Abstract
Non-small-cell lung cancer (NSCLC) remains the leading cause of cancer-related death worldwide. Accumulating research has highlighted the ability of exosome-encapsulated microRNAs (miRNAs or miRs) as potential circulating biomarkers for lung cancer. The current study aimed to evaluate the clinical significance of serum-derived exosomal miR-let-7e as a biomarker in the metastasis of NSCLC. Initially, the expression of miR-let-7e, SUV39H2, and CDH1 in human NSCLC tissues and exosomes isolated from the serum of NSCLC patients was determined by RT-qPCR, demonstrating that miR-let-7e was downregulated in NSCLC tissues and serum-derived exosomes, while SUV39H2 was upregulated in NSCLC tissues. Kaplan–Meier method revealed that both lower miR-let-7e expression and higher SUV39H2 expression were correlated with a lower survival rate of NSCLC patients. Next, SUV39H2 was predicted and validated to be a target of miR-let-7e using dual-luciferase reporter assay. NSCLC H1299 cells following ectopic expression and depletion experiments of miR-let-7e and SUV39H2 were treated with serum-derived exosomes, after which the viability, migration, and invasion of H1299 cells were detected using CCK-8 and Transwell assays. Further, in vivo experiments were conducted to elucidate the effect of exosomal miR-let-7e on tumorigenesis. Results revealed that miR-let-7e overexpression in serum-derived exosomes inhibited SUV39H2, resulting in impaired cell viability, migration, and invasion in vitro as well as delayed tumor growth in vivo. In conclusion, the key findings of the current study demonstrate that exosomal miR-let-7e from serum possesses anticarcinogenic properties against NSCLC via the SUV39H2/LSD1/CDH1 axis.
Similar content being viewed by others
Introduction
As one of the foremost types of cancer, lung cancer has a particularly high mortality in China, with non-small-cell lung cancer (NSCLC) accounting for the majority of lung cancer cases [1,2,3]. Current therapeutic approaches aimed at tackling NSCLC are mainly composed of systemic therapy, radiotherapy, chemotherapy, and immunotherapy [4, 5]. Furthermore, it has been proposed that exosome-encapsulated microRNAs (miRNAs or miRs) can serve as predictive biomarkers for lung cancer malignancy, owing to their ability to transfer biological information to recipient cells [6]. Thus, it is of great important to adequately elucidate the mechanism by which exosomal-miRs inhibit the viability and invasion of NSCLC cells.
Exosomes have emerged as significant cell–cell communication entities both in physiological and pathological processes by serving as carriers of a wide range of biomolecules, including lipids, proteins, DNAs, messenger RNAs, and miRNAs [7]. Thus, they have the potential to be utilized as disease/cancer-specific biomarkers [8, 9]. In addition, the diagnostic potential of miR-let-7e has been explored with studies indicating its prognostic significance in NSCLC as an independent prognostic factor for patients succumbed to NSCLC [10]. The depletion of miR-let-7e expression has been shown to be strongly correlated with poor overall survival rates in patients with squamous-cell lung carcinomas and lung adenocarcinomas [11]. During the current study, we explored the StarBase website (http://starbase.sysu.edu.cn/index.php) to identify a binding site between miR-let-7e and the 3′ untranslated region (3′UTR) of suppressor of variegation 3–9 homolog 2 (SUV39H2). SUV39H2 has been reported to exhibit dysregulated expression in lung adenocarcinoma and has been identified as an oncogene in lung adenocarcinoma involved in the mediation of tumorigenesis and metastasis [12]. A recent study has reported the potential of SUV39H2 to methylate and stabilize lysine specific demethylase 1 (LSD1), a histone lysine demethylase that is often highly expressed in various types of human cancers, by inhibiting the level of LSD1 polyubiquitination in human cancer cells [13]. LSD1 is a tumor-promoting factor for NSCLC whereby its overexpression has been linked with unfavorable prognosis in NSCLC patients and stimulates proliferation, migration, and invasion of tumor cells [14]. The inhibition of LSD1 has been reported to induce the loss of cell–cell adhesion gene cadherin 1 (CDH1) expression potentially via upregulation of TRIM37 in luminal breast cancer cells [15]. Lung adenocarcinoma cells in the presence of overexpressed CDH1 or reduced CDH1 methylation present with delayed tumorigenicity, which is manifested by reduced tumor size and weight in vivo [16]. Thus, we reasoned that serum exosome containing miR-let-7e could play a functional role in the progression of NSCLC via the SUV39H2/LSD1/CDH1 axis.
Materials and methods
Ethics statement
The current study was performed with the approval of the Ethics Committee of the First Hospital of Qinhuangdao. Informed written consent was obtained from each participant prior to the study. All experiments in the present study were conducted in strict accordance with the Declaration of Helsinki. All animal experiments were performed according to the protocol approved by the Institutional Animal Care and Use Committee of the First Hospital of Qinhuangdao. Extensive efforts were made to ensure minimal animal suffering and usage during the study.
Study subjects
A total of 105 primary NSCLC-patients who had received treatment at the First Hospital of Qinhuangdao since 2009 as well as 50 healthy donors were randomly selected and enrolled into our study. Peripheral blood samples were collected and used for subsequent exosome extraction. Detailed clinicopathological characteristics, including smoking history, alcohol consumption and tumor stage, were collected by medical interview. Primary NSCLC and normal lung tissues (more than 5 cm away from the margin of primary NSCLC tissues) were harvested by surgical resection or biopsy, and then promptly frozen in liquid nitrogen in the same pathology laboratory. Patient survival was analyzed by the Kaplan–Meier method using an online survival analysis tool Kaplan–Meier plotter (www.kmplot.com.). The clinical stages of the NSCLC patients are listed in Supplementary Table S1.
Isolation and labeling of serum exosomes
All qualified blood samples were coagulated at room temperature and centrifuged at 3000 g for a period of 10 min. The serum was then centrifuged at 3000 g, 4 °C for 10 min and at 10,000 g, 4 °C for 30 min to remove cell debris. The particles were then precipitated by 1 mL supernatant ultracentrifugation with a Beckman 50.4 Ti Rotor (Beckman Coulter, Brea, CA, USA) at 100,000 g and 4 °C for 2 h. The particles were then washed with phosphate-buffered saline (PBS), filtered through a 0.22-μm filter, and ultra-centrifuged again at 100,000 g and 4 °C for 2 h to precipitate the exosomes, prior to resuspension of the particles in 100 μL PBS and quantification by bicinchonininc acid (BCA) protein assay. A total of 10 μg exosomes were used in each in vitro experiment. For in vivo growth and metastasis assays, 1 μg/g exosomes were used three times a week. Afterwards, fluorescent dye Dil (Sigma-Aldrich Chemical Company, St Louis, MO, USA) was used to label exosomes. Briefly, the exosomes were incubated with Dil (1: 2000) for 2 h and then washed with PBS. Finally, receptor cell endocytosis was observed under a confocal fluorescence microscopy (LSM 7DUO, Zeiss, Jena, Germany).
