Abstract
This study aims to explore the molecular regulation mechanism of ubiquitination-specific protease 7 (USP7) in facilitating the stemness properties of hepatocellular carcinoma (HCC). Gain-of-function and loss-of-function assays were conducted in SK-Hep1 and HepG2 cells transfected with USP7 overexpression/knockdown plasmids and USP7 inhibitor P22077. The proliferation, migration, invasion, and self-renewal capacity of hepatocellular carcinoma cells were detected by CCK-8, colony formation, Transwell, scratch, and tumor sphere formation, respectively. MS was performed to identify the potential substrate of USP7 following P22077 treatment. Co-IP assay was used to verify the interaction between USP7 and basic transcription factor 3 (BTF3) in HCC cells. The overexpression of USP7 could promote the proliferation, migration, invasion, and colony formation capacity of SK-Hep1 and HepG2 cells. Additionally, ectopic UPS7 enhanced the epithelial-mesenchymal transition (EMT) and stem-like characteristics of the HCC cells. In contrast, USP7 depletion by knockdown of USP7 or administrating inhibitor P22077 significantly inhibited these malignant phenotypes of SK-Hep1 and HepG2 cells. Following MS analysis, BTF3 was identified as a potential substrate for USP7. USP7 could interact with BTF3 and upregulate its protein level, while USP7 depletion significantly upregulated the ubiquitination levels. Overexpression of BTF3 partially rescue the inhibitory effects of USP7 depletion on the malignant phenotypes and stemness properties of SK-Hep1 and HepG2 cells. USP7 can promote the stemness and malignant phenotype of HCC by stabilizing BTF3.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is a common malignant tumor with a high fatality rate worldwide (Siegel et al. 2023). Currently, surgical resection and chemotherapy are common treatments for liver cancer. However, only 5–15% of patients at early stages can be benefit from surgery (Anwanwan et al. 2020). By decades, increasing therapeutic methods have been applied in clinic, including immunotherapy, transhepatic arterial chemotherapy and embolization (TACE), and targeted therapy. Due to the high heterogeneity, the efficacy of current therapeutic strategies is limited (Bruix et al. 2014). Therefore, it is of great importance to identifying novel targets and therapeutics for liver cancer.
Numerous studies have suggested cancer stem cells (CSCs) as the main culprits of recurrence and metastasis for liver cancer (Fang et al. 2022). With the potential of self-renewal and differentiation, CSCs play a decisive role in cancer recurrence and metastasis (Atashzar et al. 2020; Liu et al. 2020). In addition, CSCs facilitate drug resistance and metastasis characteristics, which may be the main reason leading to poor therapy efficacy and high recurrence rate of liver cancer (Najafi et al. 2019). Previous studies have shown that CSCs have abnormally expressed markers, including sex-determining regions Y-box 2 (SOX-2), SOX9, and octamer-bound transcription factor 4 (OCT-4) (Chen et al. 2016). Epithelial-mesenchymal transformation (EMT) is a hallmark of CSC, which is associated not only with cancer invasion and drug resistance, but also with stemness (Celia-Terrassa and Jolly 2020). Therefore, understanding the biological functions and molecular regulatory mechanisms of liver cancer CSCs can provide a robust therapeutic strategy for the treatment of liver cancer.
Ubiquitination is a common post-translation modification, including ubiquitination and deubiquitination. Deubiquitination modification enzymes include ubiquitin-specific protease (USP), ubiquitin-C-terminal hydrolase (UCH), ovarian tumor protease (OTU), Machado-Joseph domain protease (MJDs), and JAMM/MPN domain-related metal isopeptidase (JAMM) (Mevissen and Komander 2017). Of them, the USP family is a major member of DUBs, and various studies have suggested that the USP family has been involved in multiple malignancies. Ubiquitin-specific protease 7 (USP7) is a deubiquitination enzyme (DUB), which has been reported to play a promoting role in the tumor progression (Fan et al. 2013; Zhao et al. 2015). For instance, USP7 promoted proliferation and invasion of head and neck squamous cell cancer cells by removing the K48-connected ubiquitination chain of Hippo signal effector TAZ (Li et al. 2022). USP7 rendered resistance of pancreatic cancer cells to PARP inhibitors by deubiquitinating fructose-1, 6-diphosphatase 1 (Cheng et al. 2022). Our previous studies have found that FEN1-regulated USP7 could inactivate P53 pathway by stabilizing MDM2, thereby promoting the development of liver cancer (Bian et al. 2022). However, the role and regulatory mechanisms of USP7 in liver cancer have not been fully elucidated. In this study, we further investigated the role and underlying mechanisms of USP7 in maintaining the malignant phenotype and stemness of liver cancer cells, providing new targets for the treatment of liver cancer.
