, Nininahazwe L, Xu H, Liu B, He C, Zheng YC, et al. Recent advances on the intervention sites targeting USP7-MDM2-p53 in cancer therapy. Bioorg Chem. 2021;116:105273.
Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–7.
Tavana O, Li D, Dai C, Lopez G, Banerjee D, Kon N, et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat Med. 2016;22:1180–6.
Bronner C. Control of DNMT1 abundance in epigenetic inheritance by acetylation, ubiquitylation, and the histone code. Sci Signal. 2011;4:pe3.
Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27.
Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–9.
Kon N, Zhong J, Kobayashi Y, Li M, Szabolcs M, Ludwig T, et al. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ. 2011;18:1366–75.
Bhattacharya S, Chakraborty D, Basu M, Ghosh MK. Emerging insights into HAUSP (USP7) in physiology, cancer and other diseases. Signal Transduct Target Ther. 2018;3:17.
Valles GJ, Bezsonova I, Woodgate R, Ashton NW. USP7 is a master regulator of genome stability. Front Cell Develop Biol. 2020;8:717.
Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13:482–95.
Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94.
Chen X, Muller GA, Quaas M, Fischer M, Han N, Stutchbury B, et al. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol. 2013;33:227–36.
Koo CY, Muir KW, Lam EW. FOXM1: from cancer initiation to progression and treatment. Biochim Biophys Acta. 2012;1819:28–37.
Wang IC, Chen YJ, Hughes DE, Ackerson T, Major ML, Kalinichenko VV, et al. FoxM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J Biol Chem. 2008;283:20770–8.
Huang C, Qiu Z, Wang L, Peng Z, Jia Z, Logsdon CD, et al. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res. 2012;72:655–65.
Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell. 2011;20:427–42.
Yao S, Fan LY, Lam EW. The FOXO3-FOXM1 axis: a key cancer drug target and a modulator of cancer drug resistance. Semin Cancer Biol. 2018;50:77–89.
Nilsson MB, Sun H, Robichaux J, Pfeifer M, McDermott U, Travers J, et al. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci Transl Med. 2020;12:eaaz4589.
Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N. Engl J Med. 2010;363:1938–48.
Dietze EC, Sistrunk C, Miranda-Carboni G, O’Regan R, Seewaldt VL. Triple-negative breast cancer in African-American women: disparities versus biology. Nat Rev Cancer. 2015;15:248–54.
Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90.
Geenen JJJ, Linn SC, Beijnen JH, Schellens JHM. PARP inhibitors in the treatment of triple-negative breast cancer. Clin Pharmacokinet. 2018;57:427–37.
Gonzalez-Angulo AM, Timms KM, Liu S, Chen H, Litton JK, Potter J, et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17:1082–9.
Wong-Brown MW, Meldrum CJ, Carpenter JE, Clarke CL, Narod SA, Jakubowska A, et al. Prevalence of BRCA1 and BRCA2 germline mutations in patients with triple-negative breast cancer. Breast Cancer Res Treat. 2015;150:71–80.
Pogoda K, Niwinska A, Sarnowska E, Nowakowska D, Jagiello-Gruszfeld A, Siedlecki J, et al. Effects of BRCA germline mutations on triple-negative breast cancer prognosis. J Oncol. 2020;2020:8545643.
Qi SM, Cheng G, Cheng XD, Xu Z, Xu B, Zhang WD, et al. Targeting USP7-mediated deubiquitination of MDM2/MDMX-p53 pathway for cancer therapy: are we there yet? Front Cell Develop Biol. 2020;8:233.
Lin YT, Lin J, Liu YE, Chen YC, Liu ST, Hsu KW, et al. USP7 induces chemoresistance in triple-negative breast cancer via deubiquitination and stabilization of ABCB1. Cells. 2022;11:3294.
Xia X, Liao Y, Huang C, Liu Y, He J, Shao Z, et al. Deubiquitination and stabilization of estrogen receptor alpha by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 2019;465:118–28.
Pandit B, Halasi M, Gartel AL. p53 negatively regulates expression of FoxM1. Cell Cycle. 2009;8:3425–7.
Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–305.
Lv C, Zhao G, Sun X, Wang P, Xie N, Luo J, et al. Acetylation of FOXM1 is essential for its transactivation and tumor growth stimulation. Oncotarget. 2016;7:60366–82.
Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–82.
Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–61.
