Abstract
Influenza A virus (IAV) is a widespread pathogen that poses a significant threat to human health, causing pandemics with high mortality and pathogenicity. Given the emergence of increasingly drug-resistant strains of IAV, currently available antiviral drugs have been reported to be inadequate to meet clinical demands. Therefore, continuous exploration of safe, effective and broad-spectrum antiviral medications is urgently required. Here, we found that the small molecule compound J1 exhibited low toxicity both in vitro and in vivo. Moreover, J1 exhibits broad-spectrum antiviral activity against enveloped viruses, including IAV, respiratory syncytial virus (RSV), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), human coronavirus OC43 (HCoV-OC43), herpes simplex virus type 1 (HSV-1) and HSV-2. In this study, we explored the inhibitory effects and mechanism of action of J1 on IAV in vivo and in vitro. The results showed that J1 inhibited infection by IAV strains, including H1N1, H7N9, H5N1 and H3N2, as well as by oseltamivir-resistant strains. Mechanistic studies have shown that J1 blocks IAV infection mainly through specific interactions with the influenza virus hemagglutinin HA2 subunit, thereby blocking membrane fusion. BALB/c mice were used to establish a model of acute lung injury (ALI) induced by IAV. Treatment with J1 increased survival rates and reduced viral titers, lung index and lung inflammatory damage in virus-infected mice. In conclusion, J1 possesses significant anti-IAV effects in vitro and in vivo, providing insights into the development of broad-spectrum antivirals against future pandemics.
Similar content being viewed by others
Introduction
In recent decades, an increasing number of viruses that can infect humans have emerged in livestock and wildlife, such as the severe acute respiratory syndrome coronavirus (SARS-CoV-2), avian influenza virus (AIV), Zika virus, and Ebola virus. The newly identified severe acute respiratory virus, SARS-CoV-2, has caused a global pandemic of viral pneumonia [1, 2]. Although 91 years have passed since the influenza virus was isolated in 1933, humans are still at constant risk of influenza pandemics. The two primary factors contributing to influenza epidemics are antigenic shifts and drift [3]. Antigenic drift is caused by point mutations in the influenza virus genome, whereas antigenic shift occurs when two or more different viral strains infect the same host cell, resulting in the recombination of genetic material [4]. Occasionally, antigenic shifts give rise to novel strains capable of causing pandemics through sustained and efficient human-to-human transmission [5]. Since the 20th century, influenza A virus (IAV) has been responsible for several widespread pandemics, such as the 1918 H1N1 Spanish flu [6], 1957 H2N2 Asian flu [7] and 1968 H3N2 Hong Kong flu, resulting in a large number of deaths [8].
Commonly used antiviral agents include M2 ion channel inhibitors (amantadine), neuraminidase (NA) inhibitors (oseltamivir and zanamivir), and polymerase acidic (PA) endonuclease inhibitors (baloxavir marboxil) [9]. M2 ion channel inhibitors are no longer recommended because of their widespread resistance to circulating influenza viruses [10, 11]. In addition, influenza viruses can develop resistance to oseltamivir by acquiring mutations in the neuraminidase active sites, such as H274Y and I223R (predominantly in H1N1 and H5N1 viruses) and E119V, R292K, or N294S (predominantly in H3N2 viruses) [12,13,14]. In 2008-2009, high rates of oseltamivir-resistant seasonal H1N1 viruses were observed worldwide, raising concerns about the effectiveness of oseltamivir [15, 16]. Baloxavir Marboxil, a novel antiviral agent against influenza, exhibits promising potential for combating the seasonal flu [17]. However, it is noteworthy that up to 11% of adults/adolescents in clinical trials have shown the emergence of baloxavir resistance during treatment, particularly among younger individuals and those infected with H3N2 subtype [18, 19]. Given the ongoing emergence of resistant strains, local epidemics, and global pandemics, there is an urgent need to develop highly effective broad-spectrum antiviral drugs to prevent and treat influenza.
IAV is a negative-stranded RNA virus belonging to the Orthomyxoviridae family. The viral surface is mainly composed of two spike proteins, hemagglutinin (HA) and NA, which play crucial roles in mediating viral entry and release [20]. The HA protein exists as a homologous trimer on the viral surface, composed of HA1 and HA2 subunits connected by disulfide bonds [21, 22]. IAV binds to sialic acid receptors on the surface of target cells via HA1, followed by trafficking of the virus to endosomes [20]. Subsequently, HA2 undergoes a conformational change in the acidic environment, exposing the fusion peptide and inserting it into the endosomal membranes [23]. This process allows the fusion of viral membranes with endosomal membranes, resulting in the release of viral ribonucleoprotein complexes (vRNPs) through the fusion pore into the cytoplasm, thereby completing the uncoating of IAV [23, 24]. The critical nature of HA makes it an attractive target for therapeutics that inhibit binding or fusion processes, including small molecules, proteins or antibodies [25]. Thus, interference with the function of the HA protein, including the HA1 and HA2 subunits, effectively prevents viral entry at an early stage and protects host cells from further infection [26]. Thus, HA inhibitors are expected to be developed as innovative anti-IAV agents.