Isolation of exosomes
Fetal bovine serum (FBS) was ultra-centrifuged at 100,000 g and 4 °C for 16 h to discard the exosomes. The FBS supernatant was collected and filtered through a 0.22-μm filter (Millipore, Billerica, MA, USA). The H1299 and A549 cells (CRL-5803, American Type Culture Collection) were subsequently transfected with miR-let-7e mimic and mimic-negative control (NC) were cultured in medium supplemented with exosome-free FBS. After 72 h of incubation, the culture medium was collected and centrifuged at 300 g for 10 min to remove the floating cells and cell debris. Next, the supernatant was centrifuged at 2000 g for 15 min, followed by centrifugation at 12,000 g for 30 min. Afterwards, the collected supernatant was filtered through a 0.22-μm filter (Millipore, Billerica, MA, USA) and subsequently filtered using a membrane filter for 1 h by centrifugation at 4000 g and 4 °C using ultrafiltration device (UFC900396, Millipore, Billerica, MA, USA). Finally, using an EXO Quick-TC™ isolation kit (EXOTC50A-1, System Biosciences, Mountain View, CA, USA), the sample was added to the concentrated solution of the supernatant at a ratio of 1:5, with the exosomes subsequently extracted.
Transmission electron microscopy (TEM)
The samples were fixed with 2% glutaraldehyde and loaded on Formvar carbon at room temperature. The samples were then negatively stained using 1% uranyl acetate at 4 °C for 3 min, and dried under an incandescent lamp for 10 min. Images were taken at 120 kV using a JEM-1400 TEM (JEOL, Tokyo, Japan).
Nanoparticle tracking analysis of exosomes
The exosomes were resuspended in PBS, adjusted to a concentration of 5 mg/ml, and diluted 100–500 times prior to the manual transfer of the sample into the chamber at room temperature. The size distribution of the exosomes was measured via tunable resistive pulse sensing with qNano Gold system (Izon Science Ltd, Christchurch, New Zealand) that combined tunable nanopores with proprietary data acquisition and analysis software Izon Control Suite version 3.3.2.2001 (Izon Science).
Determination of exosome protein concentration
The protein concentration of exosomes was determined using a Micro-BCA analysis kit (Thermo Fisher Scientific, Rockford, IL, USA). A fluorescent microplate reader (Molecular Devices, Sunnyvale, CA, USA) was used to estimate the total protein of the exosomes.
Cell culture
Human NSCLC H1299 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS, 2 mM L-glutamic acid, 50 U/mL penicillin and 50 μg/mL streptomycin (Sigma-Aldrich Chemical Company, St Louis, MO, USA).
Cell counting kit-8 (CCK-8) assay
The number of living cells was determined using trypan blue exclusion test. The CCK-8 assay was performed to evaluate cell viability. Briefly, H1299 and A549 cells were seeded in 96-well plates and cultured for 12, 24, 48, and 72 h. Next, CCK-8 solution (Dojindo Laboratories, Kumamoto, Japan) was added to each well and cultured for 1 h. The absorbance of each well at 450 nm was measured using a 96-well spectrophotometer with 30 mg of exosomes used in each well.
Transwell assay for cell migration and invasion
The uncoated and 20 μL matrix gel (0.5 mg/mL; BD Biosciences, MA, USA)-coated Transwell system (Corning, USA) were used to assess the migration and invasion ability of the H1299 and A549 cells. Briefly, 100 μL serum-free medium containing 2 × 104 cells was seeded into the apical chamber, while 500 μL medium containing 10% FBS was added to the basolateral chamber. After 50 mg of exosomes were incubated with the cells for 24 h, the Transwell chamber was washed twice with PBS with the cells that failed to migrate or invade were wiped away using cottons. The cells that successfully migrated or invaded were fixed with 1% formaldehyde for 15 min, washed twice with PBS and stained with 0.1% crystal violet for 15 min, followed by observation with a DMi3000B inverted microscope (Leica, Germany).
RNA isolation and quantitation
The total RNA was extracted using TRIzol reagent (Invitrogen Inc., Carlsbad, CA, USA) as per the manufacturer’s instructions. The extracted RNA was then reverse transcribed using the PrimeScript RT reagent kit (Promega Corporation, Madison, WI, USA). Quantitative analysis of gene expression was performed based on the instructions of the SYBR Green master mix (Life Technologies, Carlsbad, CA, USA). Next, the extracted RNA was reverse transcribed into complementary DNA (cDNA) using the Mir-XTM miRNA first strand synthesis kit (Takara, Dalian, China) for miRNA detection. Quantification of the expression of miR-let-7e was performed using a Mir-XTM miRNA TB Green qPCR Kit (Takara). The primer sequences are depicted in Table 1. With glyceraldehyde phosphate dehydrogenase (GAPDH; genes) and U6 (miRNA) serving as internal references, the relative expression of genes or miRNA was calculated based on relative quantification (the 2−ΔΔCt method). Prior to isolation of the exosomes, 25 fmol of C. elegans (cel)-miR-39 (Guangzhou RiboBio Co., Ltd., Guangdong, China) was added to normalize the corresponding expression in exosomes.
Western blot analysis
Both tissues and cells were ground and lysed with pre-cooled lysis buffer containing phenylmethylsulphonyl fluoride (PMSF) and phosphatase inhibitor in a pre-cooled glass grinder. Nuclear proteins were extracted using a Nuclear Extract Kit (Active Motif, Carlsbad, CA, USA). The protein was subsequently separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membranes were then blocked using 5% bovine serum albumin for 1 h and incubated at 4 °C overnight with the following specific primary rabbit antibodies: CD9 (1: 2000, ab92726), CD63 (1:1000, ab134045), CD81 (1:1000, ab109201), β-tubulin (1:500, ab6046), GM130 (1:1000, ab52649), SUV39H2 (1:1000, ab190870), H3 (1:1000, ab176842), Ubiquitin (1:1000, ab223613), LSD1 (1:1000, ab17721), CDH1 (1:10,000, ab181860), and GAPDH (1:10,000, ab181602). All the aforementioned antibodies were obtained from Abcam Inc. (Cambridge, UK). On the following day, the membranes were re-probed with secondary goat anti-rabbit IgG-H&L (horseradish peroxidase, HRP) (1:5000, ab205718, Abcam Inc., Cambridge, UK) at room temperature for 1 h. The bands on the membrane were visualized using enhanced chemiluminescence reagent. Finally, the data were analyzed with a Molecular Imager (Gel DocTM XR, 170–8170) and the associated software Quantity One-4.6.5 (Bio-Rad Laboratories, USA).