Kopanja D, Chand V, O’Brien E, Mukhopadhyay NK, Zappia MP, Islam A, et al. Transcriptional repression by FoxM1 suppresses tumor differentiation and promotes metastasis of breast cancer. Cancer Res. 2022;82:2458–71.
Dale B, Cheng M, Park KS, Kaniskan HU, Xiong Y, Jin J. Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer. 2021;21:638–54.
Smith BE, Wang SL, Jaime-Figueroa S, Harbin A, Wang J, Hamman BD, et al. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat Commun. 2019;10:131.
Zengerle M, Chan KH, Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10:1770–7.
Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle. 2011;10:396–405.
Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal: CCS. 2018;16:57.
Li L, Wu D, Yu Q, Li L, Wu P. Prognostic value of FOXM1 in solid tumors: a systematic review and meta-analysis. Oncotarget. 2017;8:32298–308.
Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med. 2013;19:1381–8.
Turner N, Tutt A, Ashworth A. Targeting the DNA repair defect of BRCA tumours. Curr Opin Pharm. 2005;5:388–93.
Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8.
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7.
Tung NM, Robson ME, Ventz S, Santa-Maria CA, Nanda R, Marcom PK, et al. TBCRC 048: phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol. 2020;38:4274–82.
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl J Med. 2009;361:123–34.
Dias MP, Moser SC, Ganesan S, Jonkers J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat Rev Clin Oncol. 2021;18:773–91.
Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EW. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta. 2014;1839:1316–22.
Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–16.
Fang P, Madden JA, Neums L, Moulder RK, Forrest ML, Chien J. Olaparib-induced adaptive response is disrupted by FOXM1 targeting that enhances sensitivity to PARP inhibition. Mol Cancer Res. 2018;16:961–73.
Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat Rev Clin Oncol. 2022;19:91–113.
Madhi H, Lee JS, Choi YE, Li Y, Kim MH, Choi Y, et al. FOXM1 inhibition enhances the therapeutic outcome of lung cancer immunotherapy by modulating PD-L1 expression and cell proliferation. Adv Sci (Weinh). 2022;9:e2202702.
Mittendorf EA, Philips AV, Meric-Bernstam F, Qiao N, Wu Y, Harrington S, et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol Res. 2014;2:361–70.
van Loosdregt J, Fleskens V, Fu J, Brenkman AB, Bekker CP, Pals CE, et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity. 2013;39:259–71.
Acknowledgements
We thank Dr. Richard Baer for kind support in this project by providing critical reagents. This work was supported by the National Cancer Institute of the National Institutes of Health under Award R35CA253059, RO1CA258390 and R01CA254970 to WG. We acknowledge the support from the Herbert Irving Comprehensive Cancer Center (HICCC; P30 CA13696) and thank the Molecular Pathology and Proteomics of Shared Resources of HICCC. This work utilized the NMR Spectrometer Systems at Mount Sinai acquired with funding from the NIH’s SIG grants 1S10OD025132 and 1S10OD028504. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Conception and experimental design: JY and WG. Methodology and data acquisition: JY, HL, and BC. Analysis and interpretation of data: JY, HL, BC, and WG. Manuscript writing: JY and WG.
Corresponding author
Ethics declarations
Competing interests
JJ is a cofounder and equity shareholder in Cullgen, Inc., a scientific cofounder and scientific advisory board member of Onsero Therapeutics, Inc., and a consultant for Cullgen, Inc., EpiCypher, Inc., Accent Therapeutics, Inc, and Tavotek Biotherapeutics, Inc. The Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics, Inc., Cullgen, Inc. and Cullinan Oncology, Inc. The other authors declare that they have no competing financial interests.
Ethics approval
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Columbia University under the supervision of the Institute of Comparative Medicine.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yi, J., Li, H., Chu, B. et al. Inhibition of USP7 induces p53-independent tumor growth suppression in triple-negative breast cancers by destabilizing FOXM1. Cell Death Differ 30, 1799–1810 (2023). https://doi.org/10.1038/s41418-023-01180-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01180-7
- Springer Nature Limited
This article is cited by
-
A comprehensive molecular characterization of a claudin-low luminal B breast tumor
Biology Direct (2024)
-
Ubiquitin recruiting chimera: more than just a PROTAC
Biology Direct (2024)
-
USP33 facilitates the ovarian cancer progression via deubiquitinating and stabilizing CBX2
Oncogene (2024)