Abstract
The invasion of glioblastoma usually results in the recurrence and poor prognosis in patients with glioma. However, the underlying mechanisms involved in glioma invasion remains undefined. In this study, immunohistochemistry analyses of glioma specimens demonstrated that high expression of Par6 was positively correlated with malignancy and poor prognosis of patients with glioma. Par6-overexpressing glioma cells showed much more fibroblast-like morphology, suggesting that regulation of Par6 expression might be associated with tumor invasion in glioma cells. Further study indicated that Par6 overexpression subsequently increased CD44 and N-cadherin expression to enhance glioma invasion through activating MEK/ERK/STAT3 pathway, in vivo and in vitro. Moreover, we found that LIN28/let-7d axis was involved in this process via a positive feedback loop, suggesting that MEK/ERK/LIN28/let-7d/STAT3 cascade might be essential for Par6-mediated glioma invasion. Therefore, these data highlight the roles of Par6 in glioma invasion, and Par6 may serve as a potential therapeutic target for patients with glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant glioma is the most deadly and malignant brain tumor in the central nervous system and is usually coupled with high rates of both recurrence and mortality with a 14.6-month median survival time [1]. Among malignant glioma subtypes, glioblastoma (GBM) is considered the most aggressive, with a 5-year survival rate of no more than 5% [2]. Despite the developments of surgical resection and chemotherapy, the prognoses of patients with glioma have not significantly improved due to recurrence and invasion [3, 4].
Tumor metastasis has been widely considered a continuous and complex pathological process that involves multiple factors and signaling pathways. The epithelial-to-mesenchymal transition (EMT) participates in the invasion and metastasis of most malignant tumors [5, 6]. Notably, glioma invasion is uniquely in association with infiltration of invasive glioma cells into adjacent noncancerous tissues, which involves increased activity, extracellular matrix (ECM) degradation, and glioma stem cell transformation [7, 8]. To facilitate invasion, glioma cells are known to regulate critical signaling pathways and alter the ECM in host tissues [9,10,11]. However, the underlying molecular mechanisms responsible for glioma invasion remain elusive. It is thus essential to dissect these molecular mechanisms in order to identify novel therapeutic targets and develop new modalities of treatment.
Par6 (or named Par6α) is a scaffolding molecule that was identified in Caenorhabditis elegans as a regulator of asymmetric cell division during embryonic development [12]. The Par6 polarity complex localizes to tight junctions (TJs) and is an important regulator of the morphological transitions associated with epithelial cell plasticity [13]. In addition, Par6 has also been found to regulate cell death, directional migration of astrocytes and keratinocytes, and axon specification in neurons [14,15,16,17]. Moreover, accumulating studies indicate that Par6 and its complexes are involved in the regulation of cancer cell proliferation and EMT-related tumor migration and invasion [18, 19]. Muthuswamy et al. reported that Par6 is overexpressed in breast cancer cell lines, and in both precancerous and advanced primary human breast cancers [20]. Many other studies showed that Par6 plays essential roles in tumor migration and invasion through regulating EMT progression in various human epithelial tumors. Several previous reports have demonstrated that Par6 can cooperate with the TGF-β receptor to promote TGF-β-induced EMT progression in breast cancer [21, 22]. Moreover, αPKC has been found to interact with TGF-β receptors to regulate phosphorylated Par6-dependent EMT progression and cell migration in various malignant tumors [23,24,25,26]. Here, we systematically analyzed microarray data of paired glioma tissues and noncancerous tissues in The Cancer Genome Atlas (TCGA) database, and found that Par6 was also significantly upregulated in invasive glioma specimens (data not shown), which encouraged us to further investigate the possible mechanisms of Par6 functions in glioma invasion. As glioma is a kind of malignant tumor derived from nonepithelial tissues; we expected that Par6 may play a role in glioma invasion, which is distinct from what it does in the metastasis of other epithelial tumors.
In this study, we found that high expression of Par6 was linked to poor survival prognosis in glioma patients and promotion of glioma cell invasion in vitro and in vivo. Mechanistically, we found that Par6-mediated activation of extracellular signal-regulated kinase (ERK1/2) can enhance glioma invasion through activating Rac1/mitogen-activated protein kinase MEK/ERK pathway. RNA-seq analyses and western blot revealed that the activation of STAT3 pathway was also involved in this process. In addition, our results indicated that LIN28/let-7d axis was correlated with the effects of Par6 on glioma invasion. Furthermore, we found that Par6 overexpression could promote glioma invasion in vivo and in vitro, indicating that Par6 regulates glioma invasion via an MEK/ERK/LIN28/let-7d-positive feedback loop and subsequently activates STAT3 pathway in glioma cells. Our findings indicate that the expression level of Par6 is closely in association with glioma invasion and that the aberrant Par6 expression may predict poor prognosis in patients with glioma.
Material and Methods
Cell Culture and Treatment
The human glioma cell lines U87MG, U251, the human normal astrocyte cell line HEB, the human breast cancer cell line MCF7, and the human non-small-cell lung cancer (NSCLC) cell line A549 were purchased from the China Center for Type Culture Collection (Shanghai, China). All these cell lines were cultured in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Gibco) and streptomycin/penicillin antibiotic mixture (Invitrogen, Thermo Fisher Scientific) at 37°C with 5% CO2. The cell lines U87MG and U251 were authenticated by short tandem repeat profiling of 15 loci and the amelogenin sex determination (X or XY) method according to the manufacturer’s (Promega, Madison, WI) instructions [27].
The primary cell culture was slightly modified from the previously reported protocol [28,29,30]. Two authenticated primary cell lines (GBM1 and GBM2) that derived from GBM surgical specimens were obtained from Procell Company (Wuhan, China). The primary cell lines were maintained in primary serum-free cultures grown on laminin [31]. Before each experiment, the primary glioma cells were cultured in DMEM, supplemented with 10% FBS, streptomycin/penicillin antibiotic mixture, and 2mM l-glutamine (Invitrogen) at 37°C with 5% CO2.
All the inhibitors and activators were obtained from MedChem Express (Monmouth Junction, NJ). Different groups of cells were treated with TGF-β (5 ng/ml; HY-P7118); U0126, an MEK specific inhibitor (10 μM; HY-12031); atractylenolide I (AT-1), a specific inhibitor of STAT3 pathway (50 μM; HY-N0201); or Rhosin (hydrochloride), a specific Rho inhibitor (5 mM; HY-12646) for 48 h before the following experiments.
Overexpression and Knockdown
The full-length PARD6A mRNA (NM_016948) was isolated from the U87MG cell line. The Par6-coding cDNA was then amplified by RT-PCR with a forward primer (5′tctgaattcatggcccggccg3′) and a reverse primer (5′gcgcggccgctcagaggctgaagcc3′), and cloned into the EcoRI/NotI sites of a pLVX-mCMV-ZsGreen-PGK-Puro vector (HedgehogBio, Shanghai, China). Sh-Par6 and Sh-EGFP were purchased from Cyagen (Suzhou, China). Stably infected U87MG, U251, A549, and MCF7 cell lines, and two primary glioma cell lines, were established by lentiviral infection. In brief, HEK293T cells were transfected with human Par6 cDNA constructed into pLVX-mCMV-ZsGreen-PGK-Puro or with shRNA in pLKO.1 together with 3.6 μg of pCMV-VSV-G, 6 μg of pMDLg/pRRE, and 2.4 μg of pRSV-REV by using Lipofectamine 3000 Transfection Reagent (Invitrogen) to produce pseudoviral particles. At 48-h post transfection, the supernatants were collected, and the viruses were harvested by centrifugation at 20,000 × g for 1.5 h. The virus-containing pellets were resuspended in DMEM with 10% FBS and incubated with cells at 37°C for 24 h. To obtain cells stably expressing Par6, shRNA, or negative control vector, the transduced cells were selected with 3 μg/ml puromycin for 7 days.
For overexpression or knockdown of let-7d, U87MG or U251 cells were seeded into 6-well plates and transfected with 100 nM let-7d mimic, mimic control (NC mimic), let-7d inhibitor, or inhibitor control (inhibitor NC; RiboBio, Shanghai, China) by using Lipofectamine 3000 Transfection Reagent. All the transfected cells were incubated in DMEM with 10% FBS for 48 h.