Oligonucleotide transfection
Small interfering RNA (si)-NC, si-SUV39H2, miR-let-7e mimic and mimic-NC were synthesized by Kangcheng Biotechnology Company (Guangzhou, China) and oligonucleotide transfection was performed using the Lipofectamine 2000 reagent (Invitrogen Inc., Carlsbad, CA, USA). Meanwhile, si-NC comprised of three different oligonucleotide duplexes served as the control siRNA (Cosmo Bio Co., Tokyo, Japan). During transfection, the final concentration of siRNA was 50 nM, while the concentration of miR-let-7e mimic and mimic-NC was 50 nM, and that of NC-vector (negative control plasmid for pLV-IRES-PURO) and SUV39H2-vector (SUV39H2 overexpression plasmid; The plasmid of pLV-IRES-PURO inserted into the SUV39H2 ORF box; both were obtained from Guangzhou RiboBio Co., Ltd., Guangdong, China) and used to transfect the H1299 cells. Lipofectamine 2000 (Invitrogen Inc., Carlsbad, CA, USA) was used to introduce mimic into exosomes as per the manufacturer′s instructions. Briefly, the purified exosomes were incubated with the mixture of Lipofectamine 2000 and mimic at 37 °C for 2 h, with the transfected exosomes precipitated by ultracentrifugation.
Construction of NSCLC mouse model in vivo
Thirty-two BALB/C-nu/nu nude mice (aged 4 weeks old and weighing 16–22 g, Beijing Huafukang Bioscience Company, Beijing, China) were randomly grouped with eight mice per group. Next, 2 × 106 H1299 cells were injected subcutaneously into the flanks of each nude mouse. After 8 days, when the tumor size was confirmed to have reached ~100 mm3, the purified exosomes, mimic-NC-containing exosomes (mimi-NC exo), miR-let-7e mimic-containing exosomes (miR-let-7e mimic exo) or PBS were injected to the tumor site (three times a week, 1 μg/g each time). After 20 days, the growth of the tumors was monitored and recorded every 2 days. The tumor volume was calculated according to the following formula: 0.5 × tumor length × tumor width2. As per the guidelines of animal care agency, the experiment was terminated when one of the following events transpired: (1) the size of the tumor exceeded 2 cm in any dimension, or (2) the development of the tumor caused other complications. After reaching the experimental endpoint, the mice were euthanized and the tumors were collected for analysis and further research.
Immunofluorescence assay
Next, the slides were dried, dehydrated, rehydrated, and blocked. The slides were then boiled and incubated with rabbit antibody to E-cadherin (1:200, Abcam Inc., Cambridge, UK) and mouse antibody to vimentin (1:200, Abcam Inc., Cambridge, UK) for 2 h, followed by incubation for 1 h with Alexa Fluor 488 goat anti-mouse IgG (1:800, Cell Signaling Technology, Beverly, MA, USA) and Alexa Fluor 594 goat anti-rabbit IgG (1:800, Cell Signaling Technology, Beverly, MA, USA).
Immunoprecipitation assay
The transfected HEK293T cells were washed with PBS and lysed in radio-immunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% Nonidet-P40, 0.1 mM PMSF) containing complete protease inhibitor (Roche Applied Science). The cell lysates were incubated at 4 °C with anti-FLAG M2 agarose (Sigma-Aldrich Chemical Company, St Louis, MO, USA) for 2 h. After washing the beads three times with 1 mL of TBS buffer (pH 7.6), the boiled Lane-labeled sample buffer (Thermo Fisher Scientific Inc., Waltham, MA, USA) was employed to elute the protein binding to the beads. The samples were subsequently subjected to SDS-PAGE and Western blot analysis. The antibodies used in the experiment were as follows: LSD1 (ab17721, Abcam Inc., Cambridge, UK), HDAC1 (SC7872, Santa Cruz Biotechnology, Santa Cruz, CA, USA), Ubiquitin (SC8017, Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Dual-luciferase reporter assay
The 3′UTR of SUV39H2 wild-type (WT) containing the seed sequence of miR-let-7e binding site was amplified. The seed sequence of miR-let-7e binding site was mutated as the 3′UTR of SUV39H2 mutant (MUT). Next, 100 μL HEK293T cells (GDC187, Wuhan Cell Bank, Wuhan, China) were seeded into a 96-well plate at a density of 1.5 × 104 cells/well, and cultured with 5% CO2 at 37 °C for 24 h. The SUV39H2-WT-3′UTR and SUV39H2-MUT-3′UTR were cloned into separate pGLO vectors, namely pGLO-SUV39H2-WT and pGLO-SUV39H2-MUT. The two aforementioned reporter plasmids were co-transfected with miR-let-7e mimic or mimic-NC (Shanghai Sangon Biotechnology Co. Ltd., Shanghai, China) to 293T cells, respectively. After 24 h of transfection, the supernatant was collected for detection. Dual-Luciferase® Reporter Assay System (E1910, Promega Corporation, Madison, WI, USA) was then used to detect the luciferase activity, and the relative luciferase activity was expressed as the ratio of Renilla luciferase activity to Firefly luciferase activity.
Chromatin immunoprecipitation (ChIP) assay
A ChIP kit (ab500, Abcam Inc., Cambridge, UK) was used for cell lysis in a cell lysis buffer as per the provided protocol to produce DNA fragments. Non-specific IgG (1:200, Sigma-Aldrich Chemical Company, St Louis, MO, USA), rabbit antibody against CDH1 (1:40, ab181860, Abcam Inc., Cambridge, UK) and rabbit antibody against H3K4me3 (ab8580, 2 µg was used for 25 µg of chromatin, Abcam Inc., Cambridge, UK) were subsequently incubated with the lysate to precipitate the target area. Next, isolated DNA was subjected to RT-qPCR analysis. Ten percent of the total DNA of the extract was applied as the input. Besides, the primers to detect the CDH1 promoter sequence were as follows: forward: 5′-AGTCCCACAACAGCATAGGG-3′ and reverse: 5′-TTCTGAACTCAGGCGATCCT-3′.
Statistical analysis
All data were analyzed using SPSS 21.0 statistical software (IBM Corp. Armonk, NY, USA). The measurement data were expressed as mean ± standard deviation of at least three independent tests. Data obeying normal distribution and homogeneity of variance between two groups were compared using paired t-test (paired data) or unpaired t-test (unpaired data). Differences among multiple groups were analyzed by one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test. Data at different time points were compared by repetitive measurement ANOVA, followed by Bonferroni’s post hoc test. The Kaplan–Meier method was applied to calculate patient survival rate, while a Log-rank test was used for univariate analysis. A value of p < 0.05 was considered to be indicative of statistical significance.