In this study, we assessed the antiviral activity of the small molecule compound J1 against various enveloped viruses and investigated its in vitro and in vivo anti-IAV effects and underlying mechanisms.
Materials and methods
Reagents
J1 (chemical name: 3-(4-chlorophenyl)-1,4-diphenyl-1,4,5,7-tetrahydro-6H-pyrazolo[3,4-b]pyridin-6-one) was synthesized at the Chemical Biology Laboratory of Inner Mongolia Medical University and its chemical structure is shown in Fig. 1. The molecular formula of compound J1 was C24H18ClN3O, with a molecular weight of 399.87. The procedure for the synthesis and structural characterization of J1 is shown in Supplementary Figs. S1–S3. J1 was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 20 mM and stored at −20 °C. Ribavirin, zanamivir, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), oseltamivir and TPCK-trypsin were purchased from Sigma-Aldrich (St. Louis, MO, USA). Oseltamivir phosphate (Ose) was purchased from Hoffmann-La Roche (Basel, Switzerland). Acyclovir was purchased from MedChem Express (Monmouth Junction, NJ, USA). The plasmids encoding HA of different isolate H5N1 virus (Thailand-HA, Qinghai-HA, Xinjiang-HA, Anhui-HA, Hong Kong-HA and Vietnam-HA), A/Thailand/Kan353/2004-NA and the HIV backbone plasmid which encodes Env-defective, luciferase-expressing HIV-1 (pNL4-3. luc.R-E-) were generated in our laboratory, as described previously [27].
Cells, viruses and animals
Human colorectal adenocarcinoma (HRT18), human embryonic kidney (293T), Madin-Darby canine kidney (MDCK), human epithelial carcinoma (HEp-2), and human lung carcinoma (A549) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). MDCK, 293T and HEp-2 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, ExCellBio, Shanghai, China) and 1% penicillin/streptomycin (Gibco, Carlsbad, CA, USA). HRT18 and A549 cells were maintained in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% FBS and 1% penicillin/streptomycin. Influenza A virus subtypes include A/HongKong/415742/2009 (H1N1), A/Aichi/2/68 (H3N2), A/Vietnam/1194/2004 (H5N1), A/Anhui/1/2013 (H7N9), A/WSN/1933 (H1N1), A/Puerto Rico/8/34 (H1N1) with the NA-H274Y and mouse-adapted strain of high pathogenicity influenza virus A/PR/8/34 (PR8) were propagated in 8-day-old embryonated chicken eggs at 37 °C, allantoic fluid containing the virus was stored at −80 °C. HSV-2 (333 strain) was provided by Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences (Guangzhou, China). HRSV strain HRSV-A-GZ08-0 was clinically isolated at Guangzhou Children’s Hospital (Guangzhou, China) in 2008, as previously reported [28] and presented by Associate Professor Zhang Ke (Guizhou Medical University, China). Authentically preserved SARS-CoV-2 (Beta-CoV/Wuhan/WIV04/2019, IVCAS6.7512; Delta, IVCAS6.7585) was obtained from the National Virus Resource, Wuhan Institute of Virology, Chinese Academy of Sciences. The virus titer was determined by analyzing the 50% tissue culture infective dose (TCID50) using the Reed-Muench assay [29]. SPF BALB/c mice were purchased from the Laboratory Animal Center of Southern Medical University (Guangzhou, China). The experiments reported in this study were approved by the National Institutional Animal Care and Medical Ethics Committee of Southern Medical University.
Cytotoxicity assay
The cytotoxicity of J1 was assessed using the MTT assay. Briefly, the cells were cultured in 96-well plates at a density of 2 × 105/mL and subsequently treated with the indicated concentrations of the compound. A blank group without the compound was established, consisting of three parallel wells at each concentration. After incubation at 37 °C for 48 h, the supernatant was removed and replaced with 100 μL serum-free medium containing a diluted solution of MTT (0.5 mg/mL), followed by an additional incubation at 37 °C for 4 h. Subsequently, the formazan crystals were dissolved by adding 150 µL of DMSO. Absorbance was measured at 570 nm using a microplate reader (GENios Pro, TECAN, Bedford, MA, USA). The percentage of cell viability was calculated using the following formula: cell viability (%) = Acompound/ABlank × 100.
