Abstract
Par6α encoded by PARD6A is a member of the PAR6 family and is reported to promote cancer initiation and progression. PARD6A is frequently upregulated in different types of cancers, but its regulatory role in lung cancer progression is yet to be established. In this study, we analyzed the PARD6A expression in biopsies from lung adenocarcinoma (LUAD) patients, and the survival probability using LUAD tissue microarray (TMA) and online datasets from TCGA and GEO. We conducted in vitro and in vivo assays to assess the role of PARD6A in regulating lung cancer progression, including proliferation, wound healing, transwell, RNA-seq, and subcutaneous tumor mice models. Our findings revealed that PARD6A is highly expressed in cancer tissues from LUAD patients and is associated with poor prognosis in LUAD patients. In vitro assays showed that PARD6A promoted cell proliferation, migration, and invasion. The transcriptome sequencing identified Serpina3 as one of the key downstream molecules of PARD6A. Ectopic expression of Serpina3 rescued impaired proliferation, migration, and invasion in PARD6A-knocking down H1299 cells, whereas silencing Serpina3 impeded enhanced proliferation, migration, and invasion in PARD6A-overexpressing H1975 cells. Our findings suggest that PARD6A promotes lung cancer progression by inducing Serpina3, which may be a promising therapeutic target.
Similar content being viewed by others
Introduction
Lung cancer is one of the most diagnosed cancers with high morbidity and mortality worldwide [1, 2]. Lung cancer is composed of 80–85% non-small cell lung cancer (NSCLC) and 15–20% small cell lung cancer (SCLC). NSCLC can be further categorized into three histologic subtypes, lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large cell lung cancer (LCLC). The 5-year overall survival of lung cancer is about 50–70% at stage I and about 1–5% at stage IV [3]. Although the prognosis is good at the early stage, only 20% of lung cancer patients are diagnosed at stage I but most lung cancer patients are diagnosed at a late stage with distant metastasis [4]. Therefore, metastasis is one of the leading causes of cancer-related death.
Partitioning-defective protein 6 (Par6) family compromises of three members, Par6α, Par6β, and Par6γ, which are encoded by partitioning defective 6 homolog alpha (PARD6A), partitioning defective 6 homolog beta (PARD6B), and partitioning defective 6 gamma (PARD6G), respectively. Par6 family members form the PAR protein complex by interacting with cell division cycle 42 (Cdc42), Par4, and atypical protein kinase C (αPKC), which is critical in cell polarity and contributes to cell differentiation, proliferation, and morphology [5]. Previous studies have demonstrated that Par6 proteins promote cancer progression in a polarity-dependent and independent manner and are the critical hinge of signaling pathways [6,7,8]. Par6 interacts with ErbB in breast cancer cells, thereby protecting cancer cells from apoptosis [6]. Par6 can also phosphorylate and activate extracellular signal-regulated kinase 1/2 (ERK1/2) by interacting with Cdc42, thereby promoting the proliferation of breast cancer cells [9]. In NSCLC cells, Par6 binds to epithelial cell transforming 2 (Ect2) and activates Rac, stimulating anchorage-free growth and invasion [10]. Additionally, Par6 induces expression of CD44 and N-cadherin, which subsequently enhances glioma invasion by activating the mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) /signal transducer and activator of transcription 3 (STAT3) pathway [11]. Recently, Par6α has been reported to upregulate Snail expression by regulating the integrin-β/integrin-linked kinase (ILK) axis and instigating the epithelial-mesenchymal transition (EMT) of ovarian cancer cells [12]. However, its function is not well structured in NSCLC.
Serpin family A member 3 (Serpina3), also known as α-1-antichymotrypsin (AACT, ACT), is one of the inhibitors of serine proteases and is found elevated in neurological diseases and various types of cancers, including glioblastoma, colorectal cancer, endometrial cancer, breast cancer, and melanoma [13]. It has been reported that Serpina3 promotes stemness, inaction, migration, proliferation, redox homeostasis, and drug resistance in cancer cells [14,15,16,17]. Contrarily, other studies have demonstrated that nuclear Serpina3 inhibits the proliferation of liver cancer cells and LUSC cells by inducing chromatin condensation and inhibiting NF-κB/speckle-type POZ protein (SPOP), respectively [18, 19]. Noteworthily, the role of Serpina3 in cancer progression varies in different cancers, and its regulatory function in LUAD remains unclear.
In this project, we analyzed the clinical data of NSCLC patients from lung cancer microarray, TCGA, and GEO. We found a higher expression of PARD6A (gene encodes Par6α) in cancer tissue than in adjacent normal tissues, which is associated with poor prognosis in LUAD patients. The expression of PARD6A is higher in lung cancer patients at late stages (stage IV). In vitro and in vivo assays showed that PARD6A promotes the proliferation, migration, and invasion of H1975 and H1299 cells. We identified Serpina3 as a critical downstream of PARD6A that may promote cancer progression in LUAD patients. Survival analysis revealed that LUAD patients with high levels of PARD6A and Serpina3 showed the poorest prognosis. Overall, we identified that PARD6A promotes the malignant progression of LUAD by upregulating Serpina3. Our study might provide new insights for diagnosis, prognosis, and treatment in LUAD patients.
Materials and methods
Data source
The microarray data and clinical information of lung cancer samples (GSE87340, N = 27; GSE63459, N = 65; GSE42127, N = 133; GSE41271, N = 183) were downloaded from the Gene Expression Omnibus (GEO) database. The gene expression and clinical information of lung adenocarcinoma samples were downloaded from The Cancer Genome Atlas (TCGA). The fragments per kilobase of transcript per million mapped reads (FPKM) and clinical data of LUAD patients treated with Nivolumab were downloaded from the Tumor Immune Dysfunction and Exclusion (TIDE) website (GSE126044 and GSE135222). All the clinicopathological information is listed in Tables S1–S7. Kaplan–Meier analysis of progress-free survival was performed online (http://kmplot.com) [20, 21].
Cell lines
The embryonic kidney HEK293T, human normal bronchial epithelial cells (BEAS-2B), and lung cancer cell lines (H1299, H1975, and PC9) were purchased from Kunming Cell Bank. HEK293T and H1299 were cultured in Dulbecco’s Modified Eagle Medium (Cat. 11965092, Gibco, MA, USA) supplemented with 10% fetal bovine serum (FBS, Cat. 100-106G-500, Gemini Bio, CA, USA) and 1% penicillin‑streptomycin (Cat. 15140122, Thermo Fisher, MA, USA). BEAS-2B, H1975, and PC9 were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Cat. 11875119, Gibco) supplemented with 10% FBS and 1% penicillin-streptomycin. Cell lines were authenticated every year during experiments via short-tandem repeat testing (STR, Beijing Tsingke Biotech, China). Cell lines were passaged for a period of 3 months after thawing from liquid nitrogen. Cell lines were tested for mycoplasma contamination after thawing from liquid nitrogen.
