Abstract
Increasing evidence has suggested the crucial role cyclin-dependent kinases (CDKs) in the biology of hepatocellular carcinoma (HCC), a lethal malignancy with high morbidity and mortality. Hence, this study explored the modulatory effect of the putative cyclin-dependent kinase 11B (CDK11B)-mediated ubiquitination on HCC stem cells. The expression of CDK11B, SAM pointed domain-containing ETS transcription factor (SPDEF) and DOT1-like histone lysine methyltransferase (DOT1L) was determined by RT-qPCR and western blot analysis in HCC tissues and cells. The interaction among CDK11B, SPDEF, miR-448, and DOT1L was analyzed by Co-IP, ubiquitination-IP and ChIP assays, whereas their effects on the biological characteristics of HCC stem cells were assessed by sphere formation and colony formation assays. An in vivo xenograft tumor model was developed for validating the regulation of CDK11B in oncogenicity of HCC stem cells. We characterized the aberrant upregulation of CDK11B and downregulation SPDEF in HCC tissues and cells. CDK11B degraded SPDEF through ubiquitin-proteasome pathway, whereas SPDEF could bind to the miR-448 promoter and inhibit the expression of DOT1L by activating miR-448, whereby promoting self-renewal of HCC stem cells. Knockdown of CDK11B attenuated the self-renewal capability of HCC stem cells and their oncogenicity in vivo. These findings highlighted that blocking the CDK11B-induced degradation of SPDEF and enhancing miR-448-dependent inhibition of DOT1L may delay the progression of HCC by restraining self-renewal capability of HCC stem cells, representing novel targets for HCC management.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is a heterogeneous disease and the most frequent primary liver cancer that ranks the 2nd leading cause of cancer-associated deaths globally because of multi-focal recurrence [1, 2]. The leading risk factors for this malignancy cover hepatitis B virus or hepatitis C virus chronic infections, excessive drinking, and non-alcoholic fatty liver disease [3]. Although liver transplantation represents a prominent tool for treating HCC, there remain several questions such as organ shortage, only works at certain stages and inconclusive selection criteria on candidates [4]. Therefore, in-depth knowledge of HCC is badly in need to achieve a better management on HCC and this study was designed to meet this demand from molecular levels through exploring a specific molecular mechanism of HCC and further providing a new therapeutic strategy.
Cyclin-dependent kinases (CDKs) are identified as crucial players in proliferation and growth of tumor cells and vital mediators of several targets and phosphorylate transcription factors implicated in tumorigenesis [5]. For instance, CDKs have been well-documented to be potential therapeutic targets of HCC and hence would be of outstanding values in the disease management [6]. CDK11B, as a member of CDKs, was identified to be responsible for the poor prognosis of colon cancer [7]. The CDK11p58 has been revealed to enhance ubiquitination of SAM pointed domain-containing ETS transcription factor (SPDEF) through the proteasome pathway [8]. As a member of the ETS transcription factor family, SPDEF is recently proposed as a prognostic maker in the prostate cancer [9]. A binding relationship between SPDEF and the microRNA-448 (miR-448) promoter has been proposed [10]. miRNAs are a group of short non-coding RNAs that are involved in many biological processes and development of cancers including HCC [11]. Intriguingly, miR-448 functioned as a tumor suppressor in HCC [12]. Our previous study suggested that miR-448 was a suppressor of the stemness and self-renewal in the HCC stem cells [13]. Furthermore, DOT1-like histone lysine methyltransferase (DOT1L) like histone lysine methyltransferase (DOT1L) as the sole histone H3 lysine 79 (H3K79) methyltransferase exerts key effects on the progression of malignancies [14]. Another research has suggested that DOT1L and miR-133b can interact with each other and affect colorectal cancer cell stem-like properties and chemoresistance [15]. Based on the above findings and our results of microarray-based analysis, we hypothesized that CDK11B might be involved in the development of HCC by interacting with the SPDEF/miR-448/DOT1L axis. Hence, the present study was conducted to testify this hypothesis by performing both in vitro and in vivo assays.
Material and methods
Ethics statement
The current study was performed with the approval of the ethics committee of Central South University Xiangya School of Medicine Affiliated Haikou Hospital and conformed to the Declaration of Helsinki. Written informed consent was obtained from each participant. All animal experiments were performed with the approval of the animal committee of Central South University Xiangya School of Medicine Affiliated Haikou Hospital and in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Microarray-based gene expression profiling
The Ualcan database revealed that CDK11B was highly expressed and SPDEF was lowly expressed in HCC. SPDEF is able to bind to downstream genes and further promote gene transcription as a transcription factor. Analysis by the SPDEF Chip-seq data suggested that SPDEF can bind to the miR-448 promoter, suggesting that SPDEF transcription activates miR-448. miR-448 is predicted to target DOT1L, which is highly expressed in HCC.
Study subjects
This study enrolled a total of 32 patients with HCC and 28 patients with benign liver hyperplasia who received needle aspiration biopsy and pathological diagnosis from March 2018 to September 2019 at the Central South University Xiangya School of Medicine Affiliated Haikou Hospital. Among them, the average age of 32 patients with HCC was 64.13 ± 9.03 years, and the average age of 28 patients with benign liver hyperplasia was 62.25 ± 9.63 years. The diagnosis of HCC was performed according to standards of World Health Organization. All tumor samples were pathological sections that passed pathological examination and confirmed to contain >80% of tumor cells. The inclusion criteria for selection of patients were as follows: patients received no anticancer treatments before surgery; patients with all tumor nodules completely resected confirmed by the resection of surface tumor-free tissues by pathological examination; patients with complete clinicopathological and follow-up data. Patients were excluded if they met any of the following criteria: patients died from non-HCC or accident. The clinicopathological characteristics of patients are shown in Table 1.