Acute toxicity study
According to the OECD-423 guidelines, female BALB/c mice were divided into six groups, and each group containing three mice was repeated twice (n = 6). Mice were fasted for 4 h before drug administration. J1 was dissolved in 0.5% CMC and administered orally via a gavage needle, whereas the control group received 0.5% CMC. The results were replicated for a second time to establish a grading system with an escalated dosage. The behavior, body weight, food intake, and water intake of the mice were monitored and recorded daily for 14 days. On day 14, all mice were euthanized, and blood samples were collected from the eyeballs for serum separation. Serum levels of inflammatory factors were determined using an ELISA kit (4A Biotech, Beijing, China). The weights of the heart, liver, spleen, lung and kidney were measured, and the organ coefficient was calculated as the percentage of organ weight to total weight (mg/g). The organs were preserved in 4% paraformaldehyde for histological investigation using hematoxylin and eosin (H&E) staining. Peritoneal, liver and kidney cells were isolated, and cell viability was assessed using a CountStar BioTech cell counter (Alit Biotech, Shanghai, China).
Antiviral assay
Antiviral activity against IAV
MDCK cells were seeded at 2 × 105/mL in 96-well plates and incubated for 24 h. The cells were washed twice with PBS and infected with IAV at an MOI (multiplicity of infection) of 0.01 for 1 h at 37 °C. Subsequently, the cells were cultured in serum-free DMEM containing 1 μg/mL TPCK and two-fold serial dilutions of J1 for 48 h. The antiviral activity of J1 was measured by the MTT assay, as previously mentioned. In addition, we observed protection against virus-induced cytopathic effects (CPE) using a microscope (Nikon, Tokyo, Japan).
Antiviral activity against SARS-CoV-2
An inhibition assay for live SARS-CoV-2 was performed as previously described [30]. Vero-E6 cells were infected with SARS-CoV-2 (MOI = 0.05) and treated with J1 for 24 h. Viral copies in the cell supernatants were quantified using the standard curve method and qRT-PCR analysis with primers targeting the S gene.
Antiviral activity against HCoV-OC43
HRT18 cells were cultured in 24-well plates until reached 80% confluence. Subsequently, the cells were infected with the virus (MOI = 0.1) for 1 h at 37 °C. After removing the supernatant, the compound was added and maintained for 24 h at 37 °C. Finally, total RNA was extracted for qRT-PCR.
Antiviral activity against RSV
HEp-2 cells were cultured in 24-well plates until reached 80% confluence. Subsequently, the cells were infected with the virus (MOI = 0.1) for 1 h at 37 °C. After removing the supernatant, the compound was added and maintained for 48 h at 37 °C. Finally, total RNA was extracted for qRT-PCR.
Antiviral activity against HSV
Anti-HSV effects have been previously reported [31]. Briefly, Vero cells were infected with HSV-1 (MOI = 0.1) or HSV-2 (MOI = 0.1) for 1 h at 37 °C, and serum-free DMEM containing J1 was added and maintained for 24 h at 37 °C. The supernatant was collected for the plaque assay.
Selection of resistant IAV variants
As previously reported [32], MDCK cells were seeded in 6-well plates and cultured overnight. Afterward, a mixture of 20 μM J1 and 100TCID50 of WSN prepared in DMEM containing 1 μg/mL TPCK-trypsin was added to the cells, followed by continued incubation for 72 h. After incubation, the supernatant was collected and stored at −80 °C to obtain the first generation of virus (F1). MDCK cells were seeded in a 6-well plate and infected with a combination of 20 μM J1 mixed with 10 μL F1 containing 1 μg/mL TPCK-trypsin in DMEM. Incubation was continued for another 72 h, after which the supernatant was collected as the second-generation virus (F2). This process was repeated up to the 10th passage, and an MTT assay was performed to measure the inhibitory activity of J1 on F10.
Plaque assay
Monolayer MDCK cells were infected with WSN (MOI = 0.01) or lung homogenates from acute lung injury (ALI) mice for 1 h. A semi-solid medium was prepared by combining a 4% microcrystalline cellulose solution with an equal volume of 2 ×DMEM (EallBio, Beijing, China) supplemented with 1 µg/mL TPCK-trypsin. The compound was added to the semi-solid medium, with each well receiving 3 mL, and incubated at 37 °C for 48 h. Subsequently, a semi-solid medium was removed, and cells were fixed using 4% paraformaldehyde followed by staining with 2% crystal violet (Sigma-Aldrich, St. Louis, MO, USA). The inhibitory effect of the compound on viral plaque formation was assessed by plaque quantification.