Bioinformatic analysis
The enrichment analysis aimed to identify alterations in specific signaling pathways potentially associated with the overexpression of the PARD6A gene in H1975. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were employed for this purpose, utilizing the clusterProfiler package in the R programming language (P < 0.05 and FDR < 0.25).
We downloaded the pan-cancer dataset (TCGA Pan-Cancer, PANCAN, N = 10,535, G = 60,499) from the University of California, Santa Cruz (USCS) database (https://xenabrowser.net/). We performed a log2(x + 1) transformation on the expression value of Serpina3 and visualized the expression data from 26 types of cancer by R software (version 3.6.4). The non-paired Wilcoxon Rank Sum and Signed Rank Tests were performed to calculate the significant difference.
Interactions between proteins related to PARD6A were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://cn.string-db.org/) and online software [22]. Proteins were clustered in different colors according to the K-Means clustering.
Reverse-transcription quantitative PCR (RT-qPCR)
Total RNAs from culture cells and murine tissues were purified using RNA isolater Total RNA Extraction Reagent (Cat. R401-01, Vazyme Biotech, China). cDNAs were synthesized using PrimeScript RT Master Mix (Perfect Real Time) (Cat. RR036A, Takara, Japan) according to the manufacturer’s instructions. PCR reactions were performed using ChamQ Universal SYBR qPCR Master Mix (Cat. Q711-02, Vazyme Biotech) and a Bio-Rad CFX96 Touch Deep Well Real-Time PCR Detection System. The reaction was performed using a two-step cycling protocol and quantification was performed using the 2−ΔΔCq method [23]. Relative mRNA levels were normalized to the mRNA levels of the GAPDH gene. Gene-specific primers are listed as follows:
Primer name | Primer sequence (5’-3’) |
|---|---|
Human GAPDH-Forward | GTC TCC TCT GAC TTC AAC AGC G |
Human GAPDH-Reverse | ACC ACC CTG TTG CTG TAG CCA A |
Human PARD6A-Forward | CTA TAC GGA TGC TCA TGG CGA C |
Human PARD6A-Reverse | GAG AGT TGG AGG CAA AAG CCA G |
Human SERPINA3-Forward | CCT GAA CGA CAT ACT TCT CCA GC |
Human SERPINA3-Reverse | CAT CAA GCA CAG CCT TAT GGA CC |
Human MMP3-Forward | AGT CTT CCA ATC CTA CTG TTG CT |
Human MMP3-Reverse | TCC CCG TCA CCT CCA ATC C |
Human MMP9-Forward | TGT ACC GCT ATG GTT ACA CTC G |
Human MMP9-Reverse | GGC AGG GAC AGT TGC TTC T |
Human PCNA-Forward | CCT GCT GGG ATA TTA GCT CCA |
Human PCNA-Reverse | CAG CGG TAG GTG TCG AAG C |
Human VTCN1-Forward | CTC ACA GAT GCT GGC ACC TAC A |
Human VTCN1-Reverse | GCA AGG TCT CTG AGC TGG CAT T |
Human ZSCAN4-Forward | GAT GAC AGC ATA AAT CCA CCT GC |
Human ZSCAN4-Reverse | TTG CTT CTC TTG TGG TTT GGG CA |
Human DLEU7-Forward | CCA GAG GAG GAG GTA GTG CGA |
Human DLEU7-Reverse | TGA CCA GCT CCG AAG TCG AGT C |
Human TP53AIP1-Forward | AGA CCA GAA CCT CTC GGT GAT G |
Human TP53AIP1-Reverse | ACC ACG GTG AGA GCA GAG TCT G |
Western blotting (WB)
Cells were washed with pre-cool phosphate-buffered saline (PBS, pH 7.8) three times and lysed with RIPA lysis buffer (Cat. 89900, Thermo Fisher, USA) supplemented with protease and phosphatase inhibitors (Cat. A32953, Thermo Fisher). The protein was quantified using Bio-Rad protein assay (Cat. 5000002, Bio-Rad, USA). Lysates (40 μg) were resolved by SDS-PAGE gel, transferred to 0.45 μm PVDF membrane (Cat. IPFL00010, Millipore, MA, USA), and incubated with primary antibodies at 4 °C overnight, followed by incubation with horse radish peroxidase-conjugated secondary antibodies (Cat. 31460 and Cat. 31430, Thermo Fisher) for 1 h at room temperature. Immunoreactive bands were visualized by chemiluminescence (Cat. E412-01, Vazyme Biotech, or Cat. 1856194, Thermo Fisher). The band size of blots was analyzed by using ImageJ software [24]. The absolute expression of a specific band was calculated as AE = TE/RE (AE represents absolute expression, TE represents the band size of the target protein, and RE represents the band size of the reference).
Antibodies are listed as follows:
Antibody | Catalog number | Manufacture |
|---|---|---|
Anti-Tublin | 250175 or R23623 | ZEN-BIOSCIENCE |
Anti-Vinculin | 66305-1-Ig | ProteinTech, IL, USA |
Anti-GAPDH | 250007 | ZEN-BIOSCIENCE |
Anti-Par6α | sc-365323 | Santa Cruz, Texas, USA |
Anti-Par6β | 13996-1-AP | ProteinTech, USA |
Anti-Par6γ | PA5-113332 | Thermo Fisher |
Anti-Serpina3 | 12192-1-AP | ProteinTech |
Anti-MMP3 | 10375-1-AP | ProteinTech |
Anti-MMP9 | 10375-2-AP | ProteinTech |
Anti-PCNA | 10205-2-AP | ProteinTech |
Small hairpin RNA (shRNA) and overexpression plasmids
PARD6A, Serpina3, and corresponding non-target control (shNT) were designed according to the cDNA sequence on the Broad Institute Website. The shRNA was constructed into pLKO-1_TRC plasmid. Par-6 and Serpina3 were constructed with the standard polymerase chain reaction (PCR) technique using Phanta HS Super-Fidelity DNA Polymerase (Cat. P502-d1, Vazyme Biotech) into pENTER vector or plvx-AcGFP plasmids. The plasmids were confirmed by sequencing from Beijing Tsingke Biotech. H1975 was transfected with pENTER empty vector of pENTER_Par-6 using the Lipo3000 DNA Transfection Reagent (Cat. L3000001, Thermo Fisher) according to the manufacturer’s instructions and screened using 2 μg/ml of puromycin (Cat. ST551, Beyotime, China). The knocking-down or overexpressing lentiviruses were produced in HEK293T cells by co-transfecting shRNA or overexpressing plasmid, psPAX2, and pMD2.G using Lipo3000 reagent. After 48 h and 72 h, the supernatant was harvested and concentrated as described before [25]. The target cells at 70–80% confluence were infected with concentrated lentiviral particles and treated with 8 μg/ml of polybrene (Cat. TR-1003-G, Sigma-Aldrich, MA, USA). The infected cells were screened by 2–4 μg/ml of puromycin for 7 days and the positive cells were verified by qRT-PCR and western blot.