Results
Bioinformatics analysis predicting the DEGs and their molecular interactions in NSCLC
Based on the microarray GSE63805 dataset (comprised of 30 normal samples and 32 NSCLC samples) retrieved in Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds) and differential analysis using R language, miR-let-7e was found to exhibit notably poor expression in NSCLC (Fig. 1a). After that, miRWalk (http://mirwalk.umm.uni-heidelberg.de; binding > 0.9, energy < −19, accessibility < 0.001), StarBase (http://starbase.sysu.edu.cn/index.php; pancancerNum > 3), miDIP (http://ophid.utoronto.ca/mirDIP/; Integrated Score > 0.3) and DIANA TOOLS (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index; miTG score > 0.5) were applied to predict the downstream target genes of miR-let-7e, with 4418, 1453, 2029 and 2952 target genes obtained, respectively. Next, the NSCLC-related GSE33532 dataset (comprised of 20 normal samples and 80 NSCLC samples) was analyzed using R language, which revealed 710 differentially expressed genes (Fig. 1b). Moreover, 700 differentially expressed genes with the smallest p values were retrieved in the TCGA database by the online tool GEPIA (http://gepia.cancer-pku.cn). Next, a Venn map was plotted to identify the intersection of the aforementioned downstream target genes and DEGs. One gene was identified within the intersection namely, SUV39H2 (Fig. 1c). Based on the exploration of the StarBase database, a binding site was identified between SUV39H2 and miR-let-7e (Fig. 1d). The GSE33532 dataset further revealed that SUV39H2 was a markedly upregulated gene in NSCLC (Fig. 1e). Accumulating evidence has indicated that SUV39H2 is capable of stabilizing LSD1 and promoting its expression through DNA methylase. Furthermore, LSD1 has been shown to inhibit CDH1 expression by removing H3K4me3 in the CDH1 promoter region and act as DNA demethylase. And, both LSD1 and CDH1 are related to NSCLC [13, 17,18,19,20]. Therefore, we hypothesized that the miR-let-7e/SUV39H2 /LSD1/CDH1 axis might play a regulatory role in the development of NSCLC.
a Boxplot of let-7e expression in the GSE63805 dataset. The left blue box indicates expression of normal samples, while the right red box indicates the expression of NSCLC samples (p = 1.587e-29). b Expression volcano map of the differentially expressed genes in the GSE33532 dataset. The red dots represent the significantly upregulated differentially expressed genes, while the green dots indicate downregulated genes. c Venn plots of the intersected genes obtained from the miRWalk, StarBase, miDIP, and DIANA TOOLS, the GSE33532 dataset and TCGA database. d Binding sites between miR-let-7e and SUV39H2 predicted by the StarBase database. e The expression of SUV39H2 in the GSE33532 dataset. The left blue box indicates the expression of normal samples, and the right red box indicates the expression of NSCLC sample (p = 3.432e-78).
Serum exosomes from NSCLC patients promotes cell viability, migration, and invasion
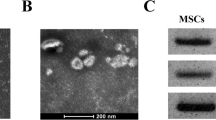
Serum exosomes were isolated from NSCLC patients and healthy donors, followed by identification. Initially, typical exosome-like round capsules were observed under TEM (Fig. 2a), with the diameter of these capsules found to be ~80 nm based on the qNano analysis (Fig. 2b). In addition, high levels of exosome surface protein markers CD9, CD81, and CD63 were detected, while β-tubulin and GM130 (used as NC) could not be detected (Fig. 2c). BCA protein analysis revealed that the concentration of total protein of exosomes in NSCLC patients was 0.361 mg/mL, which was markedly higher than that of the healthy donors (0.149 mg/mL) (Fig. 2d).
a Morphalogical observation of the purified exosomes from the serum of NSCLC patients (n = 105) and healthy donors (n = 50) by TEM (5000×). b qNano analysis of exosome size distribution in the serum of NSCLC patients and healthy donors. c Western blot analysis of CD9, CD81, CD63, β-tubulin and GM130. d The concentration of total protein exosomes in the serum of NSCLC patients and healthy donors. *p < 0.05. e The exosomes were resuspended in PBS to a concentration of 5 mg/ml, and diluted 100 times for immunofluorescence assay. The endocytic effect of the recipient cells on the exosomes observed by a confocal microscope (400×). f The viability of recipient cells detected using CCK-8 assay at 12, 24, 48, or 72 h with 30 mg of exosomes used in each well. g The number of migrated cells detected using Transwell assay after 50 mg of exosomes were incubated with cells (200×). h The number of invaded cells measured using Transwell assay after 50 mg of exosomes were incubated with cells (200×). *p < 0.05 vs. the serum exosomes from healthy donors or healthy donors. Data (mean ± standard deviation) between two groups were compared using unpaired t-test and data among multiple groups at different time points were analyzed using repeated measures ANOVA with Bonferroni multiple comparisons test. The experiment was repeated three times independently.
Next, to identify whether the serum exosomes of NSCLC patients affect the biological behavior of the recipient cells, the endocytic effect of the recipient cells on the exosomes was evaluated using a confocal microscope. As depicted in Fig. 2e, following a 2 h period of incubation, the Dil-labeled exosomes (red) were endocytosed by the H1299 cells. Subsequently, CCK-8, Transwell migration and Transwell invasion assay were employed to measure the effects of serum exosomes from NSCLC patients on the carcinogenic properties of H1299 cells, which suggested that compared with the exosomes in the serum of healthy donors, the exosomes in the serum of NSCLC patients significantly increased the viability, migration, and invasion of H1299 cells (Fig. 2f–h). Altogether, the serum exosomes of NSCLC patients exhibited the potential to promote NSCLC cell viability, migration, and invasion.
Downregulation of miR-let-7e is associated with the low survival rate and inhibits the development of NSCLC in vitro
Initially, the RT-qPCR results validated that let-7e expression in NSCLC tissues was significantly lower than that in the normal tissues (p < 0.05, Fig. 3a). Next, Kaplan–Meier analysis revealed that NSCLC patients with low miR-let-7e expression had a lower survival rate (Fig. 3b). At the same time, RT-qPCR was employed to detect the miR-let-7e expression in serum exosomes of the healthy donors as well as the NSCLC patients. As expected, the results displayed a lower miR-let-7e expression in NSCLC patients’ serum exosomes in comparison to healthy donors (Fig. 3c). Moreover, Pearson’s correlation analysis demonstrated that miR-let-7e expression in serum exosomes was positively correlated with that in NSCLC tissues (Fig. 3d).