Cell culture and transfection
Four HCC cell lines (Huh-7, HepG2, SK-Hep1, and SMMC-7721) and normal human normal liver cell line L02 were commercially available from Shanghai Institute of Life Science, Chinese Academy of Sciences (Shanghai, China) (http://www.cellbank.org.cn/index.asp). The above-mentioned cell lines were incubated in Dulbecco’s modified Eagle’s medium/Ham’s F12 medium (DMEM/F12) (Life technologies, Gaithersburg, MD) containing 10% fetal bovine serum at 37 °C with 5% CO2. HCC cells were treated with the proteasome pathway inhibitor MG132 (Sigma-Aldrich Chemical Company, St Louis, MO, USA) at a final concentration of 10 μmol/L for 16 h when cell confluence reached 50%.
For cell transfection, an appropriate amount of CD133 + CD90 + Huh-7 or CD133 + CD90 + SMMC-7721 cells were seeded into 24-well plates until the cell density reached 50–60%. Overexpression or gene silencing lentivirus was produced by transient transfection of 293 T cells. Then 2 × 106 TU corresponding lentivirus and 5 μg Poly-brene were added into 1 mL serum-free and antibacterial medium, mixed and then transfected. The medium was changed after 6 h of culture, and 48 h later, the cells were collected for subsequent experiments. Then the expression of green fluorescent protein in cells was detected using a fluorescence microscope. The plasmids were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). Cells were infected with lentiviruses expressing short hairpin RNA (sh)-CDK11B-1, sh-CDK11B-2, overexpression (oe)-CDK11B, oe-SPDEF + miR-488 inhibitor, oe-SPDEF + miR-488 inhibitor, and oe-SPDEF + oe-DOT1L as well as their separate controls (sh-negative control [NC], oe-NC, oe-NC + miR-488 inhibitor, oe-NC + NC inhibitor, oe-SPDEF + NC inhibitor, oe-NC + miR-488 inhibitor, oe-NC1 + oe-NC2, oe-SPDEF + oe-NC2, and oe-NC1 + oe-DOT1L).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Cells after different treatments were collected and lysed using TRIzol reagents (Invitrogen, Carlsbad, CA, USA) to extract total RNA. Ultraviolet-visible spectrophotometry (ND-1000, Nanodrop, Thermo Fisher Scientific, Waltham, MA) was used to detect the quality and concentration of RNA. The PrimeScript RT Reagent Kit (Takara, Dalian, Liaoning, China) was used for reverse transcription of 400 ng of total RNA. Subsequently, the complementary DNA (cDNA) was used as a template and the RT-qPCR was performed according to the instructions of SYBR® Premix Ex Taq TM II Kit (Tli RNaseH Plus, Takara, Tokyo, Japan). The primers were synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, Guangdong China) (Table 2). The expression of the mRNAs normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the miRNA normalized to U6 was determined by the 2^(−ΔΔCT) method using the formula: ΔΔCT = ΔCT (target gene)−ΔCT (reference gene).
Western blot analysis
HCC cells were washed with phosphate-buffered saline (PBS) and then incubated with cell lysis buffer (C0481, Sigma-Aldrich Chemical Company, St Louis, MO, USA) at 4 °C for 30 min. The cell lysis buffer was collected into a 1.5 mL eppendorf (EP) tube, centrifuged at 12,000 × g at 4 °C for 15 min and the supernatant was collected. The protein concentration was determined by a bicinchoninic acid (BCA) protein assay kit (Shanghai Beyotime Biotechnology Co. Ltd., Shanghai, China). The supernatant was added with protein loading buffer and after boiling for 5 min, 20 mg of protein sample was electro-transferred onto polyvinylidene fluoride membranes by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Millipore, Billerica, MA, USA) at 0.3 A and 20 V. The membrane was then blocked with 5% skimmed milk powder for 1 h, and incubated with Tris-buffered saline Tween-20 (TBST)-diluted rabbit antibodies to CDK11B (ab198813, 1: 200), SPDEF (ab53881, 1:100), DOT1L (ab72454, 1:1000), AMPK (ab32047, 1:1000), phosphorylated-AMPK (ab131357, 1:100), sex determining region Y (SRY)-box 2 (SOX-2, ab97959, 1:200), organic cation/carnitine transporter 4 (OCT-4, ab18976, 1:1000), Nanog (ab80892, 1:1000), CD133 (ab19898, 1:1000), CD90 (ab133350, 1:2000), Flag (ab1162, 1:2000), Myc (ab9106, 1:2000), Ubiquitin (ab7780, 1:1000), immunoglobulin G (IgG, ab172730, 1:2000) and GAPDH (ab37168, 1:100) at 4 °C overnight. After washing three times with TBST, the membrane was subjected to another incubation with horseradish peroxidase-labeled secondary mouse anti-rabbit (ab9482, 1:5000) for 1 h at ambient temperature. The aforementioned antibodies were all purchased from Abcam (Cambridge, UK). Images were developed by enhanced chemiluminescence (Shanghai Baoman Biotechnology Co., Ltd., Shanghai, China). GAPDH was taken as an internal control, the gray value of each band was analyzed using Image J software.
Coimmunoprecipitation (Co-IP) assay
HCC cells were washed with PBS, lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (Shanghai Beyotime Biotechnology Co. Ltd., Shanghai, China), and then incubated at 4 °C for 30 min. The RIPA cell lysis buffer was collected into a 1.5 mL EP tube and centrifuged at 12000 × g at 4 °C for 15 min, with the supernatant collected. The protein concentration was determined by a BCA protein concentration assay kit (Shanghai Beyotime Biotechnology Co. Ltd., Shanghai, China). Protein A agarose (Bio-rad, Inc., Hercules, CA, USA) was prepared. Beads were washed twice with PBS and prepared to 50% concentration with PBS. Samples were then pretreated with Protein A agarose and reacted for 2 h, followed by incubation with CDK11B rabbit antibody and SPDEF rabbit antibody and overnight reaction with the target protein (12 h). After the antibody was conjugated for 12 h, Protein A agarose (30 μL/tube) was added, and the reaction was carried out for 3–6 h. After 6 h of reaction, cell lysis was centrifuged at 3000 rpm for 5 min, and then the supernatant was discarded, after which the pellet was kept in a 1.5 mL EP tube. The pellet was washed three times with pre-chilled RIPA lysis buffer, and added with 1 mL of lysis buffer for 10 min of reaction at 4 °C. The precipitate was collected, added with loading, boiled in water for 5 min, and then placed on ice. After cooling to room temperature, cells were centrifuged at 12000 rpm for 10 min for Western blot analysis.