Proliferation and colony formation assays
Cells were seeded at a density of 3000 cells/well in 96-well plates (six wells per group) and cultured at 37 °C, 5% CO2 incubator until cells were fully attached. Cell Counting Kit 8 (CCK8, Cat. HY-L0301, MCE, USA) was used to detect cell proliferation viability. At 0, 24, 48, 72, 96, 120, and 144 h, 90 μl of pre-warm PBS and 10 μl of CCK8 reagent were mixed and added to each well, followed by incubation at 37 °C, 5% CO2 incubator for 1 h. The absorbance was measured at a wavelength of 450 nm using a SpectraMax® M3 Multi-Mode Microplate Reader (Molecular Devices, CA, USA). Each group has five replicates, and every experiment repeats at least three times.
Alive cells were counted using trypan blue and seeded at a density of 200 or 300 cells/well in 6-well plates. The cells were incubated for 2–3 weeks to allow the colony formation. Cells were stained with crystal violet for 30 min at room temperature. Crystal violet solution was removed, and the plates were washed under tap water before being dried for 24 h. Each group has three replicates, and every experiment repeats at least three times.
Wound healing assay
Alive cells were counted using trypan blue and seeded at a density of 6 × 105 cells/well in 6-well plates. When cells reached 70–80% confluence, the medium was changed into the serum-free medium to allow the cells to synchronize for 6 h. One 200 μl pipette tip was used to scratch a wound. The cells were washed with warm PBS three times and cultured in the complete medium. At 0, 24, and 36 h, the wound closure was observed and imaged under a microscope. The wound closure rates were calculated and presented in the bar graph. Each group has three replicates, and every experiment repeats at least three times.
Transwell assay
Transwell assays were conducted to detect the migration and invasion of cells with 8 μm-pore size transwell chamber (Cat. 3412, Corning, NY, USA) coated without or with growth factor reduced Matrigel (Cat. 354230, Corning), respectively. Alive cells were counted using trypan blue and seeded at a density of 2E + 4 cells in 200 μl of the serum-free medium in the upper chamber. In the lower chamber, 700 μl of the medium was added. After 24-h incubation, the inserted chambers were fixed with 4% paraformaldehyde for 20 min and then stained with crystal violet for 20 min. Crystal violet solution was removed and the chamber was carefully washed using cotton tips. Five random fields of migrated or invasive cells were imaged under a microscope. The average number of migrated or invasive cells was calculated and presented in the bar graph. Each group has three replicates, and every experiment repeats at least three times.
Cell-derived xenograft
All animal experiments were conducted under the project license (No. 2023-80) granted by the Sichuan Provincial People’s Hospital Committee on Animal Care and Use, in compliance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals and the institutional guidelines [26]. Cell-derived xenografts were established through subcutaneous inoculation with 1E + 06 of indicated LUAD cells into both flanks of BALB/c-Nude mice (male, 4 ± 1 weeks of age, weighing 18 ± 2 g; Huafukang Co., China). Each group included five nude mice. All mice were ethically sacrificed 4 weeks after inoculation and tumors were weighed and harvested for further analysis. Tumor volume was calculated as V = a × b2/2 (a and b represent length and width, respectively).
Immunohistochemistry staining and scoring
Tumor tissues from cell-derived xenografts were fixed in formalin and embedded in paraffin. The paraffin-embedded tumors were sliced into 4-μm sections for further analysis. The LUAD tissue microarray (TMA) (HLug-Ade180Sur-01) was purchased from Outdo Biotech Co., Ltd. (China). The TMA included 82 cases of LUAD and matched adjacent normal tissue. The clinical information was provided by Outdo Biotech and listed in Table S8. Immunohistochemical staining (IHC) was performed as previously described [27]. Briefly, the tissue sections were first incubated at 60 °C for 2 h before xylene deparaffinization and hydration with an ethanol gradient (100–75%). The sections were immersed in 10 mM citrate (pH 6.0) buffer and boiled for 15 min, and subsequently cooled down to room temperature and quenched by using 3% H2O2 for 15 min. The sections were incubated with anti-PARD6A (1:50, Cat. R382316, ZEN-BIOSCIENCE), anti-Serpina3 (1:400, Cat. 12192-1-AP, Proteintech), and anti-Ki67 (1:200, Cat. 16667, Abcam, MA, USA) at 4 °C overnight. After three washes with PBS, the sections were incubated with the anti-rabbit/mouse radish peroxidase (HRP)-conjugated secondary antibody (Cat. PV-8000, ORIGENE, China) at 37 °C for 30 min. The signals were detected with 3,3’-diaminobenzidine (DAB) and the nucleus was stained with hematoxylin. The sections were mounted with Cytoseal XYL Mounting Medium (Cat. 83104, Epredia, Japan). The sections were detected with the Olympus microscope (Japan). The DAB-positive cells and nuclear Ki67 were determined using Image-Pro Plus Software and ImageJ IHC Toolbox, respectively (Media Cybernetics, MD, USA).
Statistical analysis
Student’s t tests, Wilcoxon test, and analysis of variance (ANOVA) were performed to assess the statistical difference between two groups and multiple groups. The log-rank test was performed to assess the statistical difference between the two groups in survival analysis. Error bars for all figures represent SD unless otherwise stated. All figures are representative of at least three independent experiments. All statistical tests were two-sided and a p value < 0.05 was considered statistically significant wherein *P < 0.05, **P < 0.01, ***P < 0.001.