Materials and methods
Cell lines and cell culture
SK-Hep1 (catalog number, #ZQ0030) and HepG2 (#ZQ0022) were purchased from Zhong Qiao Xin Zhou Biotechnology (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, CA, USA) medium supplemented with 10% fetal bovine serum (FBS; Gibco, CA, USA) and penicillin/streptomycin at 37 °C under the condition of 5% CO2. For cell transfection, cells were incubated in the 6-well plate at the density of 5 × 103 cells/well. Subsequently, BTF3 overexpression plasmids (pCDNA3.1-FLAG-USP7; TransheepBio-Tech, Shanghai, China) or knockdown SgRNAs were transfected into cell lines by using Lipofectamine 3000 reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. The sequence of knockdown group was listed as follows: Kd-USP7#1, F-ccgGGGAATGTGGCCCTGAGTGA, R-aacTCACTCAGGGCCACATTCCC, and Kd-USP7#2, F-ccgGGTGTTGTGTCCATCACTCA, R-aacTGAGTGATGGACACAACACC.
Quantitative real-time PCR
Cells treated with RNA-easy Isolation Reagent extraction solution were collected. Total RNA of each group was extracted through TRIzol (Invitrogen, USA) according to the manufacturer’s instructions, then RNA concentration and quality were detected, and RNA was reverse-transcribed into cDNA using TaKaRa PrimeScript RT regent kit (TaKaRa, Japan). RT-qPCR was conducted by using The Fast SYBR Green Master Mix (Applied Biosystems Inc., MA, USA) according to the manufacturer’s instructions. The condition of PCR was set as follows: 30-s polymerase activation at 95 °C and 40 cycles at 95 °C for 5 s, followed by 60 °C for 30 s. The glyceraldehyde phosphate dehydrogenase (GAPDH) was used as a loading control and the relative expression level of target gene was calculated using the 2−ΔΔCt method.
CCK-8 assay
The cell proliferation were detected by Cell Counting Kit-8 assay (CCK-8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s protocols. The resuspension HCC cells (1 × 103 cells/well) treatment were cultured in 96-well plates according to the experimental requirements (overexpression and P22077) at 37 °C for 24 h, 48 h, and 72 h; 100 µL of CCK-8 reagent prepared by the substrate was added to each sample. Then the reaction was finished after 37 °C for 2-h culture. Finally, the absorbance of each sample at OD450 well were detected by a microplate reader (BioTeK, CA, USA). The assay was independently repeated for three times.
Colony formation assay
According to the experimental requirements, the treated cells were inoculated into a 6-well plate with 0.5 × 103 cells per well, and the plate was shaken to make the cells evenly distributed. After 14 days of culture at 37 °C, the cells were fixed with 4% paraformaldehyde solution for 45 min, and stained with crystal purple for 5 min. Finally, the staining solution was washed away and photographed for counting.
Cell scratch assay
According to the experimental requirements, the treated cells were inoculated into a 6-well plates and cultured in a 37 °C constant temperature incubator. When the growth and fusion rate of the cell reached 100%, a straight line was drawn with a 10-µL pipette head perpendicular to the plate, and the crossed cells were washed and added to serum-free medium for culture. Cell mobility was calculated at 0 h, 24 h, and 48 h. The relative distance (RD) of each group was calculated with the equation: RD = (D0 h − D24 h) ⁄ D0 h.