Quantitative Real-time PCR Assay
Total RNA was extracted from the different groups of cells by using TRIzolTM Reagent (Invitrogen) according to the manufacturer’s protocol. The RNA was reverse transcribed using quantitative real-time PCR (qRT-PCR) assay with a SYBR PrimeScript RT-PCR kit (TaKaRa Bio Inc., Japan) on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The sequences of primers are shown in Table S1. The PCR protocol included a denaturation program (95°C for 2 min) followed by 40 cycles of an amplification and quantification program (95°C for 5 s and 55–57°C for 30 s) and a melting curve program (55–95°C with a 0.5°C increment each cycle).
Western Blotting
Protein was extracted from different groups of cells with lysis buffer (Pierce, Thermo Fisher Scientific) for 1 h at 4°C, respectively. The supernatants were centrifuged, and the total protein was harvested. The samples containing 20 μg of protein were separated on 10% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocking, the blots were incubated with the appropriate primary antibody overnight at 4°C and then with the corresponding secondary antibody at room temperature for 2 h at 37°C. Then, the signals were visualized using a SuperSignal West Pico Substrate Kit (Pierce, Thermo Fisher Scientific), and the signal fluorescence intensity was measured with ImageJ software (National Institutes of Health, Bethesda, MD). The information of the antibodies is shown in Table S2. All secondary antibodies were from Jackson ImmunoResearch Laboratories Inc. (1:5000; West Grove, PA). All the experiments were repeated at least three times.
Immunohistochemistry
Four-micrometer-thick tissue sections were prepared from paraffin-embedded tissues. Following antigen retrieval and blocking, the sections were subjected to immunostaining using the primary antibody (Table S2), detected by using the avidin-biotin complex method (Dako) and visualized with DAB. The slides were lightly counterstained with hematoxylin, and the staining was evaluated with Image-Pro Plus software (Media Cybernetics., Rockville, MD).
Tissue Microarray Analysis
A paraffin-embedded tissue microarray containing 180 cases of glioma specimens was purchased from Outdo Biotech Ltd. (HbraG180Su01; Shanghai, China). The tissue microarray was stained with anti-PARD6A polyclonal antibody (1:50; TA323360, Origene, Rockville, MD) and the quantification of immunoreactive staining was analyzed with Image-Pro Plus software (Media Cybenetics), according to the previous report [32]. The clinicopathologic characteristics of the patients with glioma are summarized in Table 1. In brief, images of all tissue scores were acquired at the same time with a constant set of microscope and imaging software parameters. The images were then subjected to an optical density analysis with Image-Pro software. Adjustments of the background and color intensity range were performed on a representative image showing high immunoreactivity. The black and incident levels (representing the optical density of the maximal positive staining and white background, respectively) were determined from an immunostained region and a blank region in the image, respectively. Intensity range selection was based on a histogram, with intensity (I) and saturation (S) set to the maximum and hue (H) set to a range where most of the brown AEC staining was selected, while blue nuclear counterstaining was excluded. These settings were saved and subsequently applied to all images analyzed. After defining the area of interest, the mean optical density of the selected area (integrated optical density (IOD)/unit area) was determined with the software. The IOD represents the immunoreactivity of the candidate protein within the tumor tissue. The median value of the immunoreactive score was chosen as the cutoff criterion to define high- and low-expression subgroups.
Wound Healing Assay
All the cells were incubated in DMEM with 10% FBS and wounded by using a 200-μl pipette tip in a 6-well plate. The wound width was photographed at different post-scratch time points (0, 12, and 24 h) under a phase-contrast microscope. Data were obtained from three independent experiments.
Transwell Assay (Cell Invasion Assay)
An 8-μm pore size of FalconTM cell culture inserts (353097, BD Biosciences, Franklin Lakes, NJ), coated with 50 μl Matrigel (BD Biosciences), was placed in 24-well Corning transwell microplates (Corning Incorporated, NY). A total of 1×104 cells were seeded into the upper chamber of each plate in serum-free DMEM. The bottom chambers were filled with DMEM supplemented with 10% FBS. After 24 h of incubation, the cells on the upper surface were removed with a cotton swab, and the membranes were then fixed and stained with crystal violet. The invasive cells on the underside of the membrane were finally counted under a phase-contrast microscope. Each experiment was examined with three replicates.
RNA-seq and Data Analyses
RNA-seq analysis of Par6-overexpressing or untreated U87MG cells was performed by Sangon Biotech (Shanghai, China). Differentially expressed genes (DEGs) were identified according to the following rules: the |log2 fold change (FC)| > 1 and P-value and false discovery rate (FDR) < 0.05. A heatmap and volcano plot of the DEGs were drawn in the R platform. The top 100 overlapping DEGs based on the ǀlog2FCǀ values were subjected to further analysis.
To identify the DEG functional annotations, we performed Gene Ontology term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses with the Database for Annotation, Visualization, and Integrated Discovery (https://David.ncifcrf.gov/tools.jsp) [33]. P < 0.05 was considered to indicate statistical significance.
Orthotopic Xenografts in Nude Mice
Nude mice were purchased from the Center of Experimental Animals, Shantou University Medical College. For mouse orthotopic xenografts, dissociated cells from different groups were cultured in DMEM with 10% FBS, and then 1 × 106 dissociated cells from different groups were injected orthotopically into 4-week-old male nude mice (n = 5 for each group). At the end of 3 weeks, all mice were sacrificed after isoflurane treatment, and the tumor-bearing tissues, which were obtained from the mice transplanted with cells from different groups, were fixed in buffered formalin and subsequently subjected to immunohistochemistry (IHC) or hematoxylin-eosin (H&E) staining.
Luciferase Reporter Assay
To determine whether let-7d directly targets PARD6A-3′UTR, a dual luciferase reporter assay system (Promega) was used to determine luciferase activity according to our previously described protocol [34]. Wild-type (WT) and mutated putative let-7d-binding site were amplified and cloned into pGL3 vector, and the constructs were transfected into different groups of cells in 96-well plates by using Lipofectamine 3000. After 24 h, the cells were treated as indicated and lysed with passive lysis buffer (Promega). Measurements were obtained with an Infinite M200 Pro microplate reader (TECAN, Hombrechtikon, Switzerland).
Statistical Analysis
All experiments were carried out at least three times with triplicate samples. Statistical analyses of group differences were performed using Student’s t-test and ANOVA. The results of the cell proliferation, colony formation, and cell invasion and migration rate assays are expressed as the mean ± SD, and the means at a given time point were compared with two-tailed independent-sample t-tests. Differences in patient survival between different subgroups were assessed by using Kaplan-Meier analyses with log-rank tests. Correlation between Par6 expression and the clinicopathological features of glioma subjects was analyzed with the χ2 test or Fisher’s exact test. Multivariate analysis was performed using Cox proportional hazards regression model. All statistical analyses were performed using SPSS software (SPSS, Chicago, IL). P < 0.05 was regarded as statistically significant.
Results
Aberrant Expression of Par6 Correlates with Malignance and Prognosis in the Patients with Glioma
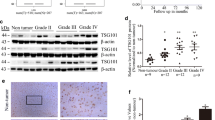
There are two known Par6 homologs in human mammary cells, including Par6α (NP_058644) and Par6β (NP_115910). To investigate whether the expression of either of them affects glioma tumorigenesis or invasion, the expressions of Par6α and Par6β were first examined in human glioma U87MG and U251 cell lines, and normal human astrocyte HEB cell line (Fig. 1A). We found strongly elevated Par6α expression in both U251 and U87MG cells compared with HEB cells. In contrast, Par6β expression was not obviously higher in U251 or U87MG cells than in HEB cells. In addition, we obtained similar results regarding the regulation of PAR6α and PAR6β mRNA levels in these three cell lines (Fig. 1B), suggesting that Par6α may play more important roles than Par6β in glioma tumorigenesis and invasion.