Results
High expression of PARD6A predicts poor prognosis of LUAD patients
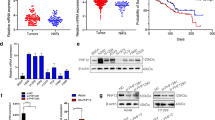
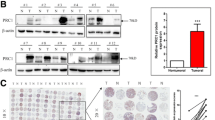
To investigate the role of PARD6A in LUAD progression, we analyzed the expression level of PARD6A in tumor tissues and adjacent normal tissues by querying the GEO (GSE87340, N = 27) database. As shown in Fig. 1A, the expression of PARD6A was significantly higher in tumor tissues compared to that in the adjacent normal tissues. To further examine the expression of Par6α in LUAD more precisely, IHC was performed on LUAD TMA containing 82 cases of LUAD and matched adjacent normal tissues. The clinicopathological characteristics of LUAD patients in TMA are listed in Table S8. The expression of Par6α was higher in tumor tissues than in adjacent normal tissues (P < 0.001, Fig. 1B). The expression of PARD6A was also examined in tumor tissues from LUAD patients at different stages. PARD6A is markedly increased in LUAD patients at stage IV (Fig. 1C). The Par6α expression elevated in LUAD patients at Stage II and III (Fig. 1D). The T3/T4 and N2/N3 stages LUAD tissues exhibited higher Par6α expression than T1/T2 and N1/N2 stages LUAD tissues, respectively (Fig. 1E, F). The Kaplan–Meier analysis of LUAD patients showed that patients with high PARD6A expression group (N = 458) have significantly lower progression-free survival (PFS) than patients with low PARD6A expression (N = 794) (Fig. 1G). Survival analysis showed that LUAD patients from TMA with high Par6α expression had a worse overall survival (OS) than those with low Par6α expression (P = 0.0079, log-rank test; Fig. 1H). Collectively, these findings suggest that Par6α is upregulated in LUAD and related to poor prognosis of LUAD patients.
A The expression of PARD6A in lung cancer tissue and corresponding adjacent normal tissues (GSE87340, N = 27); B Immunohistochemistry analysis of PARD6A in normal lung tissue and lung cancer tissue from LUAD tissue microarray (TMA). Pictures show the representative IHC of normal and corresponding LUAD tissues. Paired-line graph shows the mean integrated optical density of DAB-positive cells (N = 82); C The expression of PARD6A in cancer tissues from lung cancer patients at different stages (GSE41271, N = 183); D The expression of PARD6A in cancer tissues from lung cancer patients at different stages (TMA, N = 82); E The expression of PARD6A in cancer tissues from lung cancer patients at T1 + T2 and T3 + T4 stages (TMA, N = 82). F The expression of PARD6A in cancer tissues from lung cancer patients at N0 + N1 and N2 + N3 stages (TMA, N = 82); G The progression-free survival (PFS) of LUAD patients with high (N = 458) and low (N = 794) expression of PARD6A (N = 1252); H The overall survival (OS) of lung cancer patients with high and low expression of PARD6A (N = 8). Wilcoxon test and analysis of variance (ANOVA) were performed to assess the statistical difference between two groups and multiple groups, respectively. The log-rank test was performed to calculate the statistical comparison of two groups in survival analysis.
PARD6A enhances the malignancy of LUAD cells
To investigate the role of PARD6A in LUAD progression, we first analyzed the protein expression of Par6 family members in human normal bronchial epithelial cells (BEAS-2B), and LUAD cell lines (H1299, H1975, and PC9). The expression of Par6α in LUAD cell lines is significantly higher than it is in BEAS-2B (Fig. S1A). Then we stably knocked down PARD6A via short hairpin RNAs (shRNAs) in H1299 with high endogenous Par6α expression. The knockdown efficiency was confirmed by qRT-PCR and WB analysis (Fig. S1B, C). Our results showed that PARD6A knockdown significantly suppressed H1299 cell proliferation, clonogenic ability, cell migration, and invasion (Fig. 2A–D). We examined the cell proliferation marker proliferating cell nuclear antigen (PCNA) and invasion-related markers matrix metallopeptidase 3/9 (MMP3/9). Knockdown of PARD6A remarkedly reduced PCNA and MMP3/9 in H1299 cells (Fig. 2E). Next, we stably overexpressed PARD6A in H1975 with comparatively low endogenous Par6α expression (Fig. S1D, E). Similarly, H1975 cells with overexpressed PARD6A exhibited a remarkable increase in cell proliferation, colony formation, migration, and invasion compared to the control cells (Fig. 3A–D). PCNA and MMP3/9 were also upregulated in PARD6A overexpressing H1975 cells (Fig. 3E). We further evaluated the role of PARD6A in tumor growth in vivo by subcutaneously inoculating H1975 with overexpressed PARD6A into BALC/c nude mice. Ectopic PARD6A markedly increased tumor growth rate and weight compared to the control group (Fig. 3F–H). Therefore, PARD6A increased the proliferation of H1975 cells in nude mice and significantly increased tumor burden over time. These results were confirmed by immunohistochemical analysis of Par6α and Ki67 expression in the tumor sections (Fig. 3I).
A Proliferation analysis of PARD6A knock-down H1299 cells (N = 3/group); B Colony formation assay of PARD6A knock-down H1299 cells. The bar graph shows the average colony numbers (N = 3/group); C Wound healing assay of PARD6A knock-down H1299 cells at 0, 24, and 36 h post wounding. Pictures are taken at ×100 magnification. The bar graph shows the average wound closure rate (N = 3/group); D Transwell migration and invasion assays of PARD6A knock-down H1299 cells. Pictures are taken at ×100 magnification. The bar graph shows the average migrated or invasive cells of five random fields of per insert; E WB analysis of PARD6A knock-down H1299 cells for MMP3/9, PCNA, and Tublin. ShP-1 and shP-2 represent shPARD6A-1 and shPARD6A-2, respectively. The relative expression of the indicated blot was calculated by dividing the absolute expression of the indicated blot by the absolute expression of the blot from shNT group. The relative expression was labeled under each band. Results are expressed as mean ± SD of three independent experiments. The analysis of variance (ANOVA) was performed to assess the statistical difference between multiple groups.
A Proliferation analysis of PARD6A overexpressing H1975 cells (N = 3/group); B Colony formation assay of PARD6A overexpressing H1975 cells. The bar graph shows the average colony numbers (N = 3/group); C Wound healing assay of PARD6A overexpressing H1975 cells at 0, 24, and 36 h post wounding. Pictures are taken at ×100 magnification. The bar graph shows the average wound closure rate (N = 3/group); D Transwell migration and invasion assays of PARD6A overexpressing H1975 cells. Pictures are taken at ×100 magnification. The bar graph shows the average migrated or invasive cells of five random field of each insert; E WB analysis of PARD6A overexpressing H1975 cells for MMP3/9, PCNA, and Tublin; PARD6A overexpressing H1975 cells and control cells were subcutaneously inoculated in both flanks of Balb/c Nude mice (N = 5/group), and F the picture shows tumors taken from all mice; G tumor progression was monitored; H dot graph shows tumor weights; I the representative figures of hematoxylin-eosin staining (HE) analysis and IHC analysis of proliferative marker Ki67 and Par6α at indicated magnification. The relative expression of the indicated blot was calculated by dividing the absolute expression of the indicated blot by the absolute expression of the blot from vec group. The relative expression was labeled under each band. Results are expressed as mean ± SD of three independent experiments. The Student’s t test was performed to assess the statistical difference between two groups.