Transwell assay
Transwell assays were performed to test the effects of USP7 inhibitor P22077/overexpression on migration. First, 4 × 104 cells were pre-suspended in serum-free medium and plated in the Transwell inserts (Corning, CA, USA) for migration assay. Second, the complete culture medium was placed in the lower chamber. Following incubation for 24 h, after removing the upper layer of uninvaded cells, the cells were fixed by 4% paraformaldehyde and stained with crystal violet. After carefully rinsed with pure water, the migration cells were observed under a microscope in at least 3 fields for each sample, photographed, and counted.
Sphere formation assay
Treated suspension cells (1 × 103 cells/well) were inoculated into a 24-well ultra-low adhesion plate and cultured in stem cell medium (supplemented with B27 supplement, 27 ng/mL EGF, and 20 ng/mL bFGF serum-free RPMI1640 medium). After 7–10 days of cultivation in a 5% CO2 incubator at 37 °C, the tumor was observed under a microscope and photographed. Moreover, the size of the tumor was measured using ImageJ software and the number of sphere globules was counted.
Western blot
The collected cells were treated with RIPA lysis solution, and total proteins were extracted from the cells according to the operating requirements instructions of the kit. During the experiment, the corresponding amount of target protein was added into the sample hole of SDS-PAGE gel. After SDS-PAGE gel electrophoresis, the isolated protein was electrically transferred to the cellulose nitrate membrane (PVDF membrane) by the rotating mold device. Then the protein-loaded PVDF membrane was immersed in 5% skimmed milk powder and sealed for 2 h at room temperature. The appropriate concentration of primary antibody was added and incubated overnight at 4 °C. On the second day, the antibody was rinsed with TBS for three times and incubated with the secondary antibody for 2 h at room temperature. Finally, the bands were detected by enhanced chemiluminescence (ECL) kit (Millipore, CA, USA), with GAPDH as the internal reference.
Co-immunoprecipitation
The cells in a good growth state were added with appropriate amount of lysate. Then the cells were scraped off and continued cracking strictly on the ice for 20 min. The protein lysate was sucked out and added to the pre-cooled 1.5-mL EP tube. The appropriate proportion of Protein A + G magnetic beads was aspirated and added to the lysate, and then placed on the vertical rotary instrument for 3 h at 4 °C. Then, the rotated cracking solution was centrifuged at 1200 rpm for 5 min, and the supernatant was absorbed and added to a new EP tube. Meanwhile, 50 µL of the supernatant was taken as the input group control and temporarily stored at − 20 °C. As a result, the remaining supernatant was evenly divided into two parts, and appropriate proportions of target molecular antibodies and IgG antibodies were added, respectively. After overnight rotation at 4 °C, Protein A + G magnetic beads were added to the cracking solution again, and then rotated at 4 °C for 2.5 h. After being placed in the centrifuge at 1200 rpm again for 5 min, the supernatant was aspirated and washed twice with 1 mL of cracking solution. Finally, 5 × loading buffer was added and the supernatant of input group was removed. The corresponding loading buffer was added, mixed well and boiled in water bath for 10 min, and then stored at − 80 °C for subsequent experiments.
Ubiquitination and immunofluorescence assay
For the ubiquitination assays, P22077 (S7133, Selleck Chem, USA, concentration, 10 µM, 20 μM) treated SK-Hep1 cells for 24 h. Eight hours before collecting, 20 µM of MG132 was added for the suppression of proteasome-dependent protein degradation. Then, cells were collected and lysed for IP and IB assays. The quantitative analysis was performed by ImageJ based on the relative protein expression normalized to GAPDH. For the immunofluorescence assay, cells were fixed and permeabilized by 4% formaldehyde and 0.25% Triton X-100, then blocked with 1% BSA for 1 h at room temperature. After 12 h of incubation with the primary antibodies diluted with 1% BSA, cells were washed and probed with secondary antibodies (ABclonal Technology) and DAPI (CST), following by fluorescence microscope.
Statistical analysis
In this study, all data were collated and analyzed using IBM SPSS19.0, GraphPad Prism 7.0 (CA, USA) and R software. All data were derived from at least three repeated experiments. The comparative analysis between the two groups of data was conducted by Student’s test. Comparisons between multiple sets of data were analyzed using ANOVA. P value less than 0.05 was considered statistically significant.