Chromatin Immunoprecipitation (ChIP) assay
EZ-Magna ChIP TMA kit (Millipore, Billerica, MA) was used for ChIP assay. HCC cells in the logarithmic growth phase were cross-linked, cultured with 1% formaldehyde for 10 min, and added with 125 mM glycine at ambient temperature for 5 min to terminate the cross-linking. The cells were then washed twice with pre-chilled PBS and centrifuged at 2000 rpm for 5 min to collect the cells. Cells were resuspended in cell lysis buffer (150 mM NaCl, 50 mM Tris (pH 7.5), 5 mM ethylenediaminetetraacetic acid, 0.005% NP40, 0.01% Triton X-100) to a final cell concentration of 2 × 106 cells per 200 mL. Cells were then added with the protease inhibitor mixture, centrifuged at 5000 rpm for 5 min, resuspended in nuclear separation buffer, lysed in an ice water bath for 10 min, and sonicated to obtain 200–1000 bp chromatin fragments. Thereafter, cells were centrifuged at 14000 × g and 4 °C for 10 min and the supernatant was aspirated. Next, 100 μL of the supernatant (DNA fragment) was added to 900 μL of ChIP Dilution Buffer, 20 μL of 50× PIC and 60 μL of Protein A Agarose/Salmon Sperm DNA. After inverted and mixed at 4 °C for 1 h, cells were stand still at 4 °C for 10 min and centrifuged at 700 rpm for 1 min. The supernatant was collected and 20 μL of them were taken as Input. The supernatant was incubated with rabbit anti-SPDEF antibody in the experimental group overnight, and 1 μL of rabbit anti-IgG was added to the NC group, and 60 μL of Protein A Agarose/Salmon Sperm DNA was added to each tube for 2 h at 4 °C. After standing for 10 min, cells were centrifuged at 700 rpm for 1 min. The supernatant was discarded and the pellet was washed with 1 mL of low-salt buffer, high-salt buffer, LiCl solution, and TE (twice). Each tube was eluted twice with 250 mL ChIP wash buffer, and de-cross-linked using 20 mL of 5 M NaCl, whereupon DNA was recovered after de-cross-linking. The miR-448 promoter in the complex was quantified by fluorescent quantitative PCR. The primer sequences of miR-448 promoter are shown in Table 2.
Sphere and colony formation assays
Cells in the synchronized logarithmic growth phase were taken and trypsinized to prepare cell suspensions. Cells were triturated and dispersed to fully disperse the cells, and the single cell percentage should be above 95%. Cells were counted and the cell concentration war adjusted with gradient dilution of the medium according to the cell status and proliferation ability. According to the concentration of 50, 100, and 200 cells per dish, 5 mL of cell suspension was inoculated into an ultra-low adherent culture dish (Corning Life Science, Jiangsu, China), and the dish was gently shaken in a cross direction to spread the cells evenly. Tumor spheres were cultured in serum-free DMEM/F12 (Invitrogen, Carlsbad, California, USA) supplemented with 2% B27, 20 ng/mL epidermal growth factor, 20 ng/mL basic fibroblast growth factor, 5 μg/mL insulin and 0.4% bovine serum albumin. The culture dish was cultured at 37 °C with 5% CO2 for 2–3 weeks. During the period, the formation of stem cell microspheres was observed, and the fresh culture solution was replaced in time according to the pH change. When clones could be observed by naked eyes in the petri dish, the culture was terminated and the culture solution was discarded. Cells were immersed carefully with PBS solution twice, air-dried, fixed with methanol for 15 min, and stained with Giemsa staining solution for 10 min. Colony formation was observed in the plate.
Dual-luciferase reporter assay
Jaspar and Targetscan websites were applied to analyze the binding site between SPDEF and miR-448 promoter (TCCCAGATGTA), miR-448 and DOT1L-3′-untranslated region (3′-UTR) (AUACGU). The wild-type (WT) and mutant type (MUT) of miR-448 promoter and DOT1L-3′-UTR-WT and DOT1L-3′-UTR-MUT fragments were constructed into pGL3-basic vector (591 bp). The correctly sequenced luciferase reporter plasmids miR-448 mimic and mimic NC were co-transfected with oe-NC and oe-SPDEF into HCC stem cells or HEK293T cells (Shanghai Beinuo Biological Technology Co., Ltd., Shanghai, China), respectively. Renilla luciferase plasmid (pRL-TK) was also transfected together as a reference. Cell lysates were harvested 48 h after transfection. The luciferase activity was then determined using dual-luciferase reporter gene assay system (Promega Corporation, Madison, WI, USA). The luciferase activity was normalized to Renilla luciferase activity.
5-ethynyl-2’-deoxyuridine (EdU) assay
Cells at the synchronized logarithmic growth phase were taken and inoculated into a 24-well plate at a density of 1 × 105 cells/well, and incubated with 50 μM EdU medium for 2 h. Then discard the EdU medium was discarded and cells were washed twice with PBS and added with 100 μL of 4% paraformaldehyde PBS each well and incubated for 30 min at ambient temperature. The fixative was discarded and cells were added with 100 μL of 2 mg/mL glycine each well, and incubated in a shaker for 5 min. Glycine was then discarded and cells were added with 200 μL permeate (0.5% Triton X-100 PBS buffer) each well and incubated for 10 min, 200 μL of 1× Apollo staining solution (C10812-1, Guangzhou RiboBio Co., Ltd., Guangzhou, Guangdong China) at ambient temperature in dark box for 30 min, the reaction solution was then discarded and wash three times with 200 μL permeate per well. The permeate was discarded and cells were washed 5 min with methanol. Then cells were added with 200 μL 1× Hoechst33342 reaction solution (DA0014, Leagene, Beijing, China) per well and incubated in a light-proof shaker for 30 min, washed twice with PBS and observed under a fluorescence microscope. EdU fluorescence signal was red, and Hoechst33342 fluorescence signal was blue. Cells with pink fluorescence were proliferative cells and with blue fluorescence were non-proliferative cells.