Serpina3 is upregulated in PARD6A overexpressing H1975 cells
To understand the signaling pathways involved in PARD6A-enhanced proliferation, migration, and invasion, RNA sequencing (RNA-seq) was performed to analyze differentially expressed genes (DEGs) in H1975 cells with vector or ectopic PARD6A. We found 631 upregulated and 226 downregulated genes in PARD6A overexpressing H1975 cells in comparison to control cells (Table S9 and Fig. 4A). Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was conducted to further explore the biological processes and signaling pathways modulated by PARD6A. The most enriched biological progress (GO-BP) included negative regulation of locomotion (GO:0040013), leukocyte chemotaxis (GO:0030595), negative regulation of chemotaxis (GO:0050922), positive regulation of cell projection organization (GO:0031346), and granulocyte chemotaxis (GO:0071621) (Table S10 and Fig. 4B). In the cellular component (CC) ontology, the most enriched terms were apical plasma membrane (GO:0016324), apical part of cell (GO:0045177), sarcoplasmic reticulum membrane (GO:0033017), specific granule (GO:0042581), and platelet alpha granule (GO:0031091) (Table S11 and Fig. 4B). As for molecular function (MF) ontology, the highly enriched terms were chemorepellent activity (GO:0045499), receptor-ligand activity (GO:0048018), signaling receptor activator activity (GO:0030546), semaphoring receptor binding (GO:0030215), and cytokine activity (GO:0005125) (Table S12 and Fig. 4B). The most abundant KEGG pathways compromised Axin guidance (hsa04360), Efferocytosis (hsa04148), complement and coagulation cascades (hsa04610), AGE-RAGE signaling pathway in diabetic complications (hsa04933), and Cytokine-cytokine receptor interaction (hsa04060) (Table S13 and Fig. 4C). The expression of the most upregulated gene except PARD6A was then validated in PARD6A overexpressing H1975 and knockdown H1299 cells by qRT-PCR. As shown in Fig. 4D, E, a significant increase and reduction in the expression of Serpina3 were found in cells stably expressing PARD6A and cells with PARD6A knockdown, respectively. The protein level of Serpina3 was further confirmed. WB analysis illustrated the elevation and demotion of Serpina3 in PARD6A overexpressing H1975 and knockdown H1299 cells (Fig. 4F, G). Additionally, we used STRING to analyze the potential physical and functional associations between Serpina3 and PARD6A (Tables S14 and S15). As shown in Fig. 4H, PARD6A might regulate Serpina3 through ERK1 and serum amyloid A1 (SAA1) signaling. These results indicate that Serpina3 is one of the main downstream of PARD6A in LUAD cells.
A Waterfall plot and heatmap of differentially expressed genes (DEGs) in PARD6A overexpressing H1975 cells compared to controls (N = 3/group); B GO enrichment analysis of DEGs. The top five enriched Go terms are shown. BP, CC, and MF represent biological process, cellular component, and molecular function, respectively; C KEGG-Circle plot showing the top 10 KEGG pathway based on the adjusted p value; D qRT-PCR analysis of Serpina3 in PARD6A overexpressing H1975 cells (N = 3/group); E qRT-PCR analysis of Serpina3 in PARD6A knockdown H1299 cells (N = 3/group); F Representative of WB analysis of Serpina3 in PARD6A overexpressing H1975 cells; G WB analysis of Serpina3 PARD6A knockdown H1299 cells. H STRING analysis of the Protein-protein interaction network. The nodes indicate proteins and were clustered by k-means clustering. The line thickness indicates the strength of data support. The interactions with a medium confidence score of ≥0.4 are shown. The relative expression of the indicated blot was calculated by dividing the absolute expression of the indicated blot by the absolute expression of the blot from shNT or vec group. The relative expression was labeled under each band. Results are expressed as mean ± SD of three independent experiments. Student’s t test and analysis of variance (ANOVA) were performed to assess the statistical difference.
PARD6A stimulates proliferation, migration, and invasion through Serpina3
We hypothesized that PARD6A enhances malignancy via Sepina3 in LUAD cells. To test this hypothesis, we first rescued Serpina3 in PARD6A knockdown H1299 cells (Fig. S2A, B). As shown in Fig. 5A–C, the addition of exogenous Serpina3 recovered the proliferation, migration, and invasion curbed by PARD6A knockdown in H1299 cells. Ectopic Serpina3 also led to the upregulation of PCNA and MMP3/9 (Fig. 5D). Meanwhile, we utilized shRNAs to specifically target Serpina3 expression in PARD6A overexpressing H1975 cells (Fig. S2C, D). After blocking Serpina3, the PARD6A-elevated proliferation rate was hampered (Fig. 6A). Knockdown of Serpina3 also restrained migration and invasion in PARD6A overexpressing H1975 cells (Fig. 6B, C). The increasing PCNA and MMP3/9 also declined after silencing Serpina3 (Fig. 6D). To further investigate the role of the PARD6A/Serpina3 axis in LUAD progression, the PARD6A overexpressing H1975 cells with or without Serpina3 knockdown were subcutaneously inoculated in Balb/c Nude mice. The tumor volume curve and weight displayed that knockdown of Serpina3 significantly suppressed tumor growth (Fig. 6E–G). The Serpina3 and Ki67 expression was decreased in the PARD6A overexpression H1975 cells with Serpina3 knockdown groups compared with the vec and PARD6A overexpressing H1975 without Serpina3 knockdown groups as determined by immunochemical staining (Fig. 6H). These data indicate that Serpina3 is a potent downstream of PARD6A in LUAD cells.
A Proliferation analysis of PARD6A knock-down H1299 cells with or without exogenous Serpina3 (N = 3/group); B Wound healing assay of PARD6A knock-down H1299 cells with or without exogenous Serpina3 at 0, 24, and 36 h post wounding. Pictures are taken at ×100 magnification. The bar graph shows the average wound closure rate (N = 3/group); C Transwell migration and invasion assays of PARD6A knock-down H1299 cells with or without exogenous Serpina3. Pictures are taken at ×100 magnification. The bar graph shows the average migrated or invasive cells of five random field per insert; D WB analysis of PARD6A knock-down H1299 cells with or without exogenous Serpina3 for MMP3/9, PCNA, and GAPDH. ShP-1 and shP-2 represent shPARD6A-1 and shPARD6A-2, respectively. The relative expression of the indicated blot was calculated by dividing the absolute expression of the indicated blot by the absolute expression of the blot from shNT group. The relative expression was labeled under each band. Results are expressed as mean ± SD of three independent experiments. The analysis of variance (ANOVA) was performed to assess the statistical difference.