Result
USP7 promotes the malignant phenotypes of HCC cells
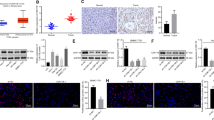
In order to investigate the effect of USP7 on the malignant phenotype of hepatocellular carcinoma cells, we transfected USP7 overexpression plasmid (OE-USP7) into SK-Hep1 and HepG2 cell lines to construct gain-of-function assays. Subsequently, CCK-8, clone formation, scratch, and Transwell tests were used to detect the proliferation, migration, and invasion ability of hepatoma cells. The results showed that the activities of HCC cells in OE-USP7 group were significantly enhanced in contrast to the control group (Fig. 1A–E). Western blot results showed that overexpression of USP7 significantly increased the protein expressions of N-cadherin and Vimentin, with the downregulated levels of E-cadherin (Fig. 1F). These data indicated that USP7 promoted the malignant phenotypes and EMT process of HCC cells.
USP7 enhanced the malignant phenotype of HCC cells. A–E The malignant phenotypes of SK-Hep1 and HepG2 cells were detected by CCK-8, clone formation, scratch, and Transwell assays, respectively. F The protein levels of EMT-related markers were detected by Western blot, including N-cadherin, E-cadherin, and Vimentin. *P < 0.05, **P < 0.01
Low expression of USP7 reduces the aggressive behaviors of HCC cells
To further demonstrate the effects of USP7 on the malignant phenotype of hepatocellular carcinoma cells, SK-Hep1 and HepG2 cells were treated with USP7 inhibitor P22077 at gradient concentrations of 0 μM, 10 μM, and 20 μM, as well as genetic depletion of USP7. As shown in Fig. 2A–D, pharmacological or genetic depletion of USP7 could inhibit the proliferation, migration, and invasion capacity of SK-Hep1 and HepG2 cells. Consistent with the observation above, EMT process was also hindered by P22077 treatment or knockdown of USP7 in SK-Hep1 and HepG2 cells, characterized by downregulation of N-cadherin/Vimentin and upregulation of E-cadherin (Fig. 2E). These data also suggested that pharmacological or genetic depletion of USP7 could inhibit the EMT process and aggressive behaviors of HCC cells.
The USP7 inhibitor P22077 reduces the malignant phenotype of HCC cells. A–D The effects of USP7 knockdown and USP7 inhibitor P22077 on HCC cells were detected by CCK-8, colony formation, scratch, and Transwell experiments, respectively. E EMT-related markers were detected by Western blot in SK-Hep1 and HepG2 cell lines. *P < 0.05, **P < 0.01, and ***P < 0.001
USP7 upregulates the stem-like properties of HCC cells
Subsequently, the effects of abnormal USP7 expression on hepatocellular carcinoma cell stem cell properties were analyzed. The tumor sphere formation assay showed that the stem cell-like properties of HCC cells were significantly enhanced with USP7 overexpression. In contrast, with the administration of USP7 inhibitor P22077, the self-renewal capacity of SK-Hep1 and HepG2 cells was dramatically reduced (Fig. 3A and B). Western blot data showed that USP7 overexpression promoted the expression of OCT4, SOX2, and SOX9 proteins, while knockdown of USP7 or P22077 administration inhibited the expression of OCT4, SOX2, and SOX9 proteins (Fig. 3C and D).