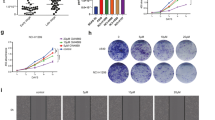
a Expression of miR-let-7e in NSCLC and normal tissues detected by RT-qPCR. N = 105. b Kaplan–Meier analysis of survival rate of patients with high or low miR-let-7e expression. N = 105. c Expression of miR-let-7e in the serum exosomes of healthy donors (n = 50) and NSCLC patients (n = 105) detected by RT-qPCR. d Pearson’s correlation coefficient of miR-let-7e expression in serum exosomes with that in NSCLC tissues. N = 105. e Expression of miR-let-7e in the exosomes detected by RT-qPCR. f Expression of miR-let-7e in H1299 cells detected by RT-qPCR. g The H1299 cell viability measured using CCK-8 assay at 12, 24, 48, or 72 h with 30 mg of exosomes used in each well. h Migrated H1299 cell number measured using Transwell assay after 50 mg of exosomes were incubated with cells (200×). i Invaded H1299 cell number measured using Transwell assay after 50 mg of exosomes were incubated with cells (200×). *p < 0.05 vs. normal tissues, exosome treatment, mimic-NC-treated exosomes or PBS-treated H1299 cells. #p < 0.05 vs. H1299 cells treated with mimic-NC exo. Data (mean ± standard deviation) between two groups were compared using unpaired t-test and data among multiple groups were analyzed using one-way ANOVA with Tukey’s multiple comparisons test. Data among multiple groups at different time points were analyzed using repeated measures ANOVA with Bonferroni multiple comparisons test. The experiment was repeated three times independently.
Next, to further verify the correlation between the promoting effect of serum exosomes on NSCLC cell migration and invasion and the low miR-let-7e expression, miR-let-7e mimic was transferred to H1299 cells to achieve miR-let-7e upregulation in exosomes. RT-qPCR results clarified that miR-let-7e expression in the exosomes was elevated following treatment with miR-let-7e mimic (Fig. 3e). The H1299 and A549 cells were then incubated with the transfected exosomes, followed by detection of miR-let-7e expression in the cells via RT-qPCR. The results revealed that the expression of miR-let-7e was slightly upregulated in exosome-treated H1299 cells and A549 cells when compared to H1299 cells and A549 cells treated with PBS. However, compared with mimic-NC exo, miR-let-7e mimic exo treatment resulted in higher miR-let-7e expression in H1299 cells and A549 cells (Fig. 3f, Supplementary Fig. S1a). Subsequent results demonstrated an enhancement in H1299 and A549 cell viability, migration, and invasion following exosome treatment. In addition, H1299 and A549 cells treated exosomes and mimic-NC exosomes exhibited no significant changes in the viability, migration, and invasion. However, miR-let-7e mimic exo led to significantly reduced H1299 and A549 cell viability, migration, and invasion (Fig. 3g–i, Supplementary Fig. S1b–d). Thus, low expression of miR-let-7e was associated with the low survival rate of NSCLC patients, and miR-let-7e could mediate exosomes to inhibit the cell viability, migration, and invasion of A549 and H1299 cells.
High expression of SUV39H2/LSD1 is associated with the low survival rate of NSCLC patients
Next, the expression of SUV39H2 in NSCLC tissues was evaluated using RT-qPCR and Western blot analysis, the results of which revealed that the expression of SUV39H2 in NSCLC tissues was evidently higher than that in normal tissues (Fig. 4a, b). The Kaplan–Meier survival curve illustrated that the survival rate of patients with high levels of SUV39H2 expression was poor (Fig. 4c). Moreover, the Starbase database revealed the presence of binding sites between the 3′UTR of SUV39H2 and miR-let-7e (Fig. 4d). In addition, miR-let-7e expression was inversely related to the mRNA expression of SUV39H2 in NSCLC tissues (Fig. 4e). Furthermore, dual-luciferase reporter assay results indicated that miR-let-7e mimic reduced the luciferase activity of SUV39H2 3′UTR in HEK293T cells, providing evidence of a targeting relationship between miR-let-7e and SUV39H2 (Fig. 4f). Next, RT-qPCR results demonstrated that transfection with miR-let-7e mimic caused upregulated miR-let-7e expression but downregulated SUV39H2 mRNA expression in H1299 cells (Fig. 4g). Furthermore, the protein expression of SUV39H2 was inhibited in the miR-let-7e mimic-transfected H1299 cells (Fig. 4h). Existing literature has documented the ability of SUV39H2 to stabilize LSD1 and promote its expression through the function of DNA methylase [13]. Besides, LSD1 induces the occurrence of NSCLC [18]. Therefore, LSD1 expression was subsequently detected, the result of which indicated that the expression of LSD1 was amplified in the NSCLC tissues (Fig. 4i, j). Pearson’s correlation analysis also demonstrated the positive correlation between SUV39H2 and LSD1 (Fig. 4k). In addition, once the expression of SUV39H2 was decreased, the level of polyubiquitination of LSD1 in H1299 cells increased (Fig. 4l). As reflected by Fig. 4, Kaplan–Meier survival curves illustrated that NSCLC patients with high expression of LSD1 exhibited low survival rates (p < 0.0001). These findings indicated that high expression of SUV39H2/LSD1 was correlated to diminished survival rate of NSCLC patients.
a SUV39H2 mRNA expression in NSCLC and normal tissues detected by RT-qPCR. N = 105. b, Western blot analysis of SUV39H2 protein in NSCLC and normal tissues. N = 105. c Kaplan-Meier analysis of survival rate of patients with high or low expression of SUV39H2. N = 105. d Predicted binding sites between SUV39H2 and miR-let-7e by the Starbase database. e Correlation analysis of SUV39H2 and miR-let-7e expression in NSCLC tissues. f Binding of miR-let-7e to SUV39H2 confirmed by dual-luciferase reporter assay in HEK293T cells. g mRNA expression of SUV39H2 in H1299 cells detected by RT-qPCR. h Western blot analysis of SUV39H2 protein in H1299 cells. i mRNA expression of LSD1 in NSCLC and normal tissues detected by RT-qPCR. j Western blot analysis of LSD1 protein in NSCLC and normal tissues. k Correlation analysis of SUV39H2 and LSD1 expression in NSCLC tissues. The protein expression of SUV39H2 and LSD1 in cancer tissues was significantly positively correlated. l The level of polyubiquitination of LSD1 in si-SUV39H2-transfected H1299 cells treated with 5 μM MG115 or 10 μM MG132. The Kaplan–Meier analysis of survival rate of patients with high or low expression of LSD1. N = 105. *p < 0.05 vs. normal tissues or cells transfected with mimic-NC. #p < 0.05 vs. H1299 cells treated with mimic-NC exosomes. Data (mean ± standard deviation) between two groups were compared using unpaired t-test. The experiment was repeated three times independently.