Flow cytometry
First, 1 × 106 cells were resuspended in 100 μL PBS containing 20 μL of FcR blocker (Miltenyi biotec, Bergisch Gladbach, Germany) and placed on ice for 10 min to block Fc receptors. After 30 min of incubation with anti-human antibodies: PE-CD90 (ab95700, Abcam Inc., Cambridge, UK) and PE-CD133 (ab252128, Abcam Inc., Cambridge, UK) in the dark, the cells were washed twice with 1 mL of cold PBS. The collected cells were resuspended in 300 μL PBS and detected by a FACSCanto II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). The isotype-matched mouse antibody was used as the control.
Xenograft tumor in nude mice
The stably transfected HCC stem cells were constructed using lentiviral vectors and cultured for 3–7 days in vitro to prepare 50 μL of 1 × 106 single cell suspension. The cell suspension and 50 μL of Matrigel solution were injected into the right upper back region of severe combined immune deficient mice. The day of inoculation was recorded as day 0. Since the 7th day after inoculation, the tumor size was monitored with vernier calipers every other week, and the tumor volume was calculated using the formula: tumor volume (V) = 0.8 × 2/3 × D12 × D2 (D1, D2 are the longest diameter and shortest diameter, respectively, which are perpendicular to each other). At the 28th day following SMMC-7721 cell inoculation, mice were killed by intravenous injection (threefold concentration) of 3% pentobarbital sodium (P3761, Sigma-Aldrich Chemical Company, St Louis, MO, USA). The formed tumors were excised and weighed. The metastatic lymph nodes from the ipsilateral side of the xenograft tumors were excised and RNA was extracted for RT-qPCR.
Immunohistochemical staining
Tumor tissues and lymph node tissues were obtained from the killed mice, and fixed in 4% paraformaldehyde, paraffin-embedded, sliced into 4-μm-thick sections, and routinely dewaxed. The streptavidin-perosidase (SP) method was used for routine operation. Microwave-stimulated antigen retrieval was conducted and upon being boiled, the heating was stopped for 5 min, followed by another heating. After cooling to normal temperature, the sections were washed with PBS, and blocked with normal goat serum blocking solution. The oe-NC group was used as a NC during the experiment, and HistostainTM SP-9000 immunochemical staining kit (Zymed Laboratories, San Francisco, CA) was used for staining. Then the sections were probed with primary antibodies (CDK11B, DOT1L, AMPK, P-AMPK, OCT-4, SOX-2, Nanog) at 4 °C overnight. The next day, the sections were re-probed with corresponding secondary rabbit antibody at 37 °C for 30 min, added with horseradish-labeled working solution and incubated, followed by development using DAB for 5–10 min. The staining time was adjusted under a microscope. After being counterstained by hematoxylin for 1 min, the sections were sealed and photographed. Next, five representative high-power fields (upright optical microscope, NIKON, Tokyo, Japan) were selected for observation and counting. The cells with brown and yellow cytoplasm were considered to be positive. The slides were imaged on an Olympus BX53 fluorescence microscope using ×20 objective (Olympus, Central Valley, PA, USA), and the intensity measurement was conducted using Olympus Cellsens Dimension 1.7 image analysis software (Olympus) [16].
Statistical analysis
The Statistic Package for Social Science 21.0 statistical software (IBM Corp, Armonk, NY, USA) was utilized for statistical analysis. The measurement data were expressed as mean±standard deviation. Data between two groups were compared by unpaired t test while data among multiple groups were analyzed using one-way analysis of variance, followed by Tukey’s test. A p < 0.05 was considered statistically significant.
Results
CDK11B presents high expression in HCC and its silencing restrains HCC cell proliferation
Candidate proteins that highly expressed in HCC were screened using the Ualcan database, which included a multifunctional human CDK11B (Fig. 1a). In order to further study the expression of CDK11B in HCC, the expression of CDK11B was determined by RT-qPCR and immunochemistry in HCC tissue samples and normal liver tissue samples, and the results showed that CDK11B was abnormally highly expressed in HCC samples (Fig. 1b, c). Further, western blot analysis revealed that the expression of CDK11B was much higher in HCC cell lines (Huh-7, HepG2, SK-Hep1, and SMMC-7721) than in normal liver cells L02 (Fig. 1d). HCC cell lines SMMC-7721 and Huh-7 presented with the highest CDK11B expression and were thus selected for subsequent experiments. In order to further study the functional mechanism of CDK11B, three CDK11B shRNAs (sh-CDK11B-1, sh-CDK11B-2, and sh-CDK11B-3) were designed and synthesized to knockdown the expression of CDK11B. The three shRNAs were transfected into SMMC-7721 and Huh-7 cells, and the results of RT-qPCR and western blot analysis demonstrated that the three shRNAs could all knockdown CDK11B expression obviously (Fig. 1e, f), with sh-CDK11B-1 showing the lowest CDK11B expression and selected for subsequent studies. The proliferation of HCC cells after CDK11B silencing was detected by EdU assay, which revealed that the proliferation of SMMC-7721 and Huh-7 cells was reduced after CDK11B silencing (Fig. 1g, h). Hence, CDK11B expressed abundantly in HCC tissues and cells, whereas its knockdown might arrest the proliferation of HCC cells.
a CDK11B expression in HCC tissues analyzed by the Ualcan database. b The expression of CDK11B in HCC tissue samples (n = 32) and normal liver tissue samples (n = 28) determined by RT-qPCR. c The positive expression of CDK11B protein in HCC tissue samples (n = 32) and normal liver tissue samples (n = 28) determined by immunochemistry (scale bar = 25 μm). d Representative western blots of CDK11B protein and its quantitation in HCC cell lines and normal liver cells. e The CDK11B expression in SMMC-7721 cells transfected with sh-CDK11B-1, sh-CDK11B-2, and sh-CDK11B-3 determined by RT-qPCR and western blot analysis. f The CDK11B expression in Huh-7 cells transfected with sh-CDK11B-1, sh-CDK11B-2, and sh-CDK11B-3 determined by RT-qPCR and western blot analysis. g The EdU-positive SMMC-7721 cells following CDK11B silencing (scale bar = 25 μm). h The EdU-positive Huh-7 cells following CDK11B silencing (scale bar = 25 μm). *p < 0.05 vs. normal samples, L02 cells or cells transfected with sh-NC. Unpaired t test was used for data analysis between two groups. One-way ANOVA was used for data analysis among multiple groups and followed by Tukey’s test. Cell experiments were repeated three times independently.