Measurement of Par6 expression in clinical glioma specimens. A, B The expression of Par6α and Par6β in U87MG, U251, and HEB cells at translational (A) and transcriptional (B) levels. C Representative images at normal (upper panel) and higher magnification (lower panel) show Par6 expression in low- and high-grade glioma specimens. Scale bar, 200 μm and 50 μm. D The correlation between Par6 expression and malignancy of the glioma specimens. E, F Disease-free (E) and overall (F) survival rates indicated that the expressoin level of Par6 was correlated with a poor prognosis in the patients with gliomas. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Our previous bioinformatic analyses demonstrated PAR6 expression probably correlative with malignancy and prognosis in glioma patients of TCGA and GDS1962 datasets [35]. To precisely characterize the correlation between Par6 expression and the prognosis in glioma patients, we performed IHC to determine Par6 expression in a paraffin-embedded glioma tissue microarray that contained 180 glioma specimens with prognostic information (Table 1). The results demonstrated that Par6 expression was significantly upregulated in high-grade gliomas (III and IV) compared with low-grade gliomas (I and II), indicating that Par6 expression was positively associated with malignancy in patients with glioma (Fig. 1C, D). Notably, the prognostic analyses demonstrated that higher Par6 expression was strongly associated with poorer disease-free and overall survival rates in glioma patients (Fig. 1E, F). Further multivariate analysis showed that Par6 expression was evaluated as an independent predictor of prognosis in the patients with glioma (Table 2).
Par6 Overexpression Induces Glioma Invasion In Vitro and In Vivo
To further illustrate the functions of Par6 in glioma, we established stably Par6-overexpressing (Par6-OE) and Par6-silenced (Par6-KD) U87MG and U251 cell lines. Western blot and qRT-PCR assays indicated that Par6 was overexpressed or silenced in Par6-OE or Par6-KD U87MG and U251 cells, respectively (Fig. 2A, B). Intriguingly, we noticed that Par6 overexpression significantly promoted a fibroblast-like morphology (red arrows), whereas the inhibition of Par6 triggered the formation of a cobblestone-like appearance (yellow arrowheads) in U87MG and U251 cells (Fig. 2C), suggesting the potential mesenchymal-promoting role of Par6 in glioma and other malignant tumors [36,37,38,39].
The morphological changes of U87MG and U251 cells upon Par6 overexpression. A, B Par6 expression in Par6-OE or Par6-KD U87MG and U251 cells at translational (A) and transcriptional (B) levels. C The representative images and quantification of the fibroblast-like or cobblestone-like morphologies formed in different groups of U87MG and U251 cells (red arrow, fibroblast-like morphology; yellow arrowhead, cobblestone-like morphology; n = 10 random fields of view each group). Scale bar, 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
In this context, we next performed wound healing and transwell assays to determine the effects of Par6 on invasion and migration in glioma cells. As expected, our results indicated that the migration activity was much higher in Par6-OE U251 cells than in Par6-KD U251 cells (Fig. 3A). Transwell assay also revealed that Par6 overexpression could significantly enhance invasive capability in U251 cells (Fig. 3B), indicating that the elevated Par6 expression can markedly induce glioma invasion. In addition, we measured several key regulators of glioma invasion, including CD44, N-cadherin, MMP9, MMP2, and MT1-MMP in different groups of U87MG cells. Western blot analyses revealed Par6 positively regulates the expression of CD44 and N-cadherin, whereas MMP2, MMP9, or MT1-MMP expression was not significantly affected regardless of Par6 regulation in U87MG and U251 cells (Fig. 3C, D). Moreover, similar results were obtained with regard to tumor invasion in nude mice orthotopically xenografted with Par6-OE and control U87MG cells (n = 5 for each group). The morphology of cell invasion into adjacent tissues was detected in all Par6-OE groups (5/5; white broken line), but not in the control group (0/5) of U87MG-xenografted nude mice (Fig. 3E). Notably, the enhanced CD44 and N-cadherin expression was also observed in orthotopic tumors, whereas the expressions of MMP2 and MMP9 were not statistically altered in Par6-OE U87MG cells, suggesting that MMP2 and MMP9 might not be involved in Par6-mediated glioma invasion (Fig. 3E, F). Our results also showed that overexpression of Par6 increased mortality in U87MG cell-xenografted nude mice (Fig. 3G), which was consistent with the prognostic analyses in glioma patients (Fig. 1E, F).
The elevated Par6 expression is essential for glioma invasion. A Wound healing for cell migration of U251 cells. Representative images and quantification of cell migration into the wounded area at 0, 12, and 24 h (n = 3 for each group). Scale bar, 200 μm. B Comparison of invasive capability in different groups of U251 cells evaluated by transwell assay (n = 3 random fields of view each group). Representative images of invading cells visualized by crystal violet staining, and quantification of cell invading capacity at 24 h. Scale bar, 200 μm. C, D The expression (C) and quantification (D) of CD44, N-cadherin, MMP9, MMP2, and MT1-MMP in different groups of U251 and U87MG cells. E Representative images of orthotopical implantation subjected to H&E and IHC stainings demonstrated the glioma invasion and expression of Par6, CD44, N-cadherin, MMP2, and MMP9 in tumor-bearing mice (n = 5 for each group). Scale bar, 50 μm. F Quantitative analysis of the expression of Par6, CD44, N-cadherin, MMP2, and MMP9 in tumors derived from the transplantation of Par6-OE and control U87MG cells in tumor-bearing mice. G Survival curve of tumor-bearing mice orthotopically xenografted with Par6-OE and control U87MG cells. *P < 0.05, **P < 0.01, ***P < 0.001; ns, no significance
Par6 Promotes Glioma Invasion by Activating MEK/ERK Pathway
To clarify the possible mechanisms by which Par6 mediated glioma invasion, RNA-seq analysis was performed on Par6-OE and non-treated U87MG cells (Fig. 4A). Our heatmap showed that 1420 upregulated DEGs were identified, whereas 1259 DEGs were downregulated in Par6-OE U87MG cells (Fig. 4B). In KEGG pathway analysis, we found that Par6 was dramatically enriched in MAPK signaling pathway (Fig. 4C), suggesting that activation of the MAPK pathway might be correlative to Par6-mediated glioma invasion.
The activation of MEK/ERK signaling pathway is involved in Par6-mediated glioma invasion. A Heatmap of DEG in Par6-OE and control U87MG cells. DEGs with a log2FC > 1 were indicated in red, and DEGs with a log2FC < -1 were indicated in green. B Volcano plot of DEGs in Par6-OE and control U87MG cells. DEGs with a a log2FC > 1.5 were indicated in red, and DEGs with a log2FC < -1.5 were indicated in green. C KEGG pathway enrichment analysis for Par6-OE and control U87MG cells. D The most significant regulated genes of MAPK signaling pathway in Par6-OE and control U87MG cells. E The expression of MEK, phosphorylated-MEK (pMEK), ERK1/2, and phosphorylated-ERK1/2 (pERK1/2) in Par6-OE and control U251 cells. F Immunohistochemistry demonstrated the upregulation of pMEK and pERK expression in Par6-OE U87MG cell-derived tumors in orthotopic tumor-bearing mice (n = 5 for each group). Scale bar, 50 μm. G The expression of MEK, pMEK, ERK1/2, pERK1/2, CD44, and N-cadherin in Par6-OE U251 cells with or without U0126 treatment. H, I The treatment of U0126 can signficantly regulate Par6-mediated glioma migration and invasion by wound healing (H) and transwell assays (I) in Par6-OE U251 cells. *P < 0.05, **P < 0.01, ***P < 0.001
To further investigate the possible regulators involved in this process, we next identified key factors in the MAPK pathway by analyzing the DEGs in Par6-OE and control U87MG cells. Our results showed that MEK and ERK1/2 were the most significantly regulated genes in MAPK pathways (Fig. 4D). Further western blot analyses confirmed that both MEK and ERK1/2 signaling pathways were activated in Par6-OE U251 cells, indicating that Par6 overexpression may induce glioma invasion by activating MEK/ERK signaling pathway in glioma cells (Fig. 4E). In addition, we also noticed that the tumors derived from mice receiving Par6-OE U87MG cells (n = 5 for each group) showed high expression of phosphorylated MEK and ERK1/2, suggesting that the elevated Par6 expression can also induce activation of MEK/ERK signaling pathways in vivo (Fig. 4F).