USP7 upregulates stem-like properties of HCC cells. A and B Tumor sphere formation assay was conducted in SK-Hep1 and HepG2 cells with USP7 overexpression or P22077. C Western blot assay was performed to detect the expression of stemness-related markers in SK-Hep1 and HepG2 cells. D Western blot assay was performed to detect the expression of stemness-related markers following knockdown of USP7 or P22077 treatment in SK-Hep1 and HepG2 cells. *P < 0.05, **P < 0.01, and ***P < 0.001
BTF3 is a potential substrate for USP7
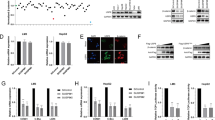
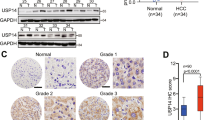
USP7 is a deubiquitination enzyme that commonly plays crucial roles by stabilizing substrates. To further investigate the potential mechanisms, mass spectrometry was performed to identify the differentially expressed proteins with statistical significance (fold change > 1 and P < 0.05) following P22077 administration. Of them, BTF3, as a stemness/EMT-related protein correlated with HCC phenotypes, was chosen for further validations (Fig. 4A). Co-immunoprecipitation (Co-IP) assay verified the interaction between USP7 and BTF3 in SK-Hep1 cells (Fig. 4B). Consistently, co-localization of the two proteins could be observed by immunofluorescence (Fig. 4C). P22077 treatment significantly enhanced the ubiquitination level of BTF3 in HCC cells (Fig. 4D). In addition, overexpression of USP7 significantly upregulated the expression of BTF3 protein, while knockdown group and P22077 inhibitor group significantly inhibited the level of BTF3 protein (Fig. 4E and F). However, quantitative real-time PCR (qRT-PCR) results showed that there was no significant difference of BTF3 mRNA levels in SK-Hep1 and HepG2 cells with USP7 overexpression/knockdown or P22077 treatment (Fig. 4G and H). Subsequently, the protease inhibitor MG132 could rescue the BTF3 expression downregulated by USP7 knockdown group or P22077 administration in SK-Hep1 and HepG2 cells, and this phenomenon was reversed (Fig. 4I). The results above indicated that BTF3 might be a potential substrate of USP7 in HCC cells.
Identifying BTF3 as a potential substrate of USP7. A Mass spectrometry analysis identified BTF3 as a potential substrate of USP7. B Co-IP assay was conducted to validate the interaction between BTF3 and USP7. C The ubiquitination level of BTF3 was evaluated in HCC cells treated by P22077. D The co-localization of USP7 and BTF3 was detected by immunofluorescence in SK-Hep1 and HepG2 cells. E Western blot assay was performed to detect expression at protein level after overexpression USP7 in SK-Hep1 and HepG2 cells. F Western blot assay was performed to detect USP7 and BTF3 expression at protein level following knockdown of USP7 or P22077 treatment. G and H qRT-PCR was performed to detect BTF3 expression at mRNA levels in SK-Hep1 and HepG2 cells. I Western blot assay was conducted to investigate the effects of MG132 on BTF3 expression with P22077 treatment or USP7 knockdown. *P < 0.05, **P < 0.01, and ***P < 0.001; NS, no significance
USP7 upregulates the malignant phenotype and stemness of HCC cells via BTF3
To further analyze the role of USP7-BTF3 axis in regulating malignant phenotypes of HCC cells, we conducted rescue assays in SK-Hep1 and HepG2 cell lines. Compared with the control group, P22077 significantly inhibited the proliferation, migration, invasion, and EMT of SK-Hep1 and HepG2 cells, while BTF3 overexpression restored the aggressive behaviors of HCC cells (Fig. 5A–C). In addition, BTF3 significantly rescued the self-renewal ability of HCC cells, which was impaired by P22077 (Fig. 5D). Consistently, western blot assay showed that overexpression of BTF3 restored the expression of EMT markers and stemness-related markers (Fig. 5E and F). It suggests that USP7 facilitated HCC phenotypes by stabilizing BTF3 protein (Fig. 6).
USP7 upregulates the malignant phenotype and stemness of HCC cells via BTF3. A–C CCK-8, Transwell, and scratch assays were performed to evaluated the phenotypes of HCCs. D The tumor sphere formation assay was conducted to test the self-renewal capacity of HCC cells. E and F Expression levels of EMT-related proteins and stemness markers in SK-Hep1 and HepG2 cells were detected by western blot. *P < 0.05, **P < 0.01, and ***P < 0.001
Discussion
Liver cancer is one of the leading causes of death worldwide. Although great progress has been made in the treatment of liver cancer, the effective approaches remain limited due to the heterogeneity of liver cancer. As a result, relapse and metastasis are still the difficulties in the treatment of liver cancer. In recent years, the CSC has been recognized as playing an important role in chemical resistance, recurrence, and metastasis of various cancers. Therefore, identifying novel signaling pathways that regulate cancer cell stemness may enhance the efficacy of cancer therapy. In this study, we found that USP7 could promote the proliferation, migration, invasion, and EMT of liver cancer cells; increase the self-renewal ability of liver cancer cells; and promote the expression of stem markers like OCT4, SOX2, and SOX9. The observations above suggest that USP7 might play an important role in facilitating the stemness of liver cancer cells.