CDK11B downregulates SPDEF expression by elevating SPDEF phosphorylation and ubiquitination-dependent degradation in HCC cells
CDK11B is able to interact with SPDEF according to National Center of Biotechnology Information (NCBI) database analysis. SPDEF is an epithelial-specific ETS transcription factor and plays an important role in tumor proliferation [9]. A recent study has identified the downregulation of SPDEF in HCC [17]. Meanwhile, the expression of SPDEF in HCC and normal liver tissues determined by RT-qPCR showed a downregulation in HCC tissues relative to normal liver tissues (Fig. 2a). Immunochemistry results also confirmed that the positive expression of SPDEF protein was higher in normal liver tissues than in HCC tissues (Fig. 2b). The expression of SPDEF in HCC cell line (SMMC-7721 and Huh-7) and normal liver cells L02 was measured by western blot analysis, and the results presented aberrant downregulation of SPDEF in HCC cell lines (Fig. 2c). In order to further explore the relationship between CDK11B and SPDEF, SMMC-7721, and Huh-7 cells were transfected with sh-CDK11B and sh-NC, respectively, and the expression of SPDEF was subsequently measured by RT-qPCR and western blot analysis. The results illustrated no changes in SPDEF mRNA expression, whereas SPDEF protein expression was upregulated in HCC cells after transfection with sh-CDK11B (Fig. 2d). Co-IP assay verified a direct interaction between CDK11B and SPDEF (Fig. 2e). The IP experimental results demonstrated that after co-transfection of oe-CDK11B and oe-SPDEF, the band showed the most significant diffuse shape, indicating that CDK11B could increase the ubiquitination level of SPDEF (Fig. 2f). After treatment with the proteasome pathway inhibitor MG132, the diffuseness of the bands weakened (Fig. 2f), suggesting that CDK11B degraded SPDEF through the ubiquitin-proteasome pathway. To sum up, CDK11B could downregulate SPDEF expression by stimulating its ubiquitination and degradation.
a The expression of SPDEF in HCC tissue samples (n = 32) and normal liver tissue samples (n = 28) determined by RT-qPCR. b The positive expression of SPDEF protein in HCC tissue samples (n = 32) and normal liver tissue samples (n = 28) determined by immunochemical staining (scale bar = 25 μm). c Representative western blots and quantitation of SPDEF protein in SMMC-7721, Huh-7, and L02 cells. d The expression of SPDEF in HCC cells transfected with sh-CDK11B determined by RT-qPCR and western blot analysis. e The interactions between SPDEF and CDK11B in HCC cells detected by Co-IP. f The SPDEF ubiquitination regulated by CDK11B revealed by ubiquitination-IP experiment. *p < 0.05 vs. normal samples, L02 cells, or cells transfected with sh-NC. Data analysis between two groups was conducted by unpaired t test, and data analysis among multiple groups was conducted by one-way ANOVA and Tukey’s test. Cell experiments were repeated three times independently.
SPDEF upregulates miR-448 expression while downregulating DOT1L expression in HCC stem cells
Further, the expression of miR-448 in HCC tissues and normal liver tissues was measured by RT-qPCR, and miR-448 was evidenced to be poorly expressed in HCC tissues (Fig. 3a). Consistently, miR-448 expression was much lower in SMMC-7721 and Huh-7 cells than in normal liver cells L02 (Fig. 3b). Analysis using the Targetscan website suggested that DOT1L may be a downstream gene of miR-448 (Supplementary Fig. 1A), and meanwhile, dual-luciferase reporter assay confirmed that miR-448 targeted DOT1L (Supplementary Fig. 1B). In addition, DOT1L mRNA and protein expression was found to be inhibited in SMMC-7721 and Huh-7 cells overexpressing miR-448 (Supplementary Fig. 1C, D). In silico analysis further identified that DOT1L was an upregulated gene in liver cancer (Fig. 3c). Our results obtained from RT-qPCR and western blot analysis consistently suggested the upregulation of DOT1L in clinical HCC tissues relative to normal liver tissues (Fig. 3d), as well as in SMMC-7721 and Huh-7 cells relative to normal liver cells L02 (Fig. 3e). Sphere formation analyses showed that compared with parental cells, the percentage of CD90+ and CD133+ was increased in sphere-derived cells (Fig. 3f), and SMMC-7721 and Huh-7 sphere cells were thereby selected as HCC stem cell models for subsequent experiments.
a The expression of miR-448 in HCC tissue samples (n = 32) and normal liver tissue samples (n = 28) determined by RT-qPCR. b The expression of miR-448 in HCC cells and normal liver cells determined by RT-qPCR. c The expression of DOT1L in HCC identified by the Ualcan database. d The expression of DOT1L in HCC tissue samples (n = 32) and normal liver tissue samples (n = 28) determined by RT-qPCR. e The expression of DOT1L in HCC SMMC-7721 and Huh-7 cells determined by RT-qPCR and western blot analysis. f The proportion of stem cell markers CD90 and CD133 positive in HCC cells determined by sphere formation assay (scale bar = 50 μm). g The expression of DOT1L determined by RT-qPCR and western blot analysis in sphere and parental cells. h Binding sites between SPDEF and miR-448 promoter predicted by the Jaspar website. i The binding of SPDEF to the miR-448 promoter confirmed by dual-luciferase reporter gene assay. j The binding relationship between SPDEF and miR-448 detected by ChIP assay. k The expression of miR-448 and DOT1L determined by RT-qPCR in SMMC-7721 and Huh-7 cells transfected with oe-SPDEF, miR-448 inhibitor, or both. l Representative western blots and quantitation of DOT1L and SPDEF proteins in SMMC-7721 and Huh-7 cells transfected with oe-SPDEF, miR-448 inhibitor, or both. *p < 0.05 vs normal samples, L02 cells, parental cells, or cells treated with oe-NC, or oe-NC + NC inhibitor. Data analysis between two groups was conducted using unpaired t test, and data among multiple groups were analyzed using one-way ANOVA and followed by Tukey’s test. Cell experiments were repeated three times independently.