We next determined the effects of Par6 on glioma invasion through inhibiting MEK pathway in Par6-OE U251 cells. We inhibited MEK pathways by treating Par6-OE U251 cells with U0126, and western blot results showed that the phosphorylation levels of MEK and ERK1/2 were decreased in U0126-treated U251 cells. In addition, the expression of CD44 and N-cadherin was also significantly downregulated after the inhibition of MEK/ERK signaling pathway in Par6-OE U251 cells (Fig. 4G). Furthermore, we assessed tumor migration and invasion in U0126-treated Par6-OE U251 cells. Our results showed that the inhibition of MEK/ERK pathway could affect tumor migration or invasion induced by Par6 regulation in Par6-OE U251 cells (Fig. 4H, I; Fig. S1A and B), suggesting that activation of MEK/ERK pathway was essential for Par6-mediated invasion in glioma cells. Taken together, all these results indicated that MEK/ERK signaling pathway was involved in Par6-mediated invasion in glioma cells.
STAT3 Signaling Pathway Is Involved in Par6-Mediated Glioma Invasion
To investigate the downstream factors associated with Par6-mediated glioma invasion, we next identified the most significantly regulated signaling pathways, including STAT3, HDAC, AKT, VEGF, CDK1, and PTN pathways, in Par6-OE U87MG cells through DEG analysis (Fig. 5A). Our results indicated that STAT3 pathway was the most markedly regulated pathway, suggesting that the activation of STAT3 signaling pathway is probably in association with Par6-mediated glioma invasion. To confirm our hypothesis, we performed western blot to detect the expression of STAT3 and phosphorylated STAT3 in the Par6-OE and control groups of U87MG and U251 cells. The results indicated that the levels of phosphorylated STAT3 were significantly upregulated in both Par6-OE U87MG and U251 cells, suggesting that the activation of STAT3 signaling pathway can be induced in Par6-OE U87MG and U251 cells (Fig. 5B).
STAT3 signaling pathway is associated with regulation of Par6-mediated glioma invasion. A Go term enrichment analysis for Par6-OE and control U87MG cells. B The expression of STAT3 and phosphorylated-STAT3 (pSTAT3) in Par6-OE and control U87MG and U251 cells. C The expression of CD44 and N-cadherin in Par6-OE and control U251 cells with or without AT-1 treatment. D, E AT-1 treatment can inhibit Par6-mediated glioma migration and invasion by wound healing (D) and transwell assay (E) in Par6-OE and control U87MG cells (n = 3 for each goup). Scale bar, 200 μm. *P < 0.05, **P < 0.01
Next, to examine the potential correlation between the activation of STAT3 signaling pathway and Par6-mediated glioma invasion, we measured N-cadherin and CD44 expression in Par6-OE and control U87MG cells with or without AT-1 treatment. Interestingly, western blot analysis showed that the expression of both N-cadherin and CD44 was significantly inhibited in both AT-1-treated Par6-OE and control U87MG cells (Fig. 5C). Moreover, wound healing and transwell assays indicated that the tumor invasion and migration could be inhibited through the inactivation of STAT3 signaling pathway, which was specifically treated with AT-1 in Par6-OE and control U87MG cells (Fig. 5D, E).
Par6 Activates LIN28/let-7d Axis to Promote Glioma Invasion Via ERK-Dependent Mechanisms
A previous report revealed that ERK1/2 can regulate STAT3 via regulating LIN28/let7 axis in glioblastoma [40]. We therefore expected whether similar mechanism might be involved in Par6-mediated glioma invasion. We first examined LIN28 expression in different groups of U87MG and U251 cells. As shown in Fig. 6A, the inhibition of Par6 expression could significantly downregulate LIN28 expression in U87MG and U251 cells. Notably, the inhibition of LIN28 expression markedly reduced migration and invasion in Par6-OE and control U251 cells (Fig. 6B, C) through negatively regulating CD44 and N-cadherin in Par6-OE U87MG and U251 cells (Fig. 6D).
The regulation of LIN28/let-7d axis is related to Par6-mediated glioma invasion. A LIN28 expression in different groups of U87MG and U251 cells. B, C The inhibition of LIN28 significantly downregulates glioma migration and invasion by wound healing (B) and transwell assays (C) in Par6-OE and control U251 cells (n = 3 for each group). Scale bar, 200 μm. D Inhibition of LIN28 reduces the expression of LIN28, CD44, and N-cadherin in Par6-OE U251 and U87MG cells. E qPCR assay was performed to confirm the most significantly regulated miRNAs, including let-7f-3p, miR-17-5p, miR-106-6p, miR-9-3p, let-7a-6p, miR-205-5p, let-7d-5p, miR-1-3p, or miR-106-5p, which may be correlative with the activation of STAT3 pathway, in Par6-OE and control U251 cells. F The levels of let-7d in Par6-OE and control U87MG cells. G The treatment of U0126 significantly regulates let-7d in Par-OE U87MG cells. H, I The inhibition of let-7d expression remarkably rescues Par6-mediated glioma migration and invasionin by wound healing (H) and transwell assays (I) in Par6-KD and control U251 cells (n = 3 for each group). Scale bar, 200 μm. J The inhibition of let-7d upregulates the expression of CD44 and N-cadherin in Par6-KD U87MG and U251 cells. K The expression of STAT3 and pSTAT3 in Par6-KD U87MG and U251 cells with the treatment of let-7d inhibitor and control, respectively. *P < 0.05, **P < 0.01, ***P < 0.001; ns, no significance
We then identified the top 10 differentially regulated miRNAs, which may be linked to the activation of STAT3 pathway, from miRbase dataset (Data not shown). qPCR assay was performed to confirm these predicted miRNAs, including the upregulation of let-7f-3p, miR-17-5p, miR-106-6p, miR-9-3p, and let-7a-6p, and downregulation of let-7d-5p, miR-1-3p, and miR-106-5p, in Par6-OE and control U251 cells (Fig. 6E). Notably, we noticed that let-7d was not only the most significantly downregulated miRNA in Par6-OE U251 cells, which may activate STAT3 signaling pathway not only via MEK/ERK-regulated LIN28/let7 axis [40], but also via the exact complementary base pairing with PARD6A-3′UTR binding site (Fig. 7B, C). In this context, we found that let-7d expression was downregulated in Par6-OE U87MG cells, and upregulated after inactivating MEK/ERK signaling pathway in U0126-treated Par6-OE U87MG cells (Fig. 6F, G). Moreover, the inhibition of let-7d could significantly promote the migration and invasion of Par-KD and control U251 glioma cells (Fig. 6H, I), and upregulate CD44 and N-cadherin expression (Fig. 6J) through activating STAT3 pathway (Fig. 6K) in Par6-KD U87MG and U251 cells.
let-7d negatively regulates Par6 expression through directly targeting PARD6A-3′UTR binding site. A Par6 expression in U87MG cells with the treatment of NC mimic, has-let-7d mimic, inhibitor NC, or let-7d inhibitor. B Predicted binding site of wildtype (WT) and mutant (Mut) sequences of let-7d in the 3′-UTR of PARD6A. C Luciferase reporter assay was performed in U87MG cells with co-transfection of indicated WT or mutant 3′-UTR constructs and has-let-7d mimics or NC mimics. *P < 0.05, **P < 0.01; ns, no significance
let-7d Directly Targets PARD6A-3′UTR Binding Site to Negatively Regulate Par6 Expression in Glioma Cells
Previous studies have indicated that LIN28/let-7 axis plays important roles in the regulation of key factors in some tumor-related signaling pathways, including VEGF, Oct4, IGF1R, STAT3, and Akt, with positive feedback loop [40, 41]. We therefore sought to determine whether LIN28/let-7d axis not only can activate STAT3 signaling pathway but also can negatively regulates PAR6 expression in glioma cells. Western blot was performed to examine Par6 expression in U87MG cells treated with a hsa-let-7d mimic, a let-7d inhibitor, or their respective control constructs (NC mimic), respectively. It is noted that Par6 expression was reduced in hsa-let-7d mimic-treated, while being increased in let-7d inhibitor-treated U87MG cells (Fig. 7A), suggesting that let-7d can inhibit Par6 expression in glioma cells. Thereafter, luciferase reporter assay was performed to evaluate whether Par6 is the direct target of let-7d after predicting the interactions between let-7d and the binding site on the 3′UTR of PARD6A gene (Fig. 7B). The results showed that relative luciferase activity was increased upon treatment with the hsa-let-7d mimic in U87MG cells transfected with Par6 plasmids containing the WT 3′UTR sequence of PARD6A gene, whereas mutation of the 3′UTR of PARD6A gene attenuated the suppressive effect of let-7d in U87MG cells (Fig. 7C). Taken together, all these results suggested that the 3′UTR of PARD6A gene is a direct binding target of let7d in glioma cells.