Some studies have shown that liver cancer originates from liver stem cells in adult liver tissue. Moreover, these cells in the liver with stem cell properties are often referred to as liver cancer stem cells. Current treatments for liver cancer include intravenous chemotherapy, arterial embolization, surgical resection, local radiotherapy, and radiofrequency ablation. Although the therapeutics shown positive efficiency for improving the prognosis of HCC patients, the overall outcome remains dismal due to the failure in eliminating liver cancer stem cells (Walcher et al. 2020). CSCs are defined by several markers that may represent potentially important therapeutic targets that may have important functions for CSCs, making them more attractive as therapeutic targets. OCT4 is one of the stem cell factors necessary for embryogenesis and pluripotency. Also, OCT4 is highly expressed in CSCs of various cancers. Although most CSC-related studies have reported that OCT4 expression is associated with chemotherapy resistance and clinical prognosis (Mohiuddin et al. 2020), increasing evidence suggest that OCT4 and SOX2 are core factors in the network of pluripotent genes involved in the induction, maintenance, and loss of pluripotent genes (Li and Belmonte 2017). Overexpression of SOX9 increased the self-renewal ability of glioma cells and induces the formation of more tumors (Garros-Regulez et al. 2016). The results of this study confirmed that USP7 significantly enhanced the expression of OCT4, SOX2, and SOX9, as well as promoted the self-renewal ability of HCC cells. In contrast, its inhibitor P22077 or knockdown of USP7 significantly inhibited these phenotypes. It is supposed that P22077 is a robust targeted therapy agent. As is known, P22077 could also inhibit the activity of USP47, which plays an essential role in liver regeneration. However, the association between USP47 and HCC remain unclear. Additionally, another important property of CSCs is their high capacity of invasive and metastasis, which are implicated in the EMT processes. During EMT, epithelial cells transdifferentiate and lose their transverse connections with maintaining mesenchymal phenotypes (Grunert et al. 2003). Therefore, these cells play an important role in the invasive and migrate processes of cancer cells (Dongre and Weinberg 2019). In this study, USP7 could play a promoting role in the process of EMT, characterized as upregulated protein levels of N-cadherin/Vimentin, and downregulated E-cadherin expression. Studies have reported that EMT and CSCs are the main factors boosting cancer cell metastasis, and the proteins or signaling pathways involved in regulating these two processes are the promising therapeutic targets for the treatment of metastasis (Babaei et al. 2021).
Protein ubiquitination regulates a variety of cellular biological processes, such as protein degradation, transcriptional activation or inhibition, and immune signaling transduction pathways, by affecting the stability, localization, and function of substrate proteins (Nakamura 2018). Deubiquitinases (DUBs) are divided into five subfamilies. In addition, USP family is the most abundant DUBs in all subfamilies. An increasing number of studies have found that USP is involved in the occurrence and development of cancer. For example, USP2 is characterized as an oncogene by stabilizing various oncoproteins (Zhao et al. 2018). USP10 and USP13 can guide the ubiquitination of S-phase kinase-associated protein 2 (SKP2) and participate in the regulation of cancer development (Chen et al. 2011; Liao et al. 2019). Besides, Ching et al. have found that USP7 can stabilize E1B55K protein expression by interacting with E1B55K at N-terminal domain (Ching et al. 2013). USP7 is a DUB that can remove ubiquitin from the substrate, protect the substrate from degradation, and then release free ubiquitin for cycloubiquitination reactions (Cheng et al. 2019). Moreover, overexpression of USP7 accelerates HCC cell growth by forming a complex with thyroid hormone receptor interacting protein 12 (TRIP12) and stabilizing its induction of constitutive p14 (ARF) ubiquitination (Cai et al. 2015). Previous studies found that USP7 was highly expressed in HCC tissues, and that high levels of USP7 can enhance the growth of HCC cells in vitro and in vivo. In this study, we further investigated the key substrate for the enhanced stemness futures induced by USP7. Following the analysis of mass, BTF3 was identified as a candidate substrate of USP7. Overexpression of USP7 increased the protein level of BTF3 with no obvious alterations at the mRNA level. However, protease inhibitor MG132 could partially restore the inhibitory effects of P22077 or USP7 knockdown on the protein level of BTF3, as well as the aggressive behaviors of HCC cells. Taken above, USP7 can stabilize the expression of BTF3 protein through deubiquitination, and contribute to malignant phenotype, EMT, and self-renewal of HCC.