Furthermore, the expression of SPDEF detected by RT-qPCR and western blot analysis exhibited an upregulation in sphere cells relative to parental cells (Fig. 3g). Next, we aimed to investigate whether SPDEF activated miR-448 transcription by binding to the miR-448 promoter region. The Jaspar website predicted the presence of binding sites between SPDEF and miR-448 promoter (Fig. 3h). Moreover, dual-luciferase reporter assay indicated that luciferase activity was enhanced in oe-SPDEF-transfected cells (Fig. 3i). The results of ChIP assay revealed that relative SPDEF occupancy at the miR-448 promoter was increased in SMMC-7721 and Huh-7 cells overexpressing SPDEF, suggesting that SPDEF was able to bind to the miR-448 promoter region as a protein nucleic acid (Fig. 3j). In addition, the HCC stem cells were transfected with oe-SPDEF or miR-448-inhibitor, and RT-qPCR and western blot analysis exhibited that SPDEF overexpression inhibited the expression of DOT1L by activating miR-448 and the inhibitory effect of oe-SPDEF on DOT1L was counteracted by miR-448 inhibitor (Fig. 3k, l). In short, SPDEF had the potential to activate miR-448 transcription and further inhibit DOT1L expression.
SPDEF downregulates DOT1L and further inhibits the self-renewal of HCC stem cells
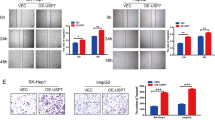
To further analyze the implications of SPDEF/miR-448/DOT1L axis on the self-renewal of HCC stem cells, oe-SPDEF, and oe-DOT1L vectors were constructed and transfected into SMMC-7721 and Huh-7 cells, respectively. The overexpression efficiency of SPDEF and DOT1L was determined by RT-qPCR and western blot analysis, which revealed that overexpression of SPDEF downregulated the expression of DOT1L (Fig. 4a, b, Supplementary Fig. 2A, B). Furthermore, the protein levels of HCC stem cell markers CD133, CD90, AMPK, p-AMPK, OCT-4, SOX-2, and Nanog were measured by western blot analysis to show the stemness. SPDEF overexpression was observed to downregulate the levels of OCT-4, SOX-2, and Nanog as well as the extent of AMPK phosphorylation, which was reversed following DOT1L overexpression (Fig. 4c, Supplementary Fig. 2C). In addition to those molecular levels, the results of sphere and colony formation assay consistently demonstrated that SPDEF overexpression weakened the sphere and colony formation potentials of HCC stem cells. Overexpression of DOT1L could promote the formation of sphere of HCC stem cells. Meanwhile, overexpression of SPDEF was found to reverse the promotion of DOT1L overexpression in maintaining colony formation potentials of HCC stem cells and stem cell markers (Fig. 4d, Supplementary Fig. 2D). Therefore, SPDEF inhibited DOT1L expression and thus impeded the self-renewal of HCC stem cells.
a The overexpression efficiency of SPDEF and DOT1L in SMMC-7721 stem cells determined by RT-qPCR. b The overexpression efficiency of SPDEF and DOT1L in SMMC-7721 stem cells determined by Western blot analysis. c Quantitation of levels of AMPK, OCT-4, SOX-2, and Nanog proteins by western blot assay in SMMC-7721 cells transfected with oe-SPDEF, oe-DOT1L or both. d SMMC-7721 stem cell self-renewal detected by microsphere formation and colony formation experiments following transfection with oe-SPDEF, oe-DOT1L, or both (scale bar = 25 μm). *p < 0.05 vs. SMMC-7721 cells transfected with oe-NC1 oe-NC2. Data among multiple groups were analyzed by one-way ANOVA and followed by Tukey’s test. Cell experiments were repeated three times independently.
Knockdown of CDK11B represses the oncogenic potential of HCC stem cells in vivo
As CDK11B could degrade SPDEF, and the inhibition of SPDEF elevated the expression of DOT1L through downregulation of miR-448, thus inducing enhanced self-renewal of HCC stem cells, we developed xenograft tumors in nude mice to further verify the effects of CDK11B and DOT1L on HCC stem cells. SMMC-7721 stem cells were stably transfected with sh-CDK11B and oe-DOT1L and injected into nude mice. The data obtained from RT-qPCR and western blot analysis suggested successful downregulation of CDK11B by sh-CDK11B and overexpression of DOT1L by oe-DOT1L in vivo. Correspondingly, SPDEF and miR-448 were elevated while DOT1L was reduced by downregulation of CDK11B (Fig. 5a, b). Besides, HE staining revealed that CDK11B silencing and DOT1L overexpression did not cause obvious pathological changes in the tumor tissues (Supplementary Fig. 3A). The results of immunochemical staining also confirmed the successful knockdown of CDK11B by sh-CDK11B and overexpression of DOT1L by oe-DOT1L in vivo (Supplementary Fig. 3A). Also, immunochemical staining revealed no changes in AMPK expression after CDK11B silencing or DOT1L overexpression. However, CDK11B silencing downregulated the levels of OCT-4, SOX-2, and Nanog proteins as well as the extent of AMPK phosphorylation, whereas DOT1L overexpression could restore the levels of those proteins and AMPK phosphorylation (Supplementary Fig. 3B). By measuring the weight and volume of the tumor, it was visible that CDK11B silencing resulted in an inhibitory effect on tumor formation, and DOT1L overexpression reversed the inhibitory effect of CDK11B silencing on tumor formation (Fig. 5c, d, Supplementary Fig. 3C). Taken together, CDK11B knockdown restrained the oncogenic and self-renewal potentials of HCC stem cells by mediating the SPDEF/miR-448/DOT1L axis.
a The expression of CDK11B, DOT1L, and SPDEF mRNAs and miR-448 in HCC stem cells transfected with sh-CDK11B, oe-DOT1L or both determined by RT-qPCR. b Representative western blots and quantitation of CDK11B, DOT1L, and SPDEF proteins in HCC stem cells transfected with sh-CDK11B, oe-DOT1L, or both. c The tumor weight of mice treated with sh-CDK11B, oe-DOT1L, or both. d The tumor volume of mice treated with sh-CDK11B, oe-DOT1L or both. *p < 0.05 vs. nude mice injected with HCC stem cells transfected with sh-NC or oe-NC. Data among multiple groups were analyzed by one-way ANOVA and followed by Tukey’s test. n = 15 mice in each group. Cell experiments were repeated three times independently.