Conservation of Par6-Mediated Tumor Invasion Via MEK/ERK/LIN28/let-7d/STAT3 Mechanism in Glioma Cells
In the context of the Par6/αPKC complex in the TGF-β-induced EMT in various malignant epithelial tumors, including NSCLC, breast cancer, and prostate cancer [22, 24,25,26, 42, 43], we next explored whether TGF-β can play a role in Par6-mediated glioma invasion. We detected the mRNA and protein expression of Par6 upon treatment with TGF-β at different time points (from 0 to 48 h) in glioma cells. The results showed that Par6 expression was significantly upregulated 8 h after TGF-β stimulation (Fig. S2A), and increased in a time-dependent manner in glioma cells (Fig. S2B). In addition, TGF-β treatment could also increase the expression of CD44 and N-cadherin, and activate MEK/ERK and STAT3 signaling pathways (Fig. S2C-E). Further, cell invasion assay showed that TGF-β stimulation can significantly enhance the invasive capability (Fig. S2F), which can be inhibited by AT-1 treatment (Fig. S2G) in glioma cells, suggesting TGF-β signaling might be involved in glioma invasion through upregulating Par6 expression. TGF-β-stimulated U87MG or U251 cells were co-treated with AT-1 or Rhosin to specifically block Par6/αPKC/Smurf1/RhoA or Par6/MEK/ERK/LIN28/let-7d/STAT3 signaling pathways, respectively.
Since glioblastoma is a highly heterogeneous tumor with regard to both inter- and intra-tumor heterogeneity, we next investigated the significance of Par6-enhanced glioma invasive capability in patient-like glioblastoma cell lines. Additional glioma cell lines from two primary glioma specimens were used to examine the effects of Par6 on glioma invasion. Similar results were determined that the expressions of CD44 and N-cadherin were upregulated, whereas MMP2 and MMP9 expression were inapparently affected in primary glioma cell lines (Fig. S3A). Moreover, further wound healing and transwell assays indicated the capacities of migration and invasion can be enhanced in the Par6-OE group, or inhibited in the Par6-KD group in primary glioma cells (Fig. S3B and C). In addition, we next examined whether the downstream effectors LIN28 and let-7d were also existed in the primary glioma cells. As we expected, LIN28 expression was positively correlative with Par6 expression in primary glioma cells (Fig. S4A). The results also showed that the inhibition of LIN28 expression could suppress the migration and invasion (Fig. S4B and C) through downregulating CD44 and N-cadherin in Par6-OE and control glioma primary cells (Fig. S4D). Furthermore, compared with the control group, let-7d expression was significantly downregulated in the Par6-OE group (Fig. S4E), and can be rescued with U0126 treatment (Fig. S4F). In the Par6-KD group, the inhibition of let-7d could enhance the migration and invasion capacities (Fig. S4G and H), upregulate CD44 and N-cadherin expression (Fig. S4I), and activate STAT3 pathway (Fig. S4J) in primary glioma cells, indicating that Par6-mediated glioma migration and invasion can be determined and concluded valid for in both glioma cell lines and primary glioma specimens.
To confirm that Par6/MEK/ERK/LIN28/let-7d/STAT3 pathways also exist in epithelial tumor cells, western blot was performed to measure the expression of N-cadherin and CD44 in Par6-OE and control A549 and MCF7 cells. We found that either CD44 or N-cadherin expression was unaffected in these cell lines (Fig. 8A). In addition, the inhibition of LIN28 failed to regulate Par6, CD44, or N-cadherin expression in A549 and MCF7 cells (Fig. 8B), which was different from the regulation of their expression in shRNA-LIN28-transfected U87MG and U251 cells (Fig. 6D). In contrast, we also found that the expression of E-cadherin, N-cadherin, and CD44 were inhibited in AT-1-treated groups, whereas they were unaffected in Rhosin-treated groups of TGF-β-stimulated glioma cells (Fig. 8C), excluding the existence of TGF-β-activated Par6/αPKC/Smurf1/RhoA pathways in glioma cells. Therefore, these findings indicated that the mechanisms of Par6-mediated tumor invasion and migration via MEK/ERK/LIN28/let-7d/STAT3 pathways in glioma cells (right panel, Fig. 8D) were different from those via Par6/αPKC/Smurf1/RhoA pathways in other epithelial tumor cells (left panel, Fig. 8D).
The expression of the regulatory factors between Par6/αPKC/Smurf1/RhoA and Par6/MEK/ERK/LIN28/let-7d/STAT3 pathways in tumor invasion and migration of epithelial tumors or glioma cells. A The expression of Par6, CD44, and N-cadherin in Par6-OE and control A549 and MCF7 cells. B Western blot was performed to determine the expression of LIN28, Par-6, CD44, and N-cadherin in shRNA-NC or shRNA-LIN28 transfected A549 or MCF7 cells. C Western blot determination of different groups treated with or without TGF-β, AT-1, and/or Rhosin in U87MG and U251 cells, respectively. D Schematic diagram of the possible mechanisms of Par6 functions in tumor invasion and migration in epithelial tumor cells (left panel) and glioma cells (right panel)
Discussion
Tumor invasion and migration constitute major problems in the treatment of cancer patients. Approximately 30% of patients with newly diagnosed solid tumors already have clinically detectable metastases. In the most common type of primary brain tumor, glioma, aggressive invasion by malignant glioma cells into surrounding normal brain tissues has been recognized as an important cause for relapse after surgical excision [44], and the poor prognoses of patients with glioma are partly caused by the highly invasive nature of the cancer [45]. Unlike malignant epithelial tumors, whose metastasis is usually associated with the EMT, glioma has a unique mechanism involving degradation of the ECM. Matrix metalloproteinases (MMPs), especially MMP2 and MMP9, play important roles in glioma invasion and progression [46]. Additionally, CD44 and N-cadherin, key multifunctional cell-cell adhesion molecules, have been found to be closely associated with glioma invasion processes, and their expressions are associated with the prognoses of patients with glioma [47,48,49,50]. Although the mechanisms of glioma invasion and migration have been recognized as key drivers of the malignant progression of gliomas, the signals that promote these processes are still unclear.
Par6 is an adaptor molecule for the polarity complex that works in concert with the complex to control apical-basal cell polarity, directional cell polarization, migration, and cell proliferation [51, 52]. Additional evidence has indicated that Par6 is associated with tumor invasion and migration in various malignant tumors. Phosphorylated Par6 mediates the EMT by recruiting Smurf1, which in turn leads to ubiquitin-dependent degradation of RhoA [21]. In this study, we found high expression of Par6 in advanced glioma specimens, and the expressions were correlated with the prognoses of patients with glioma (Fig. 1), suggesting that the aberration of Par6 expression may be associated with glioma invasion and progression. Our results also demonstrated that Par6 activates MEK/ERK pathway in glioma cells in vivo and in vitro and that the activation or inhibition of Par6/MEK/ERK pathway significantly regulates invasion and migration in U87MG and U251 glioma cells (Fig. 4). Nolan and colleagues found that Par6 complexes containing PKCα and Cdc42 induce cell proliferation by increasing the phosphorylated form of ERK protein in breast cancer cells [20]. In a pancreatic cancer cell line, Par6/PKCα complex has been found to increase transformed growth and invasion by activating ERK pathway [53]. Thus, our data imply that Par6-mediated activation of MEK/ERK pathway might be essential for glioma invasion.