A Proliferation analysis of PARD6A overexpressing H1975 cells with or without Serpina3 knockdown (N = 3/group); B Wound healing assay of PARD6A overexpressing H1975 cells with or without Serpina3 knockdown at 0, 24, and 36 h post wounding. Pictures are taken at ×100 magnification. The bar graph shows the average wound closure rate (N = 3/group); C Transwell migration and invasion assays of PARD6A overexpressing H1975 cells with or without Serpina3 knockdown. Pictures are taken at ×100 magnification. Bar graph shows the average migrated or invasive cells of five random fields per insert; D WB analysis of PARD6A overexpressing H1975 cells with or without Serpina3 knockdown for MMP3/9, PCNA, and Tublin; PARD6A overexpressing H1975 cells with or without Serpina3 knockdown and control cells were subcutaneously inoculated in both flanks of Balb/c Nude mice (N = 5/group), and E the picture shows tumors taken from all mice; F tumor progression was monitored; G dot graph shows tumor weights; H the representative figures of hematoxylin-eosin staining (HE) analysis and IHC analysis of proliferative marker Ki67, Par6α, and Serpina3 at indicated magnification. ShS-1 and shS-2 represent shSerpina3-1 and shSerpina3-2, respectively. The relative expression of the indicated blot was calculated by dividing the absolute expression of the indicated blot by the absolute expression of the blot from vec group. The relative expression was labeled under each band. Results are expressed as mean ± SD of three independent experiments. The analysis of variance (ANOVA) was performed to assess the statistical difference.
Additionally, we analyzed the expression of Serpina3 in tumors and its paired normal tissues of LUAD patients from the TCGA and GEO databases. As shown in Fig. S3A, Serpina3 in tumor tissues is higher than that in adjacent normal tissues (GSE87340, N = 27). However, the expression of Serpina3 in tumors and adjacent normal tissues showed no significant difference from the TCGA dataset (Fig. S3B). The Kaplan–Meier analysis of LUAD patients showed that patients with a high Serpina3 expression group (N = 488) have significantly lower PFS than patients with a low Serpina3 expression (N = 1415) (Fig. S3C). Moreover, we analyzed the prognosis of LUAD patients with high levels of PARD6A and Serpina3 from GEO databases (GSE63459 and GSE42127, N = 207). As shown in Fig. S4A, LUAD patients with high levels of PARD6A and low levels of Serpina3 comprised 72% Stage I (N = 29), 15% Stage II (N = 6), and 12% Stage III (N = 5). LUAD patients with high levels of both PARD6A and Serpina3 included a lower percentage of patients at Stage I (46%, N = 12), and a higher percentage of patients at Stage II (31%, N = 8) and Stage III (19%, N = 5), and Stage IV (4%, N = 1). LUAD patients with low levels of both PARD6A and Serpina3 consist of 69% Stage I (N = 29), 16% Stage II (N = 6), and 15% Stage III (N = 5). Additionally, LUAD patients with low levels of PARD6A and high levels of Serpina3 are all at Stage I (N = 5). As shown in Fig. S3B–D, LUAD patients with high levels of PARD6A or Serpina3 have worse survival probability. The LUAD patients with both higher PARD6A and Serpina3 have the worst survival probability. Overall, these data indicate that PARD6A accelerates LUAD progression by inducing Serpina3.
Discussion
Lung cancer is the most common malignancy with the highest mortality worldwide. Metastasis and recurrence are the main cause of cancer-related death. Understanding the underlying mechanisms is essential for successful cancer treatment. Par6 is one of the portioning-defective genes identified in C. elegans and plays a critical role in asymmetric embryonic cell division [28]. In epithelial cells, the Par6 family has been reported to interact with Par3, CDC42, and αPKC at tight junctions, and regulate axon specification, directional migration, and other cell activity related to cellular asymmetry [29]. Previous studies have demonstrated that the cell polarity protein Par6 participates in regulating cancer progression in various types of cancer, including breast cancer, glioblastoma, lung cancer, and so forth [30, 31]. In breast cancer cells, Par6/αPKC has been found to induce cell proliferation and protect cancer cells from apoptosis [32, 33]. Moreover, most evidence has shown that Par6 encourages tumor invasion and migration by modulating cell polarity and motion [31]. In the present study, we first uncovered that a high level of PARD6A is associated with poor prognosis in LUAD patients. Mechanically, we did not observe the morphological change after manipulating PARD6A expression in lung cancer cells (data not shown). Moreover, Par6 was found to localize in both the nucleus and cytoplasm (data not shown), which might indicate the polarity-independent function of Par6 in regulating tumor progression.
Serine proteinase inhibitors (SPIs) are ubiquitous regulators of serine proteinase and involve protein folding, inflammation, cell migration, ECM remodeling, etc. They have been reported to participate in the progression of various diseases, such as diabetes, neurological disease, and cancers. The most studied SPIs in cancer metastasis include Serpina5 (PCI), Serpinb2 (PAI-2), Serpinb5 (Maspin), Serpinb9, SerpinE1 (PAI-1), and others. Serpina3 (alpha-1-antichymorypsin, ACT) belongs to α1-globulin fraction of serum proteins. It is an anti-inflammatory protein that has been proven to inhibit the activity of chymotrypsin, cathepsin G, and alpha-chymase, obstacle the chemotaxis of neutrophils, and be involved in tumor progression. Analysis of Serpina3 expression in tumors and normal tissues showed that Serpina3 is highly expressed in specific types of tumors, such as glioblastoma and breast invasive carcinoma (Fig. S3D). Previous studies have demonstrated that the high expression of Serpina3 is closely related to poor prognosis in various types of cancers, including melanoma, colorectal cancer, breast cancer, liver cancer, glioblastoma, gastric cancer, and lung cancer [34,35,36,37]. The GlcNAcyl-Serpina3 in serum has also been found promising in early diagnosis of lung cancer [38]. In vitro, Serpina3 has been reported to be transcriptionally regulated by Oncostatin M (OSM), NF-κB/AP1, Nur77, and STAT3 and promotes metastasis by inducing the expression of MMPs and enhancer of zeste homolog 2 (EZH2) or activating PI3K/AKT and MAPK pathways [14, 39,40,41,42,43,44]. Serpina3 protein has a unique double DNA-binding domain without the inhibitory activity of its serine protease. In hepatocellular carcinoma cells, N-glycosylated Serpina3 transports into the nucleus by interacting with importin α/β protein and binds to DNA, which leads to chromatin condensation and the cell cycle arrest at the G0/G1 phase [18]. The reactive oxygen species (ROS)-dependent Serpina3 can function as a transcription enhancer and form a complex with hnRNPK, which induces the transcription of Ubiquitin-like containing PHD and Ring Finger domain 1 (UHRF1) and switches on PI3Kδ signaling [16]. Additionally, Serpina3 could enhance the phosphorylation of AKT and ribosomal protein S6, thereby inducing the proliferation of colon cancer cells [45]. The tumorigenic role of Serpina3 suggests the potential as a treatment target, whereas, the context-dependent mechanisms underlying remain obscure. However, the most recent study in LUSC cells reported that Serpina3 suppresses tumor progression by inhibiting NF-κB signaling, which suggests the divergent role of Serpina3 in tumor progression in a histology-dependent manner. In LUAD cells, we found that Serpina3 is a main effector of PARD6A and advances proliferation, migration, and migration. We predicted the promising physical or functional connection between Serpina3 and PARD6A using STRING and found that PARD6A might modulate Serpina3 through ERK1 signaling. Previous studies demonstrated that PARD6A activates the MEK/ERK/STAT3 pathway and Serpina3 is one of the targets of STAT3 [11, 43]. Therefore, we assumed PARD6A might induce Serpina3 through the ERK/STAT3 pathway. Nevertheless, further experiments are required to decipher more detailed mechanisms.