Discussion
Increasing studies have demonstrated the crucial role of CDKs in the pathogenesis and progression of HCC [6, 18]. However, the functionality and downstream mechanism of CDK11B in HCC are still undefined. The current study mainly demonstrated that CDK11B promoted self-renewal of HCC stem cells and the tumorigenesis of HCC. CDK11B-induced phosphorylation and ubiquitination of SPDEF, and ultimately blocked the activation of miR-448 and its mediated inhibition of DOT1L.
This study revealed robust expression of CDK11B in HCC tissue and cell samples, whereas CDK11B silencing inhibited the self-renewal of HCC cells. Repression of CDKs has been confirmed as inhibitors of HCC cell clonogenic survival by arresting cell cycles and inducing cell apoptosis [19]. For instance, HCC tissues exhibited abnormally high expression and activity of CDK5, whereas ablation of CDK5 was able to suppress HCC cell proliferation and clonogenicity [20]. A more recent study also identified the increased expression of CDK8 in HCC tissues and its vital role as a prognostic factor for predicting the overall survival and disease-free survival of HCC patients [21]. More importantly, the abnormal expression of CDK11B was also demonstrated to be responsible for cell cycle arrest and cell proliferation suppression [22]. Hence, the downregulation of CDK11B in HCC cells was indicated to potentially play an inhibitory role in the HCC progression, which was substantiated by the in vivo data.
Subsequently, the current study that CDK11B could inhibit SPDEF expression by promoting its phosphorylation and ubiquitination, which was consistent with the previous study reported by Tamura and et al. [8]. SPDEF is a member of the ETS transcription factor family that has been proved as a key regulator of malignancies such as prostate cancer, breast cancer, and colorectal cancer [9, 23, 24]. Degradation of SPDEF was also suggested as an enhancer of cancer cell migration, invasion, and metastasis [8]. Moreover, the current study further demonstrated that SPDEF could decrease DOT1L expression by increasing the expression of miR-448, thereby inhibiting the self-renewal of HCC stem cells and HCC progression. miRNAs are a group of small non-coding RNAs that capable of regulating gene expression at multiple levels and their aberrant expression is responsible for cancer development [25]. miR-448 overexpression resulted in suppression of growth and migrating ability of non-small-cell lung cancer cells [26]. Meanwhile, miR-448 has also been evidenced to be a suppressor of colon cancer progression [27]. Besides, miR-448 overexpression is capable of weakening the biological functions of HCC cells, such as sphere formation, colony formation, cell migration, invasion, the stemness maintenance and self-renewal [13]. miRNAs can modulate gene expression post-transcriptionally by interacting with the 3′-UTR of specific target mRNAs [28]. In this study, the biological prediction website and luciferase reporter assay identified that miR-448 bound to the 3′-UTR of DOT1L mRNA and could negatively regulate its expression. DOT1L, also known as KMT4, is a histone methyltransferase implicated in the mediation of cell development, cell cycle progression and DNA damage repair [29, 30]. A previous research has demonstrated that DOT1L acts as a tumor-promotive gene in head and neck cancer by strengthening self-renewal of cancer stem cells, cancer cell invasion, and chemotherapy resistance [31]. In addition, DOT1L can mediate H3K79me2 modification of stem cell makers and its upregulation reverses the inhibiting effects of miR-133b on colorectal cancer stemness and chemoresistance [15]. Interestingly, our results suggested that CDK11B elevated DOT1L expression and downregulated miR-448 expression by inducing SPDEF phosphorylation and ubiquitination.
In summary, this study provided evidence demonstrating that CDK11B silencing could prevent HCC progression via attenuating the potential of HCC stem cells. After CDK11B knockdown, ubiquitination of SPDEF was weakened and hence SPDEF expression was restored, which consequently upregulated the expression of miR-448 and enhanced miR-448-targeted inhibition of DOT1L (Fig. 6). These findings may aid in the in-depth understanding of pathogenic mechanisms involving HCC stem cells. However, this study lacks clinical verification and hence we would like to pay more attention on the future clinical applications of gene target strategies. In spite of that, this study validated the tumor-suppressing effect of CDK11B downregulation, which may be a promising clinically viable target in HCC treatment.
References
Shimada S, Mogushi K, Akiyama Y, Furuyama T, Watanabe S, Ogura T, et al. Comprehensive molecular and immunological characterization of hepatocellular carcinoma. EBioMedicine 2019;40:457–70.
Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301–14.
Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology 2019;156:477–91. e1.
Santopaolo F, Lenci I, Milana M, Manzia TM, Baiocchi L. Liver transplantation for hepatocellular carcinoma: where do we stand? World J Gastroenterol. 2019;25:2591–602.
Liao Y, Feng Y, Shen J, Hornicek FJ, Duan Z. The roles and therapeutic potential of cyclin-dependent kinases (CDKs) in sarcoma. Cancer Metastasis Rev. 2016;35:151–63.
Shen S, Dean DC, Yu Z, Duan Z. Role of cyclin-dependent kinases (CDKs) in hepatocellular carcinoma: therapeutic potential of targeting the CDK signaling pathway. Hepatol Res. 2019;49:1097–108.
Alonso MH, Ausso S, Lopez-Doriga A, Cordero D, Guino E, Sole X, et al. Comprehensive analysis of copy number aberrations in microsatellite stable colon cancer in view of stromal component. Br J Cancer. 2017;117:421–31.
Tamura RE, Paccez JD, Duncan KC, Morale MG, Simabuco FM, Dillon S, et al. GADD45alpha and gamma interaction with CDK11p58 regulates SPDEF protein stability and SPDEF-mediated effects on cancer cell migration. Oncotarget 2016;7:13865–79.