STAT3 pathway, which contains key factors downstream of MEK/ERK pathway, has been found to be related to tumor growth, apoptosis, and metastasis in various malignant epithelial tumors [54,55,56,57,58]. In addition, activation of STAT3 signaling pathway can promote growth, metastasis, chemoresistance, immune escape, and stemness in tumors [59]. STAT3 is usually phosphorylated and activated by JAK2 or Src, and phosphorylated STAT3 can undergo dimerization and subsequently translocate from the cytoplasm into the nucleus. The activated STAT3 further binds to DNA and induces the transcription of downstream target genes [59]. Previous studies have indicated that STAT3 activation can regulate tumor metastatic factors, including CD44, N-cadherin, and E-cadherin [60,61,62,63]. Notably, Mu et al. demonstrated that activation of STAT3 is involved in glioma proliferation and invasion [64]. Therefore, given our results that overexpression of Par6 induces glioma invasion through regulation of CD44 and N-cadherin expression (Fig. 3), the mechanism of Par6-mediated glioma invasion may be associated with MEK/ERK-activated STAT3 pathway, binding of phosphorylated STAT3 to specific DNA sequences, and activation of the transcription of tumor-related genes (e.g., N-cadherin and CD44, right panel, Fig. 8D).
LIN28, first discovered in Caenorhabditis elegans as an RNA-binding protein [65], plays critical roles in many biological processes, including cell proliferation, metastasis, resistance to chemotherapy and radiotherapy, and stem cell differentiation [66, 67]. Overexpression of LIN28 resulting in dysregulation of let-7 is an essential pathological event affecting the tumorigenesis, progression, and prognosis of many malignant human cancers [68]. Additionally, LIN28 can influence let-7 target translation by inhibiting let-7-mediated downstream effects and then can promote the progression of malignant tumors [69]. Several mechanisms that contribute to the functions of LIN28/let-7 have been identified in malignant tumors. Let-7 can bind to the 3′UTRs of some key tumor-related genes, including c-myc, E2F2, cyclin D, Ras, and STAT3 [40, 70,71,72]. Intriguingly, we noticed that let-7f can regulate Par6 expression by directly targeting the 3′UTR of PARD6A in rat bone marrow mesenchymal stem cells [73], indicating that LIN28/let-7 axis might regulate Par6 expression via a positive feedback loop in this process. In our present study, we also showed that Par6 can indirectly regulate LIN28/let-7d axis through activation of MEK/ERK signaling pathway and that let-7d negatively regulates Par6 expression by subsequently targeting its 3′UTR binding site (Fig. 7).
Previous studies have been elucidated that TGF-β can induce the EMT by phosphorylating Par6 and recruiting PKCα-bound phosphorylated Par6 in epithelial cancer cells (left panel, Fig. 8D). The phosphorylated Par6 recruits the ubiquitin ligase Smurf1 to mediate the degradation of RhoA, which results in rearrangement of the actin cytoskeleton and cell migration during the TGF-β-induced EMT in cancer cells [22, 25, 42]. In this study, our results indicated the conservation of Par6-mediated tumor migration and invasion exists in glioma cells, not in other epithelial cancer cells, suggesting that TGF-β-regulated Par6 expression may be essential for tumor invasion or tumorigenesis in glioma cells. It is known that high expression of autocrine TGF-β is in association with tumor invasion and poor prognosis in glioma [74,75,76]. Also, TGF-β is usually regarded as a key regulator of the mesenchymal transition in glioma via inducing the expression of mesenchymal genes, including N-cadherin, ZEB1, Slug, and CD44 [77,78,79]. Theoretically, autocrine TGF-β signaling probably induced tumor invasion through upregulation of Par6 expression in glioma cells (right panel, Fig. 8D), which may partially illustrate the mechanism of TGF-β-related poor prognosis in glioma.
In this present study, we have identified Par6 as an upstream regulatory factor for promoting glioma invasion through MEK/ERK signaling pathway. Although our results indicated that Par6 overexpression significantly enhances the migratory ability of glioma cells, the direct experimental evidence of Par6-induced tumor invasion in patients with glioma is still limited because of the lack of glioma specimens from the tumor patients. In addition, it is noted that the mechanisms of the regulatory network in glioma invasion are very complicated. The crosstalk of different signaling pathways and the regulation of the tumor microenvironment were involved in this process. We mainly manipulated Par6 expression to investigate Par6-induced regulation of glioma invasion in glioma cell lines and orthotopic xenografts in nude mice. However, the correlation between Par6 expression and glioma microenvironment, especially for the regulation of ECM, in the process of Par6-induced glioma invasion, needs to be further studied.
In summary, we found that the expression of Par6 is positively correlated to malignancy and prognosis of the patients with glioma. Further investigations revealed that Par6 overexpression could promote glioma invasion via activating MEK/ERK signaling pathway. In addition, our results indicated that LIN28/let-7d axis is critical for regulating Par6-mediated glioma invasion through a LIN28/let-7d positive feedback loop, which directly targets the binding site of 3′UTR of PARD6A gene, to negatively regulate the Par6 expression in glioma cells. Therefore, these data highlight a novel Par6 regulatory mechanism and could be a promising therapeutic target for the therapy of gliomas.
Data Availability
All routine analysis methods are included in the “Methods” section. Sequencing data that supporting the findings of this study have been deposited in the Gene Expression Omnibus database under accession code SUB6244767. The data that support the findings of this study are presented in the paper, and all raw data are available from the corresponding author upon reasonable request. The published miRbase dataset used in this study were obtained from the publicly available database.
Abbreviations
- CNS:
-
Central nervous system
- GBM:
-
Glioblastoma
- EMT:
-
Epithelial-to-mesenchymal transition
- ECM:
-
Extracellular matrix
- TJs:
-
Tight junctions
- TCGA:
-
The Cancer Genome Atlas
- ERK:
-
Extracellular signal-regulated kinase
- NSCLC:
-
Non-small-cell lung cancer
- STR:
-
Short tandem repeat
- IHC:
-
Immunohistochemistry
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- H&E:
-
Hematoxylin-eosin
- TGF-β:
-
Transforming growth factor-β
- TMA:
-
Tissue microarray
- MMP:
-
Matrix metalloproteinase
- DEGs:
-
Differentially expressed genes.