Although the current evidence is promising, this study has certain limitations. The effects of USP7 on the growth and metastasis of liver cancer were observed in vitro, while future studies should clarify the underlying roles in multiple models. In addition, the exact regulatory axis for USP7-BTF3 still needs further relevant molecular assays to verify and elucidate. Furthermore, the stem cells and EMT of cancer cells involve multiple signals, such as TGF-β signaling, WNT signaling, Notch signaling, and other signaling pathways. It may provide novel targets for chemotherapy and even immunotherapy (Barbato et al. 2019; Patra et al. 2023). Further studies should also focus on the effects of USP7 on the multiple pathway networks that synergistically facilitate HCC progression.
In summary, this study suggests that USP7 advance malignant phenotypes of HCC cells by stabilizing BTF3 protein level, providing potential targets for HCC therapy.
Data availability
No datasets were generated or analysed during the current study.
References
Anwanwan D, Singh SK, Singh S, Saikam V, Singh R (2020) Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer 1873(1):188314
Atashzar MR, Baharlou R, Karami J, Abdollahi H, Rezaei R, Pourramezan F et al (2020) Cancer stem cells: a review from origin to therapeutic implications. J Cell Physiol 235(2):790–803
Babaei G, Aziz SG, Jaghi NZZ (2021) EMT, cancer stem cells and autophagy; the three main axes of metastasis. Biomed Pharmacother 133:110909
Barbato L, Bocchetti M, Di Biase A, Regad T (2019) Cancer stem cells and targeting strategies. Cells 8(8):926
Bian S, Ni W, Zhu M, Zhang X, Qiang Y, Zhang J et al (2022) Flap endonuclease 1 facilitated hepatocellular carcinoma progression by enhancing USP7/MDM2-mediated P53 inactivation. Int J Biol Sci 18(3):1022–1038
Bruix J, Gores GJ, Mazzaferro V (2014) Hepatocellular carcinoma: clinical frontiers and perspectives. Gut 63(5):844–855
Cai JB, Shi GM, Dong ZR, Ke AW, Ma HH, Gao Q et al (2015) Ubiquitin-specific protease 7 accelerates p14(ARF) degradation by deubiquitinating thyroid hormone receptor-interacting protein 12 and promotes hepatocellular carcinoma progression. Hepatology 61(5):1603–1614
Celia-Terrassa T, Jolly MK (2020) Cancer stem cells and epithelial-to-mesenchymal transition in cancer metastasis. Cold Spring Harb Perspect Med 10(7)
Chen M, Gutierrez GJ, Ronai ZA (2011) Ubiquitin-recognition protein Ufd1 couples the endoplasmic reticulum (ER) stress response to cell cycle control. Proc Natl Acad Sci USA 108(22):9119–9124
Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF (2016) LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun 7:12598
Cheng J, Guo J, North BJ, Wang B, Cui CP, Li H et al (2019) Functional analysis of deubiquitylating enzymes in tumorigenesis and development. Biochim Biophys Acta Rev Cancer 1872(2):188312
Cheng X, Zhang B, Guo F, Wu H, Jin X (2022) Deubiquitination of FBP1 by USP7 blocks FBP1-DNMT1 interaction and decreases the sensitivity of pancreatic cancer cells to PARP inhibitors. Mol Oncol 16(7):1591–1607
Ching W, Koyuncu E, Singh S, Arbelo-Roman C, Hartl B, Kremmer E et al (2013) A ubiquitin-specific protease possesses a decisive role for adenovirus replication and oncogene-mediated transformation. PLoS Pathog 9(3):e1003273
Dongre A, Weinberg RA (2019) New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 20(2):69–84
Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH et al (2013) USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis 4(10):e867
Fang X, Yan Q, Liu S, Guan XY (2022) Cancer stem cells in hepatocellular carcinoma: intrinsic and extrinsic molecular mechanisms in stemness regulation. Int J Mol Sci 23(20):12327
Garros-Regulez L, Aldaz P, Arrizabalaga O, Moncho-Amor V, Carrasco-Garcia E, Manterola L et al (2016) mTOR inhibition decreases SOX2-SOX9 mediated glioma stem cell activity and temozolomide resistance. Expert Opin Ther Targets 20(4):393–405
Grunert S, Jechlinger M, Beug H (2003) Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol 4(8):657–665
Li M, Belmonte JC (2017) Ground rules of the pluripotency gene regulatory network. Nat Rev Genet 18(3):180–191
Li J, Dai Y, Ge H, Guo S, Zhang W, Wang Y et al (2022) The deubiquitinase USP7 promotes HNSCC progression via deubiquitinating and stabilizing TAZ. Cell Death Dis 13(8):677
Liao Y, Liu N, Xia X, Guo Z, Li Y, Jiang L et al (2019) USP10 modulates the SKP2/Bcr-Abl axis via stabilizing SKP2 in chronic myeloid leukemia. Cell Discov 5:24
Liu YC, Yeh CT, Lin KH (2020) Cancer stem cell functions in hepatocellular carcinoma and comprehensive therapeutic strategies. Cells 9(6):1331
Mevissen TET, Komander D (2017) Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem 86:159–192
Mohiuddin IS, Wei SJ, Kang MH (2020) Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim Biophys Acta Mol Basis Dis 1866(4):165432
Najafi M, Farhood B, Mortezaee K (2019) Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol 234(6):8381–8395
Nakamura N (2018) Ubiquitin system. Int J Mol Sci 19(4):1080
Patra T, Cunningham DM, Meyer K, Toth K, Ray RB, Heczey A et al (2023) Targeting Lin28 axis enhances glypican-3-CAR T cell efficacy against hepatic tumor initiating cell population. Mol Ther 31(3):715–728
Siegel RL, Miller KD, Wagle NS, Jemal A (2023) Cancer statistics, 2023. CA Cancer J Clin 73(1):17–48
Walcher L, Kistenmacher AK, Suo H, Kitte R, Dluczek S, Strauss A et al (2020) Cancer stem cells—origins and biomarkers: perspectives for targeted personalized therapies. Front Immunol 11:1280
Zhao GY, Lin ZW, Lu CL, Gu J, Yuan YF, Xu FK et al (2015) USP7 overexpression predicts a poor prognosis in lung squamous cell carcinoma and large cell carcinoma. Tumour Biol 36(3):1721–1729
Zhao Y, Wang X, Wang Q, Deng Y, Li K, Zhang M et al (2018) USP2a supports metastasis by tuning TGF-beta signaling. Cell Rep 22(9):2442–2454
Funding
This study was supported by grants from National Natural Science Foundation (82272839, 81702419) and Postdoctoral Science Foundation of Jiangsu Province (2021K243B).
Author information
Authors and Affiliations
Contributions
W. Z., Z. T., and C. D. designed and supervised this study. M. H., Y. C., C. D., and S. B. conducted the experiments and drafted the manuscript. N. X., X. S., Z. L., and S. X. analyzed data and performed the statistics. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mingchao Hu, Chengchen Dai, and Xieyin Sun contributed equally to this study.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, M., Dai, C., Sun, X. et al. Ubiquitination-specific protease 7 enhances stemness of hepatocellular carcinoma by stabilizing basic transcription factor 3. Funct Integr Genomics 24, 28 (2024). https://doi.org/10.1007/s10142-024-01310-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-024-01310-5