LSD1 suppresses CDH1 expression by regulating CDH1 promoter demethylation
E-cadherin protein encoded by CDH1 gene has been reported to inhibit the migration and invasion abilities of NSCLC cells [21]. In order to determine whether LSD1 promotes the development of NSCLC by regulating the demethylation of H3K4me3 in the promoter region of CDH1, the expression of CDH1 in the clinical tissues were detected using RT-qPCR and Western blot analysis. As depicted in Fig. 5a, b, CDH1 expression in NSCLC tissues was evidently lower than that in normal tissues. In addition, the mRNA level of CDH1 was negatively associated with the protein expression of LSD1 in NSCLC tissues (Fig. 5c). Moreover, Kaplan–Meier analysis revealed that NSCLC patients with low CDH1 expression exhibited lower survival rate (Fig. 5d).
a CDH1 expression in NSCLC and normal tissues detected using RT-qPCR. N = 105. b CDH1 expression in NSCLC and normal tissues tested through Western blot analysis. N = 105. c Correlation analysis of CDH1 mRNA expression and LSD1 protein expression in NSCLC tissues. N = 105. d Kaplan–Meier analysis of the survival rate of NSCLC patients with low or high expression of CDH1. e mRNA expression of LSD1 and CDH1 in H1299 cells detected using RT-qPCR. f LSD1, CDH1 and H3K4me3 proteins in H1299 cells analyzed through Western blot analysis. g The enrichment of LSD1 and H3K4me3 in the CDH1 promoter region analyzed by ChIP. *p < 0.05 vs. normal tissues or si-NC-transfected H1299 cells. Data (mean ± standard deviation) between two groups were analyzed by unpaired t-test. The experiment was repeated three times independently.
Next, to further investigate the regulatory role of LSD1 in CDH1 expression, RT-qPCR was initially employed to identify the mRNA expression of CDH1 in H1299 cells following different treatment. The results (Fig. 5e) showed that LSD1 silencing downregulated the mRNA expression of LSD1 but upregulated that of CDH1. Western blot analysis also demonstrated that the si-LSD1 exhibited reducing effects on the protein expression of LSD1, while promoting the expression of CDH1 and H3K4me3 (Fig. 5f). ChIP assay further verified that the enrichment of LSD1 in the CDH1 promoter region was decreased while that of H3K4me3 was increased in the absence of LSD1 (Fig. 5g). The aforementioned data demonstrated that LSD1 could inhibit the expression of CDH1 through regulation of the CDH1 promoter demethylation.
Exosomal miR-let-7e hampers the viability, migration, and invasion of NSCLC cells by mediating the SUV39H2/LSD1/CDH1 axis
RT-qPCR and Western blot analysis were conducted to detect the expression of miR-let-7e, SUV39H2, LSD1, and CDH1. As illustrated in Fig. 6a, b, compared with the transfection with both mimic-NC exo and NC-vector, H1299 cells treated with miR-let-7e mimic exo + NC-vector exhibited elevated miR-let-7e expression, elevated CDH1 expression, but reduced expression of SUV39H2 and LSD1. In comparison to mimic-NC exo + NC-vector, treatment with mimic-NC exo + SUV39H2-vector resulted in notable difference in relation to the mRNA levels of miR-let-7e and LSD1, while an increase was observed in the mRNA and protein expression of SUV39H2, and the protein expression of LSD1, however the mRNA and protein expression of CDH1 declined. Moreover, similar trends were observed in the cells transfected with miR-let-7e mimic exo + SUV39H2-vector relative to the treatment with miR-let-7e mimic exo + NC-vector. At the same time, MTT and Transwell assays were conducted, which revealed that the cell viability, migration, and invasion abilities were attenuated in response to miR-let-7e mimic exo + NC-vector treatment, but promoted after the treatment with mimic-NC exo + SUV39H2-vector. Besides, when compared with miR-let-7e mimic exo + NC-vector, co-transfection with miR-let-7e mimic exo + SUV39H2-vector enhanced NSCLC cell viability, migration, and invasion (Fig. 6c–e). Altogether, exosomal miR-let-7e could inhibit NSCLC cell viability, migration, and invasion by mediating the SUV39H2/LSD1/CDH1 axis.
a mRNA expression of miR-let-7e, SUV39H2, LSD1 and CDH1 in H1299 cells detected using RT-qPCR. b the protein levels of miR-let-7e, SUV39H2, LSD1, and CDH1 proteins in H1299 cells evaluated by Western blot analysis of. c Cell viability measured using CCK-8 assay. d The number of migrated cells measured using Transwell assay (200×). e The number of invaded cells measured using Transwell assay (200 ×). *p < 0.05 vs. cells treated with mimic-NC exo + NC-vector. #p < 0.05 vs. cells treated with miR-let-7e mimic exo + NC-vector. Data (mean ± standard deviation) among multiple groups were analyzed using one-way ANOVA with Tukey’s multiple comparisons test. The experiment was repeated three times independently.
MiR-let-7e in the exosomes prevents tumor growth and metastasis in vivo
A xenograft model was successfully constructed to examine the effects of serum exosomal miR-let-7e on tumor growth and metastasis. The results showed that the mice injected with exosome-treated H1299 cells exhibited increased tumor weight and volume, while an opposite trend was observed following treatment with miR-let-7e mimic exo (Fig. 7a–c). Next, Immunofluorescence staining revealed that the expression of E-cadherin was reduced while that of Vimentin was elevated in the tumor of mice injected with exosome-treated H1299 cells. In contrast, when compared with the treatment with mimic-NC exo, the injection with let-7e mimic exo led to increased E-cadherin but reduced Vimentin (Fig. 7d). Further, Western blot analysis revealed that the protein expression of Vimentin and N-cadherin was upregulated while that of E-cadherin was reduced in the presence of exosomes. miR-let-7e mimic exo led to decreased protein expression of Vimentin and N-cadherin yet increased E-cadherin protein expression (Fig. 7e). Altogether, the aforementioned data supported the notion that miR-let-7e-mediated exosomes inhibited tumor growth and metastasis in vivo.
a Tumor size after the treatment with mimic-NC exo or miR-let-7e mimic exo. b Tumor weight after the treatment with mimic-NC exo or miR-let-7e mimic exo. c Tumor volume after the treatment with mimic-NC exo or miR-let-7e mimic exo. d Immunofluorescence staining of E-cadherin and Vimentin expression in tumor tissues (400×). e Western blot analysis of E-cadherin, Vimentin and N-cadherin in tumor tissues. *p < 0.05 vs. PBS-treated mice. #p < 0.05 vs. mimic-NC-exo-treated mice. Data (mean ± standard deviation) between two groups were analyzed by unpaired t-test, while data among multiple groups at different time points were analyzed using repeated measurement ANOVA, followed by Bonferroni’s multiple post hoc test. The experiment was repeated three times independently. N = 8.