Meiners J, Schulz K, Moller K, Hoflmayer D, Burdelski C, Hube-Magg C, et al. Upregulation of SPDEF is associated with poor prognosis in prostate cancer. Oncol Lett. 2019;18:5107–18.
Wu J, Qin W, Wang Y, Sadik A, Liu J, Wang Y, et al. SPDEF is overexpressed in gastric cancer and triggers cell proliferation by forming a positive regulation loop with FoxM1. J Cell Biochem. 2018;119:9042–54.
Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314.
Zhu H, Zhou X, Ma C, Chang H, Li H, Liu F, et al. Low expression of miR-448 induces EMT and promotes invasion by regulating ROCK2 in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36:487–98.
Guo JC, Yang YJ, Zhang JQ, Guo M, Xiang L, Yu SF, et al. microRNA-448 inhibits stemness maintenance and self-renewal of hepatocellular carcinoma stem cells through the MAGEA6-mediated AMPK signaling pathway. J Cell Physiol. 2019;234:23461–74.
Gibbons GS, Chakraborty A, Grigsby SM, Umeano AC, Liao C, Moukha-Chafiq O, et al. Identification of DOT1L inhibitors by structure-based virtual screening adapted from a nucleoside-focused library. Eur J Med Chem. 2020;189:112023.
Lv L, Li Q, Chen S, Zhang X, Tao X, Tang X, et al. miR-133b suppresses colorectal cancer cell stemness and chemoresistance by targeting methyltransferase DOT1L. Exp Cell Res. 2019;385:111597.
Reish NJ, Maltare A, McKeown AS, Laszczyk AM, Kraft TW, Gross AK, et al. The age-regulating protein klotho is vital to sustain retinal function. Invest Ophthalmol Vis Sci. 2013;54:6675–85.
Chen EB, Zhou SL, Pang XG, Yin D, Miao PZ, Yang Y, et al. Prostate-derived ETS factor improves prognosis and represses proliferation and invasion in hepatocellular carcinoma. Oncotarget 2017;8:52488–500.
Shao YY, Li YS, Hsu HW, Lin H, Wang HY, Wo RR, et al. Potent activity of composite cyclin dependent kinase inhibition against hepatocellular carcinoma. Cancers (Basel). 2019;11:1433.
Haider C, Grubinger M, Reznickova E, Weiss TS, Rotheneder H, Miklos W, et al. Novel inhibitors of cyclin-dependent kinases combat hepatocellular carcinoma without inducing chemoresistance. Mol Cancer Ther. 2013;12:1947–57.
Ehrlich SM, Liebl J, Ardelt MA, Lehr T, De Toni EN, Mayr D, et al. Targeting cyclin dependent kinase 5 in hepatocellular carcinoma–a novel therapeutic approach. J Hepatol. 2015;63:102–13.
Yin T, Liu MM, Jin RT, Kong J, Wang SH, Sun WB. miR-152-3p Modulates hepatic carcinogenesis by targeting cyclin-dependent kinase 8. Pathol Res Pract. 2019;215:152406.
Liu TH, Wu YF, Dong XL, Pan CX, Du GY, Yang JG, et al. Identification and characterization of the BmCyclin L1-BmCDK11A/B complex in relation to cell cycle regulation. Cell Cycle. 2017;16:861–8.
Xiao B, Kuang Z, Zhang W, Hang J, Chen L, Lei T, et al. Glutamate ionotropic receptor kainate type subunit 3 (GRIK3) promotes epithelial-mesenchymal transition in breast cancer cells by regulating SPDEF/CDH1 signaling. Mol Carcinog. 2019;58:1314–23.
Lo YH, Noah TK, Chen MS, Zou W, Borras E, Vilar E, et al. SPDEF induces quiescence of colorectal cancer cells by changing the transcriptional targets of beta-catenin. Gastroenterology 2017;153:205–18. e8.
Motti ML, S DA, Meccariello R. MicroRNAs, cancer and diet: facts and new exciting perspectives. Curr Mol Pharm. 2018;11:90–6.
Qi H, Wang H, Pang D. miR-448 promotes progression of non-small-cell lung cancer via targeting SIRT1. Exp Ther Med. 2019;18:1907–13.
Lou Q, Liu R, Yang X, Li W, Huang L, Wei L, et al. miR-448 targets IDO1 and regulates CD8(+) T cell response in human colon cancer. J Immunother Cancer. 2019;7:210.
Ivey KN, Srivastava D. microRNAs as developmental regulators. Cold Spring Harb Perspect Biol. 2015;7:a008144.
Yang L, Lei Q, Li L, Yang J, Dong Z, Cui H. Silencing or inhibition of H3K79 methyltransferase DOT1L induces cell cycle arrest by epigenetically modulating c-Myc expression in colorectal cancer. Clin Epigenetics. 2019;11:199.
Sarno F, Nebbioso A, Altucci L. DOT1L: a key target in normal chromatin remodelling and in mixed-lineage leukaemia treatment. Epigenetics. 2019;15:439–53.
Bourguignon LY, Wong G, Shiina M. Up-regulation of histone methyltransferase, DOT1L, by matrix hyaluronan promotes MicroRNA-10 expression leading to tumor cell invasion and chemoresistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem. 2016;291:10571–85.
Acknowledgements
We acknowledge and appreciate our colleagues for their valuable suggestions and technical assistance for this study. This work was supported by the Key Research and Development Plan of Hainan Province (2019RC373) and Hainan Natural Science Foundation (818MS161).
Author information
Authors and Affiliations
Contributions
Jun-cheng Guo and Yi-jun Yang designed the study. Min Guo, Jian-quan Zhang, and Jin-fang Zheng collated the data, carried out data analyses and produced the initial draft of the manuscript. Zhuo Liu contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Guo, Jc., Yang, Yj., Guo, M. et al. Involvement of CDK11B-mediated SPDEF ubiquitination and SPDEF-mediated microRNA-448 activation in the oncogenicity and self-renewal of hepatocellular carcinoma stem cells. Cancer Gene Ther 28, 1136–1149 (2021). https://doi.org/10.1038/s41417-020-00261-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-020-00261-w
- Springer Nature America, Inc.