References
Shi J, Dong B, Cao J, Mao Y, Guan W, Peng Y et al (2017) Long non-coding RNA in glioma: signaling pathways. Oncotarget 8:27582–27592
Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS (2018) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro-Oncology 20:iv1–iv86
Jansen M, Yip S, Louis DN (2010) Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol 9:717–726
Rock K, McArdle O, Forde P, Dunne M, Fitzpatrick D, O'Neill B et al (2012) A clinical review of treatment outcomes in glioblastoma multiforme--the validation in a non-trial population of the results of a randomised Phase III clinical trial: has a more radical approach improved survival? Br J Radiol 85:e729–e733
Thiery JP (2002) Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2:442–454
Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7:131–142
Cox TR, Erler JT (2011) Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech 4:165–178
Lu P, Weaver VM, Werb Z (2012) The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 196:395–406
Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME (2007) Molecular targets of glioma invasion. Cell Mol Life Sci 64:458–478
Yuan Y, Li SL, Cao YL, Li JJ, Wang QP (2019) LKB1 suppresses glioma cell invasion via NF-kappaB/Snail signaling repression. Oncotargets Ther 12:2451–2463
Ou Y, Wu Q, Wu C, Liu X, Song Y, Zhan Q (2017) Migfilin promotes migration and invasion in glioma by driving EGFR and MMP-2 signalings: a positive feedback loop regulation. J Genet Genomics 44:557–565
Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC et al (1996) par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development 122:3133–3140
Bose R, Wrana JL (2006) Regulation of Par6 by extracellular signals. Curr Opin Cell Biol 18:206–212
Etienne-Manneville S, Hall A (2001) Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell 106:489–498
Kim M, Datta A, Brakeman P, Yu W, Mostov KE (2007) Polarity proteins PAR6 and aPKC regulate cell death through GSK-3beta in 3D epithelial morphogenesis. J Cell Sci 120:2309–2317
Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E (2003) ACF7: an essential integrator of microtubule dynamics. Cell 115:343–354
Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME (2004) Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci 7:1195–1203
Aranda V, Nolan ME, Muthuswamy SK (2008) Par complex in cancer: a regulator of normal cell polarity joins the dark side. Oncogene 27:6878–6887
Marques E, Klefstrom J (2015) Par6 family proteins in cancer. Oncoscience 2:894–895
Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC et al (2008) The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer Res 68:8201–8209
Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL (2005) Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science 307:1603–1609
Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D et al (2009) A role for the TGFbeta-Par6 polarity pathway in breast cancer progression. Proc Natl Acad Sci U S A 106:14028–14033
Avery-Cooper G, Doerr M, Gilbert RW, Youssef M, Richard A, Huether P et al (2014) Par6 is an essential mediator of apoptotic response to transforming growth factor beta in NMuMG immortalized mammary cells. Cancer Cell Int 14:19
Gunaratne A, Thai BL, Guglielmo GM (2013) Atypical protein kinase C phosphorylates Par6 and facilitates transforming growth factor beta-induced epithelial-to-mesenchymal transition. Mol Cell Biol 33:874–886
Mu Y, Zang G, Engstrom U, Busch C, Landstrom M (2015) TGFbeta-induced phosphorylation of Par6 promotes migration and invasion in prostate cancer cells. Brit J Cancer 112:1223–1231
Zhang K, Zhao H, Ji Z, Zhang C, Zhou P, Wang L et al (2016) Shp2 promotes metastasis of prostate cancer by attenuating the PAR3/PAR6/aPKC polarity protein complex and enhancing epithelial-to-mesenchymal transition. Oncogene 35:1271–1282
Barash U, Spyrou A, Liu P, Vlodavsky E, Zhu C, Luo J et al (2019) Heparanase promotes glioma progression via enhancing CD24 expression. Int J Cancer 145:1596–1608
Li Y, Wang H, Sun T, Chen J, Guo L, Shen H et al (2015) Biological characteristics of a new human glioma cell line transformed into A2B5+ stem cells. Mol Cancer 14:75
Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E et al (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483:479–483
Luo H, Chen Z, Wang S, Zhang R, Qiu W, Zhao L et al (2015) c-Myc-miR-29c-REV3L signalling pathway drives the acquisition of temozolomide resistance in glioblastoma. Brain 138:3654–3672
Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R et al (2009) Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4:568–580
Duan JJ, Wang D, Cai J, Chen JJ, Zheng XX, Chen TQ et al (2022) An aldehyde dehydrogenase 1A3 inhibitor attenuates the metastasis of human colorectal cancer. Cancer Lett 536:215662
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Duan ZH, Wang HC, Zhao DM, Ji XX, Song M, Yang XJ et al (2015) Cooperatively transcriptional and epigenetic regulation of sonic hedgehog overexpression drives malignant potential of breast cancer. Cancer Sci 106:1084–1091
Liu P, Zhu C, Luo J, Lan S, Su D, Wang Q et al (2020) Par6 regulates cell cycle progression through enhancement of Akt/PI3K/GSK-3beta signaling pathway activation in glioma. FASEB J 34:1481–1496
Baghel KS, Tewari BN, Shrivastava R, Malik SA, Lone MU, Jain NK et al (2016) Macrophages promote matrix protrusive and invasive function of breast cancer cells via MIP-1beta dependent upregulation of MYO3A gene in breast cancer cells. Oncoimmunology 5:e1196299
Fan CC, Cheng WC, Huang YC, Sher YP, Liou NJ, Chien YC et al (2017) EFHD2 promotes epithelial-to-mesenchymal transition and correlates with postsurgical recurrence of stage I lung adenocarcinoma. Sci Rep 7:14617
Li Y, Zhang H, Gong H, Yuan Y, Li Y, Wang C et al (2018) miR-182 suppresses invadopodia formation and metastasis in non-small cell lung cancer by targeting cortactin gene. J Exp Clin Cancer Res 37:141
Jones ML, Siddiqui J, Pienta KJ, Getzenberg RH (2013) Circulating fibroblast-like cells in men with metastatic prostate cancer. Prostate 73:176–181
Xu X, Bao Z, Liu Y, Jiang K, Zhi T, Wang D et al (2018) PBX3/MEK/ERK1/2/LIN28/let-7b positive feedback loop enhances mesenchymal phenotype to promote glioblastoma migration and invasion. J Exp Clin Cancer Res 37:158
Balzeau J, Menezes MR, Cao S, Hagan JP (2017) The LIN28/let-7 pathway in cancer. Front Genet 8:31
Gunaratne A, Guglielmo GM (2013) Par6 is phosphorylated by aPKC to facilitate EMT. Cell Adhes Migr 7:357–361
Viloria-Petit AM, Wrana JL (2010) The TGFbeta-Par6 polarity pathway: linking the Par complex to EMT and breast cancer progression. Cell Cycle 9:623–624
Zagzag D, Salnikow K, Chiriboga L, Yee H, Lan L, Ali MA et al (2005) Downregulation of major histocompatibility complex antigens in invading glioma cells: stealth invasion of the brain. Lab Investig 85:328–341
Lefranc F, Brotchi J, Kiss R (2005) Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol 23:2411–2422
Annabi B, Lachambre MP, Plouffe K, Sartelet H, Beliveau R (2009) Modulation of invasive properties of CD133+ glioblastoma stem cells: a role for MT1-MMP in bioactive lysophospholipid signaling. Mol Carcinog 48:910–919
Merzak A, Koocheckpour S, Pilkington GJ (1994) CD44 mediates human glioma cell adhesion and invasion in vitro. Cancer Res 54:3988–3992
Okada H, Yoshida J, Sokabe M, Wakabayashi T, Hagiwara M (1996) Suppression of CD44 expression decreases migration and invasion of human glioma cells. Int J Cancer 66:255–260
Shi Q, Song X, Wang J, Gu J, Zhang W, Hu J et al (2015) FRK inhibits migration and invasion of human glioma cells by promoting N-cadherin/beta-catenin complex formation. J Mol Neurosci 55:32–41
Velpula KK, Rehman AA, Chelluboina B, Dasari VR, Gondi CS, Rao JS et al (2012) Glioma stem cell invasion through regulation of the interconnected ERK, integrin alpha6 and N-cadherin signaling pathway. Cell Signal 24:2076–2084
Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP et al (2006) Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol 8:1235–1245
Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T (2000) A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol 2:540–547
Butler AM, Scotti Buzhardt ML, Erdogan E, Li S, Inman KS, Fields AP et al (2015) A small molecule inhibitor of atypical protein kinase C signaling inhibits pancreatic cancer cell transformed growth and invasion. Oncotarget 6:15297–15310
Fang XY, Zhang H, Zhao L, Tan S, Ren QC, Wang L et al (2018) A new xanthatin analogue 1beta-hydroxyl-5alpha-chloro-8-epi-xanthatin induces apoptosis through ROS-mediated ERK/p38 MAPK activation and JAK2/STAT3 inhibition in human hepatocellular carcinoma. Biochimie 152:43–52