Discussion
NSCLC represents the leading cause of cancer-associated mortality worldwide, with the overall cure and survival rates largely unsatisfactory despite major breakthroughs in the treatment strategies [22]. Exosomes continue to be an area of research interest owing to their ability to carry miRNAs with studies indicating their potential as prospective diagnostic markers for lung cancer [23]. The current study investigated the role of miR-let-7e from the serum-derived exosome in NSCLC and demonstrated that miR-let-7e from the serum-derived exosome could regulate the cell viability, migration, and invasion of NSCLC both in vitro and in vivo via the SUV39H2/LSD1/CDH1 axis.
Our initial observations revealed that the serum exosomes from NSCLC patients had the potential to promote NSCLC cell viability, migration, and invasion. Accumulating data have discussed exosomes as excellent biomarkers for disease monitoring and prognosis in NSCLC patients [24]. High level of exosomes has been detected among NSCLC patients and has been suggested to induce the development of NSCLC [25]. Consistent with the findings of the current study, tumor-derived exosomes from the plasma of NSCLC patients have been previously reported to be capable of promoting proliferation, invasion and metastasis abilities of NSCLC cells in vitro [26]. In the present study, miR-let-7e was found to be downregulated in NSCLC tissues and NSCLC patients with poor miR-let-7e expression had low survival rates. For example, the expression of miR-let-7e has been previously documented to be decreased in squamous-cell lung carcinomas and lung adenocarcinomas clinical tissues [11]. Existing literature has provided verification indicating that the low levels of miR-let-7e are associated with a shorter overall survival rate of NSCLC patients [10]. miR-let-7e in the serum is related to the dedifferentiation of NSCLC, which is consistent with previous reports [11, 27]. However, our study used more clinical samples to verify this, and systematically analyzed the relationship between the miR-let-7e expression and pathological parameters of NSCLC patients. Our results also suggested that miR-let-7e encapsulated in serum exosomes could inhibit the cell viability, migration, and invasion of H1299 and A549 cells. Various studies have widely investigated the role of miR-let-7e in human cancers. Consistent with our findings, the elevation of miR-let-7e impairs the cell migration and invasion of papillary thyroid carcinoma cells [28]. A prior study concluded that lncRNA FEZF1-AS1 suppressed cell epithelial–mesenchymal transition process by increased E-cadherin but declined Vimentin levels in NSCLC cells [29], which was consistent with our results in vivo whereby the treatment of miR-let-7e exo induced upregulated E-cadherin but reduced Vimentin expression.
Our results further revealed that miR-let-7e targeted SUV39H2 and miR-let-7e elevation consequently suppressed its expression. The downregulation of SUV39H2 has been reported to elevate the level of polyubiquitination of LSD1 in H1299 cells [30]. SUV39H2 acts as an oncogene in lung adenocarcinoma and its expression is upregulated in NSCLC tissues, which was mainly associated with a shorter overall survival of NSCLC patients [12]. A recent study emphasized the potential of SUV39H2 to methylate and stabilize LSD1 via disturbing the level of LSD1 polyubiquitination in human cancer cells [13]. Moreover, human NSCLC A549 and H460 cells have been reported to present with amplified LSD1 expression [18]. LSD1 has been reported to be overexpressed in human NSCLC tissues, which was in line with our detection results [19]. Furthermore, the overexpression of LSD1 has been shown to play a contributory role in the promotion of NSCLC cell proliferation and migration, thus exerting an oncogenic effect on NSCLC [18]. Therefore, high expression of SUV39H2/LSD1 is associated with the unfavorable survival rate of NSCLC patients.
Moreover, our results identified that LSD1 suppressed CDH1 expression by regulating CDH1 promoter demethylation. In agreement with our results, LSD1 has been reported to decreases H3K4 in the promoter region of CDH1 and subsequently downregulates CDH1 expression, consequently inducing colon cancer metastasis [17]. In patients with NSCLC, expression of CDH1 is diminished and its downregulation has been shown to potentiate cancer cell viability, migration, and invasion abilities [21]. The potential correlation between miR-let-7e and LSD1 and CDH1 still remains notably enigmatic. However, in the present study, we identified that NSCLC cells treated with miR-let-7e mimic exo exhibited lower LSD1 mRNA and protein expression while higher CDH1 mRNA and protein expression [6]. In our study, exosome-shuttled miR-let-7e was found to attenuate NSCLC cell viability, migration, and invasion in vitro in addition to hindering cell growth and metastasis in vivo via mediation of the SUV39H2/LSD1/CDH1 axis.
Conclusion
Overall, the key findings of our study provide evidence demonstrating that serum exosome-encapsulated miR-let-7e could potentially inhibit the progression of NSCLC by regulating the SUV39H2/LSD1/CDH1 signaling axis (Fig. 8), suggesting that miR-let-7e may be promising candidate in the development of highly sensitive biomarker for NSCLC diagnosis. Further investigation into the specific regulatory mechanisms of the miR-let-7e/SUV39H2/LSD1/CDH1 axis in NSCLC will be summarized in our next study due to the limited reports on the relationship between miR-let-7e and SUV39H2/LSD1/CDH1. Meanwhile, systematical analysis in relation to the original source of the exosomes is required so as to validate the biological effects of serum-derived exosomes on NSCLC cell lines. Moreover, primary NSCLC cells shall be adopted to validate the effects of exosomal let-7e mimic (vs. mimic-NC) in future studies.
miR-let-7e in the exosomes targets SUV39H2 that can inhibit the ubiquitination of LSD1 through methylation, thereby stabilizing the expression of LSD1. LSD1 inhibits CDH1 expression by removing the CDH1 promoter methylation through demethylase function, ultimately impairing NSCLC cell viability, migration, and invasion in vitro as well as preventing tumor growth and EMT in vivo.
Change history
01 December 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41417-021-00402-9
References
Zhou C. Lung cancer molecular epidemiology in China: recent trends. Transl Lung Cancer Res. 2014;3:270–9.
Boeckx B, Shahi RB, Smeets D, De Brakeleer S, Decoster L, Van Brussel T, et al. The genomic landscape of nonsmall cell lung carcinoma in never smokers. Int J Cancer. 2019;146:3207–18.
Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw. 2013;11:645–53. quiz 53
Li W, Yu H. Separating or combining immune checkpoint inhibitors (ICIs) and radiotherapy in the treatment of NSCLC brain metastases. J Cancer Res Clin Oncol. 2020;146:137–52.
Rebuzzi SE, Alfieri R, La Monica S, Minari R, Petronini PG, Tiseo M. Combination of EGFR-TKIs and chemotherapy in advanced EGFR mutated NSCLC: Review of the literature and future perspectives. Crit Rev Oncol Hematol. 2019;146:102820.
Fujita Y, Kuwano K, Ochiya T, Takeshita F. The impact of extracellular vesicle-encapsulated circulating microRNAs in lung cancer research. Biomed Res Int. 2014;2014:486413.
Walker S, Busatto S, Pham A, Tian M, Suh A, Carson K, et al. Extracellular vesicle-based drug delivery systems for cancer treatment. Theranostics. 2019;9:8001–17.
Couto N, Caja S, Maia J, Strano Moraes MC, Costa-Silva B. Exosomes as emerging players in cancer biology. Biochimie. 2018;155:2–10.
Zhou S, Hu T, Zhang F, Tang D, Li D, Cao J, et al. Integrated microfluidic device for accurate extracellular vesicle quantification and protein markers analysis directly from human whole blood. Anal Chem. 2020;92:1574–81.
Zhu WY, Luo B, An JY, He JY, Chen DD, Xu LY, et al. Differential expression of miR-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Investig. 2014;32:394–401.
Zhang YK, Zhu WY, He JY, Chen DD, Huang YY, Le HB, et al. miRNAs expression profiling to distinguish lung squamous-cell carcinoma from adenocarcinoma subtypes. J Cancer Res Clin Oncol. 2012;138:1641–50.
Zheng Y, Li B, Wang J, Xiong Y, Wang K, Qi Y, et al. Identification of SUV39H2 as a potential oncogene in lung adenocarcinoma. Clin Epigenetics. 2018;10:129.
Piao L, Suzuki T, Dohmae N, Nakamura Y, Hamamoto R. SUV39H2 methylates and stabilizes LSD1 by inhibiting polyubiquitination in human cancer cells. Oncotarget. 2015;6:16939–50.
Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, et al. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS ONE. 2012;7:e35065.
Hu X, Xiang D, Xie Y, Tao L, Zhang Y, Jin Y, et al. LSD1 suppresses invasion, migration and metastasis of luminal breast cancer cells via activation of GATA3 and repression of TRIM37 expression. Oncogene. 2019;38:7017–34.
Gao LM, Xu SF, Zheng Y, Wang P, Zhang L, Shi SS, et al. Long non-coding RNA H19 is responsible for the progression of lung adenocarcinoma by mediating methylation-dependent repression of CDH1 promoter. J Cell Mol Med. 2019;23:6411–28.
Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, et al. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109:994–1003.
Zhang X, Zhang X, Yu B, Hu R, Hao L. Oncogene LSD1 is epigenetically suppressed by miR-137 overexpression in human non-small cell lung cancer. Biochimie. 2017;137:12–9.
Yu S, Yang D, Ye Y, Liu P, Chen Z, Lei T, et al. Long noncoding RNA actin filament-associated protein 1 antisense RNA 1 promotes malignant phenotype through binding with lysine-specific demethylase 1 and repressing HMG box-containing protein 1 in non-small-cell lung cancer. Cancer Sci. 2019;110:2211–25.
Liu B, Sun X. miR-25 promotes invasion of human non-small cell lung cancer via CDH1. Bioengineered. 2019;10:271–81.
Zhao W, Zhang LN, Wang XL, Zhang J, Yu HX. Long noncoding RNA NSCLCAT1 increases non-small cell lung cancer cell invasion and migration through the Hippo signaling pathway by interacting with CDH1. FASEB J. 2019;33:1151–66.
Karuppasamy R, Veerappapillai S, Maiti S, Shin WH, Kihara D. Current progress and future perspectives of polypharmacology: from the view of non-small cell lung cancer. Semin Cancer Biol. 2019. (Online ahead of print).
Lin J, Wang Y, Zou YQ, Chen X, Huang B, Liu J, et al. Differential miRNA expression in pleural effusions derived from extracellular vesicles of patients with lung cancer, pulmonary tuberculosis, or pneumonia. Tumour Biol. 2016. (Online ahead of print).
Taverna S, Giallombardo M, Gil-Bazo I, Carreca AP, Castiglia M, Chacartegui J, et al. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget. 2016;7:28748–60.
Wu F, Yin Z, Yang L, Fan J, Xu J, Jin Y, et al. Smoking induced extracellular vesicles release and their distinct properties in non-small cell lung cancer. J Cancer. 2019;10:3435–43.
Zheng H, Zhan Y, Liu S, Lu J, Luo J, Feng J, et al. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. J Exp Clin Cancer Res. 2018;37:226.
Tsai CH, Lin LT, Wang CY, Chiu YW, Chou YT, Chiu SJ, et al. Over-expression of cofilin-1 suppressed growth and invasion of cancer cells is associated with up-regulation of let-7 microRNA. Biochim Biophys Acta. 2015;1852:851–61.
Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23:5311–9.
He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC). Biomed Pharmacother. 2017;95:331–8.
Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. J Biomed Sci. 2015;22:9.
Acknowledgements
The authors would like to acknowledge the helpful comments on this paper received from the reviewers.
Author information
Authors and Affiliations
Contributions
L.G. and S.X. designed the study. L.Z. and L.K. collated the data, carried out data analyses, and produced the initial draft of the manuscript. L.G. and H.X. contributed to drafting the manuscript. All authors have read and approved the final submitted manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Xu, S., Zheng, L., Kang, L. et al. microRNA-let-7e in serum-derived exosomes inhibits the metastasis of non-small-cell lung cancer in a SUV39H2/LSD1/CDH1-dependent manner. Cancer Gene Ther 28, 250–264 (2021). https://doi.org/10.1038/s41417-020-00216-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-020-00216-1
- Springer Nature America, Inc.
This article is cited by
-
Exosomal miRNAs: the tumor's trojan horse in selective metastasis
Molecular Cancer (2024)
-
Applications and advancements of nanoparticle-based drug delivery in alleviating lung cancer and chronic obstructive pulmonary disease
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Emerging role of exosomes in cancer progression and tumor microenvironment remodeling
Journal of Hematology & Oncology (2022)
-
RNA-based therapies: A cog in the wheel of lung cancer defense
Molecular Cancer (2021)
-
Tumor-associated exosomes promote lung cancer metastasis through multiple mechanisms
Molecular Cancer (2021)