Immunotherapy, especially immune checkpoint inhibitors (ICIs), has shown significant potential for improving the prognosis of LUAD patients. We also analyzed the role of PARD6A and Serpina3 in predicting response to ICIs. As shown in Fig. S5A–G, no significant difference in PARD6A or Serpina3 expression was found between LUAD patients with or without response to Nivolumab. Additionally, a relatively higher percentage of LUAD patients without ICI response was shown in LUAD patients with high PARD6A (Fig. S5B, F). However, the percentage of LUAD patients with or without ICI response in cohorts with differentially expressing Serpina3 varied in two datasets (Fig. S5D, H). Further analysis of ICI response in LUAD patients with differential expression of PARD6A and Serpina3 in two GEO datasets did not show a significant correlation (Fig. S5I, J). These data indicated that PARD6A might be a promising biomarker for ICI response prediction. However, around 50% of LUAD patients with relatively low PARD6A exhibited no response to ICIs in the GSE126044 dataset, suggesting PARD6A alone might not be a biomarker with accuracy. Moreover, the sample size of GSE126044 and GSE135222 is small, and more clinical data is needed to validate the value and precision of PARD6A and Serpina3 in predicting ICI response.
Overall, these studies indicated that (1) PARD6A is a promising prognostic biomarker in LUAD patients. The higher expression of PARD6A predicts poorer prognosis of LUAD patients, (2) PARD6A induces Serpina3, ultimately promoting proliferation, migration, and invasion of LUAD cells, and (3) Serpina3 exhibits pro-tumoral function in LUAD patients. The detailed mechanisms underlying PARD6A/Serpina3 require further investigation.
Data availability
The RNA-seq data based on the pan-Cancer Atlas run of the TCGA were obtained from the cBioPortal (https://www.cbioportal.org/). The data used for analyzing clinicopathological association are downloaded from GEO or TCGA database. The remaining data are available within the Article or Supplementary Information. Data will be made available upon request to the corresponding author.
References
Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J. 2022;135:584–90.
He S, Li H, Cao M, Sun D, Lei L, Li N, et al. Trends and risk factors of lung cancer in China. Chin J Cancer Res. 2020;32:683–94.
Li C, Lei S, Ding L, Xu Y, Wu X, Wang H, et al. Global burden and trends of lung cancer incidence and mortality. Chin Med J. 2023;136:1583–90.
Chan MV, Huo YR, Cao C, Ridley L. Survival outcomes for surgical resection versus CT-guided percutaneous ablation for stage I non-small cell lung cancer (NSCLC): a systematic review and meta-analysis. Eur Radiol. 2021;31:5421–33.
Halaoui R, McCaffrey L. Rewiring cell polarity signaling in cancer. Oncogene. 2015;34:939–50.
Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D, et al. A role for the TGFbeta-Par6 polarity pathway in breast cancer progression. Proc Natl Acad Sci USA 2009;106:14028–33.
Zhang K, Zhao H, Ji Z, Zhang C, Zhou P, Wang L, et al. Shp2 promotes metastasis of prostate cancer by attenuating the PAR3/PAR6/aPKC polarity protein complex and enhancing epithelial-to-mesenchymal transition. Oncogene. 2016;35:1271–82.
Liu P, Zhu C, Luo J, Lan S, Su D, Wang Q, et al. Par6 regulates cell cycle progression through enhancement of Akt/PI3K/GSK-3beta signaling pathway activation in glioma. FASEB J. 2020;34:1481–96.
Togawa A, Sfakianos J, Ishibe S, Suzuki S, Fujigaki Y, Kitagawa M, et al. Hepatocyte growth factor stimulated cell scattering requires ERK and Cdc42-dependent tight junction disassembly. Biochem Biophys Res Commun. 2010;400:271–7.
Elbediwy A, Zhang Y, Cobbaut M, Riou P, Tan RS, Roberts SK, et al. The Rho family GEF FARP2 is activated by aPKCiota to control tight junction formation and polarity. J Cell Sci. 2019;132:jcs223743.
Huang Y, Liu P, Luo J, Zhu C, Lu C, Zhao N, et al. Par6 enhances glioma invasion by activating MEK/ERK pathway through a LIN28/let-7d positive feedback loop. Mol Neurobiol. 2023;60:1626–44.
Lu Z, Yuan S, Ruan L, Tu Z, Liu H. Partitioning defective 6 homolog alpha (PARD6A) promotes epithelial-mesenchymal transition via integrin beta1-ILK-SNAIL1 pathway in ovarian cancer. Cell Death Dis. 2022;13:304.
Soman A, Asha Nair S. Unfolding the cascade of SERPINA3: inflammation to cancer. Biochim Biophys Acta Rev Cancer. 2022;1877:188760.
Zhang Y, Tian J, Qu C, Peng Y, Lei J, Li K, et al. Overexpression of SERPINA3 promotes tumor invasion and migration, epithelial-mesenchymal-transition in triple-negative breast cancer cells. Breast Cancer. 2021;28:859–73.