Fang Z, Tang Y, Fang J, Zhou Z, Xing Z, Guo Z et al (2013) Simvastatin inhibits renal cancer cell growth and metastasis via AKT/mTOR, ERK and JAK2/STAT3 pathway. PLoS One 8:e62823
Kim SJ, Pham TH, Bak Y, Ryu HW, Oh SR, Yoon DY (2018) Orientin inhibits invasion by suppressing MMP-9 and IL-8 expression via the PKCalpha/ ERK/AP-1/STAT3-mediated signaling pathways in TPA-treated MCF-7 breast cancer cells. Phytomedicine 50:35–42
Shi L, Wang S, Zangari M, Xu H, Cao TM, Xu C et al (2010) Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget 1:22–33
Zhang D, Liu H, Yang B, Hu J, Cheng Y (2019) L-securinine inhibits cell growth and metastasis of human androgen-independent prostate cancer DU145 cells via regulating mitochondrial and AGTR1/MEK/ERK/STAT3/PAX2 apoptotic pathways. Biosci Rep 39:BSR20190469
Qin JJ, Yan L, Zhang J, Zhang WD (2019) STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res 38:195
Kim S, Kil WH, Lee J, Oh SJ, Han J, Jeon M et al (2014) Zerumbone suppresses EGF-induced CD44 expression through the inhibition of STAT3 in breast cancer cells. Oncol Rep 32:2666–2672
Wei B, Sun X, Geng Z, Shi M, Chen Z, Chen L et al (2016) Isoproterenol regulates CD44 expression in gastric cancer cells through STAT3/MicroRNA373 cascade. Biomaterials 105:89–101
Quintanal-Villalonga A, Ojeda-Marquez L, Marrugal A, Yague P, Ponce-Aix S, Salinas A et al (2018) The FGFR4-388arg variant promotes lung cancer progression by N-cadherin induction. Sci Rep 8:2394
Pei G, Lan Y, Chen D, Ji L, Hua ZC (2017) FAK regulates E-cadherin expression via p-SrcY416/p-ERK1/2/p-Stat3Y705 and PPARgamma/miR-125b/Stat3 signaling pathway in B16F10 melanoma cells. Oncotarget 8:13898–13908
Mu P, Liu K, Lin Q, Yang W, Liu D, Lin Z et al (2019) Sirtuin 7 promotes glioma proliferation and invasion through activation of the ERK/STAT3 signaling pathway. Oncol Lett 17:1445–1452
Horvitz HR, Sulston JE (1980) Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96:435–454
Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD et al (2017) H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis 8:e2569
Shyh-Chang N, Daley GQ (2013) Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12:395–406
Wang T, Wang G, Hao D, Liu X, Wang D, Ning N et al (2015) Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol Cancer 14:125
Newman MA, Thomson JM, Hammond SM (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14:1539–1549
Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F et al (2010) MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One 5:e10147
Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P et al (2007) MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 67:9762–9770
Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan W et al (2013) Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro-Oncology 15:1491–1501
Han L, Wang Y, Wang L, Guo B, Pei S, Jia Y (2018) MicroRNA let-7f-5p regulates neuronal differentiation of rat bone marrow mesenchymal stem cells by targeting Par6alpha. Biochem Biophys Res Commun 495:1476–1481
Guo Z, Li G, Bian E, Ma CC, Wan J, Zhao B (2017) TGF-beta-mediated repression of MST1 by DNMT1 promotes glioma malignancy. Biomed Pharmacother 94:774–780
Singh SK, Fiorelli R, Kupp R, Rajan S, Szeto E, Lo Cascio C et al (2016) Post-translational modifications of OLIG2 regulate glioma invasion through the TGF-beta pathway. Cell Rep 16:950–966
Bayin NS, Ma L, Thomas C, Baitalmal R, Sure A, Fansiwala K et al (2016) Patient-specific screening using high-grade glioma explants to determine potential radiosensitization by a TGF-beta small molecule inhibitor. Neoplasia 18:795–805
Joseph JV, Conroy S, Tomar T, Eggens-Meijer E, Bhat K, Copray S et al (2014) TGF-beta is an inducer of ZEB1-dependent mesenchymal transdifferentiation in glioblastoma that is associated with tumor invasion. Cell Death Dis 5:e1443
Mahabir R, Tanino M, Elmansuri A, Wang L, Kimura T, Itoh T et al (2014) Sustained elevation of Snail promotes glial-mesenchymal transition after irradiation in malignant glioma. Neuro-Oncology 16:671–685
Zheng Y, Miu Y, Yang X, Yang X, Zhu M (2017) CCR7 mediates TGF-beta1-induced human malignant glioma invasion, migration, and epithelial-mesenchymal transition by activating MMP2/9 through the nuclear factor KappaB signaling pathway. DNA Cell Biol 36:853–861
Acknowledgements
Not applicable.
Funding
This study was supported by National Natural Science Foundation of China (81872070 and 81673652), Science and Technique Foundation of Guangdong Province (210728156901639), and Natural Science Foundation of Guangdong Province (2022A1515012424).
Author information
Authors and Affiliations
Contributions
YH, PL, JL, CZ, CL, and NZ performed experiments and collected data. WZ and WC reviewed and helped the manuscript writing. WC and XY supervised the project, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
All animal experiments in this study were approved by the Shantou University Medical College Animal Committee. Ethical approvals were from the Medical Ethics Committee of Shantou University Medical College (no. SUMC2019-002 and SUMC2020-73).
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

Fig. S1
The inhibition of MEK/ERK signaling pathways significantly inhibits Par6-mediated glioma invasion. (A,B) The determination of wound healing (A) and transwell assay (B) in Par6-OE with or without U0126 treatment (n = 3 for each group). Scale bars, 200 μm. (PNG 2345 kb)

Fig. S2
TGF-β treatment induces glioma invasion through upregulating Par6 expression. (A) Par6 expression in TGF-β stimulation in different time points (0, 1, 2, 4, and 8 h) in U87MG cells. (B) TGF-β stimulation induces the upregulation of Par6 in time-dependent manner from 12 to 48 h in U87MG and U251 cells. (C) The expression of Par6, CD44, and N-cadherin in TGF-β-treated U251 and U87MG cells. (D) The expression of MEK, pMEK, ERK1/2, and pERK1/2 in U87MG and U251 cells with or without TGF-β treatment. (E) The expression of STAT3, pSTAT3 in U251 and U87MG cells with or without TGF-β treatment. (F) The representative images and quantitative analyses of glioma invasion in U251 and U87MG cells with or without TGF-β treatment, respectively (n = 3 for each group). Scale bar, 200 μm. (G) AT-1 treatment inhibit TGF-β-stimulated glioma invasion in U87MG cells. Scale bar, 200 μm. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, no significance. (PNG 802 kb)

Fig. S3
The Par6 expression is positively correlative with the glioma invasion and migration in primary glioma cells. (A) The expression levels of CD44, N-cadherin, MMP2, and MMP9 in Par6-OE, Par6-KD, and control groups of the cell lines from two primary glioma specimens (GBM1 and GBM2). (B) Cell invasion assays in Par6-OE, Par6-KD and control groups in two primary glioma cell lines (n = 3 random fields of view each group). Representative images of invading cells visualized by crystal violet staining, and quantification of cell invading capacity at 24 h. Scale bar, 200 μm. (C) Wound healing for cell migration in Par6-OE, Par6-KD and control groups in primary glioma cells. Representative images and quantification of cell migration into the wounded area at 0, 12, and 24 h (n = 3 for each group). Scale bar, 200 μm. *P < 0.05, **P < 0.01, ***P < 0.001. (PNG 1264 kb)

Fig. S4
The validation of downstream LIN28/let-7d axis in Par6-mediated tumor invasion in primary glioma cells. (A) Western blot determination and quantification of LIN28 expression in different groups in primary glioma cells. (B,C) Wound healing and transwell assays for migration (B) and invasion (C) in Par6-OE groups in primary glioma cells with or without the inhibition of LIN28 expression (n = 3 for each group). (D) The downregulation of CD44 and N-cadherin expression in Par6-OE cells after the inhibition of LIN28 expression. (E) qPCR assays were performed to determine the levels of let-7d in Par6-OE and control cells. (F) U0126 treatment can induce the upregulation of let-7d in Par6-OE groups in primary glioma cells. (G,H) Wound healing and transwell assays for migration (G) and invasion (H) in Par6-KD groups in primary glioma cells with or without the treatment of let-7d inhibitor (n = 3 for each group). (I,J) Western blot determination and quantification of CD44, N-cadherin (I), STAT3, and pSTAT3 (J) in Par6-KD groups in primary glioma cells with or without the treatment of let-7d inhibitor. *P < 0.05, **P < 0.01, ***P < 0.001. (PNG 1179 kb)
Supplementary file 1
(DOCX 25 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, Y., Liu, P., Luo, J. et al. Par6 Enhances Glioma Invasion by Activating MEK/ERK Pathway Through a LIN28/let-7d Positive Feedback Loop. Mol Neurobiol 60, 1626–1644 (2023). https://doi.org/10.1007/s12035-022-03171-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-03171-0