Lara-Velazquez M, Zarco N, Carrano A, Phillipps J, Norton ES, Schiapparelli P, et al. Alpha 1-antichymotrypsin contributes to stem cell characteristics and enhances tumorigenicity of glioblastoma. Neuro Oncol. 2021;23:599–610.
Ko E, Kim JS, Bae JW, Kim J, Park SG, Jung G. SERPINA3 is a key modulator of HNRNP-K transcriptional activity against oxidative stress in HCC. Redox Biol. 2019;24:101217.
Zhou J, Zhu M, Wang Q, Deng Y, Liu N, Liu Y, et al. SERPINA3-ANKRD11-HDAC3 pathway induced aromatase inhibitor resistance in breast cancer can be reversed by HDAC3 inhibition. Commun Biol. 2023;6:695.
Santamaria M, Pardo-Saganta A, Alvarez-Asiain L, Di Scala M, Qian C, Prieto J, et al. Nuclear alpha1-antichymotrypsin promotes chromatin condensation and inhibits proliferation of human hepatocellular carcinoma cells. Gastroenterology. 2013;144:818–28.e814.
Jin Y, Zhang Y, Huang A, Chen Y, Wang J, Liu N, et al. Overexpression of SERPINA3 suppresses tumor progression by modulating SPOP/NF‑kappaB in lung cancer. Int J Oncol. 2023;63:96.
Gyorffy B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation. 2024;5:100625.
Gyorffy B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br J Pharm. 2024;181:362–74.
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–46.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Lo HL, Yee JK. Production of vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped retroviral vectors. Curr Protoc Hum Genet. 2007;Chapter 12:Unit 12.7.
National Research Council. 2011. Guide for the Care and Use of Laboratory Animals: Eighth Edition. Washington, DC: The National Academies Press.
Wu H, Leng X, Liu Q, Mao T, Jiang T, Liu Y, et al. Intratumoral microbiota composition regulates chemoimmunotherapy response in esophageal squamous cell carcinoma. Cancer Res. 2023;83:3131–44.
Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–20.
Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–49.
Marques E, Klefstrom J. Par6 family proteins in cancer. Oncoscience. 2015;2:894–5.
Ruan L, Shen Y, Lu Z, Shang D, Zhao Z, Lu Y, et al. Roles of partitioning-defective protein 6 (Par6) and its complexes in the proliferation, migration and invasion of cancer cells. Clin Exp Pharm Physiol. 2017;44:909–13.
Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC, et al. The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer Res. 2008;68:8201–9.
Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, et al. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol. 2006;8:1235–45.
Karashima S, Kataoka H, Itoh H, Maruyama R, Koono M. Prognostic significance of alpha-1-antitrypsin in early stage of colorectal carcinomas. Int J Cancer. 1990;45:244–50.
Hurlimann J, van Melle G. Prognostic value of serum proteins synthesized by breast carcinoma cells. Am J Clin Pathol. 1991;95:835–43.
Luo D, Chen W, Tian Y, Li J, Xu X, Chen C, et al. Serpin peptidase inhibitor, clade A member 3 (SERPINA3), is overexpressed in glioma and associated with poor prognosis in glioma patients. Onco Targets Ther. 2017;10:2173–81.
Nimbalkar VP, Kruthika BS, Sravya P, Rao S, Sugur HS, Verma BK, et al. Differential gene expression in peritumoral brain zone of glioblastoma: role of SERPINA3 in promoting invasion, stemness and radioresistance of glioma cells and association with poor patient prognosis and recurrence. J Neurooncol. 2021;152:55–65.
Jin Y, Wang J, Ye X, Su Y, Yu G, Yang Q, et al. Identification of GlcNAcylated alpha-1-antichymotrypsin as an early biomarker in human non-small-cell lung cancer by quantitative proteomic analysis with two lectins. Br J Cancer. 2016;114:532–44.
Funamoto M, Fujio Y, Kunisada K, Negoro S, Tone E, Osugi T, et al. Signal transducer and activator of transcription 3 is required for glycoprotein 130-mediated induction of vascular endothelial growth factor in cardiac myocytes. J Biol Chem. 2000;275:10561–6.
Zhao Y, Liu Y, Zheng D. Alpha 1-antichymotrypsin/SerpinA3 is a novel target of orphan nuclear receptor Nur77. FEBS J. 2008;275:1025–38.
Yang GD, Yang XM, Lu H, Ren Y, Ma MZ, Zhu LY, et al. SERPINA3 promotes endometrial cancer cells growth by regulating G2/M cell cycle checkpoint and apoptosis. Int J Clin Exp Pathol. 2014;7:1348–58.
Li Y, Dong X, Cai J, Yin S, Sun Y, Yang D, et al. SERPINA3 induced by astroglia/microglia co-culture facilitates glioblastoma stem-like cell invasion. Oncol Lett. 2018;15:285–91.
Kulesza DW, Ramji K, Maleszewska M, Mieczkowski J, Dabrowski M, Chouaib S, et al. Search for novel STAT3-dependent genes reveals SERPINA3 as a new STAT3 target that regulates invasion of human melanoma cells. Lab Invest. 2019;99:1607–21.
Zhou ML, Chen FS, Mao H. Clinical significance and role of up-regulation of SERPINA3 expression in endometrial cancer. World J Clin Cases. 2019;7:1996–2002.
Meijers WC, Maglione M, Bakker SJL, Oberhuber R, Kieneker LM, de Jong S, et al. Heart failure stimulates tumor growth by circulating factors. Circulation. 2018;138:678–91.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (Grant No. 2023YFC3402100) and the National Natural Science Foundation of China (Grant No. 92259102).
Author information
Authors and Affiliations
Contributions
LH and BT established stable knock-down and overexpressing cell lines and performed all in vitro experiments; LH and ML performed in vivo experiments; BT and DW performed bioinformatic analyses; HX and HW reconstructed knock-down and overexpression plasmids; LH, ML, BT, and XL analyzed all data and organized figures; LH, SL, and CX designed and supervised experiments; CX obtained the fundings; LH wrote the paper. All authors read and commented on the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The tumor tissue microarray (TMA) was purchased from Outdo Biotech Co., Ltd. (China). The study using the TMA was approved by the Life Sciences Ethics Committee of Outdo Biotech Co., Ltd., under the reference number: YB M-05-02. All patients and healthy donors provided informed consent to participate.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, L., Liu, M., Tang, B. et al. PRAD6A promotes lung adenocarcinoma cell proliferation and invasion through Serpina3. Cancer Gene Ther (2024). https://doi.org/10.1038/s41417-024-00829-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41417-024-00829-w
- Springer Nature America, Inc.