Western blotting assay
A549 cells were seeded in 12-well plates, cultured to 80%–90% confluence, infected with WSN virus (MOI = 0.1) for 1 h, and cultured in serum-free medium containing the compound for 24 h. Cell proteins were collected using radioimmunoprecipitation assay (RIPA, Fdbio Science, Hangzhou, China) lysis buffer. Proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Roche, Basel, Switzerland). After blocking with 5% non-fat milk (Fdbio Science, Hangzhou, China) for 1 h at room temperature (RT), membranes were incubated overnight at 4 °C with anti-NP monoclonal antibody (1:5000 dilution, GeneTex, GTX125989), anti-PB2 monoclonal antibody (1:5000 dilution, GeneTex, GTX125926), and GAPDH (1:5000 dilution, Bioss, bs-2188R). The membrane was then incubated with an anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000 dilution, Zenbio, 511203) for 1 h at RT. Following three times of membrane cleaning with Tris-buffered saline-Tween 20 (TBST), the target protein bands were visualized using a Tanon-5200Multi Automatic chemiluminescence instrument (Shanghai, China).
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells and mouse tissues using an RNA isolation kit (ForeGene, Chengdu, China). cDNA was amplified from 100 ng of RNA using the PrimeScriptTM RT Master Kit (TaKaRa, Otsu, Shiga, Japan). Gene products were analyzed by qPCR using SYBR Select Master Mix (TaKaRa, Otsu, Shiga, Japan) in a LightCycler 480 system (Roche Applied Science, Basel, Switzerland). The primer sequences are listed in Supplementary Table S1. The relative expression of the target genes was calculated and analyzed using the 2−ΔΔCT method. GAPDH was used as an endogenous control. All primers were synthesized by Tsingke Biotech Co., Ltd. (Beijing, China).
Indirect immunofluorescence assay (IFA)
MDCK cells were seeded into 48-well plates, infected with WSN (MOI = 0.01) at 37 °C for 1 h, and treated with the diluted compound in serum-free DMEM containing 1 µg/mL TPCK-trypsin for 24 h. The cells were then washed twice with cold PBS and fixed with 4% paraformaldehyde for 10 min, followed by blocking with 3% bovine serum albumin (BSA) at RT for 1 h. Subsequently, the cells were incubated overnight at 4 °C with an anti-IAV NP antibody (1:500 dilution, GeneTex, GTX125989), followed by incubation with an Alexa Fluor 488 conjugate anti-rabbit IgG antibody (1:500 dilution, Abcam, ab150077) for 1 h at RT. After washing the cells with PBST, Hoechst 33342 (Beyotime, Shanghai, China) was used to visualize the cellular nuclei. Images were captured using a Ti Eclipse inverted fluorescence microscope (Nikon, Tokyo, Japan).
Time-of-addition assay
The time-of-addition assay was performed as previously described [33]. Briefly, A549 cells were cultured in 12-well plates until reached 80%–90% confluence. Subsequently, 20 μM J1 was added at the indicated time intervals (0–2 h, 2–5 h, 5–8 h, 8–10 h and 0–10 h) after WSN (MOI = 0.1) infection. At 10 h post-infection, total proteins were extracted using RIPA lysis buffer for Western blotting to determine the expression of influenza NP protein.
Entry inhibition assays of different pseudotypes
Briefly, 60%–70% confluent 293T cells were co-transfected with one of the five HA plasmids (2 μg), NA plasmid (2 μg), and pNL4-3. luc.R-E- (3 μg) in a 6-well plate using polyjetTM transfection reagent (SignaGen, Rockville, MD, USA). After 48 h, culture supernatants were harvested and centrifuged at 1000 × g for 5 min before being stored at −80 °C until use. MDCK cells were seeded in 96-well plates and incubated overnight. Compound J1 was serially diluted two-fold in a serum-free medium and incubated with an equal volume of pseudovirus for 30 min at 37 °C. The mixture was then transferred to MDCK cells and incubated for 48 h. Pseudovirus titers were quantified using a luciferase assay (Promega, Madison, WI, USA). The VSV-G pseudovirus was used as a negative control, which was produced in the same manner by replacing the H5N1 envelope plasmid with the VSV-G plasmid. The results were expressed as percent inhibition rate, calculated using the following formula: inhibition rate (%) = (1 − Fsample/Fvirus) × 100. Here, Fsample represents the fluorescence of pseudovirus particles in the presence of J1, and Fvirus represents the fluorescence of the pseudovirus control.
MU-NANA assay
As previously described [34], the substrate 4-MUNANA (2-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid sodium) is cleaved by NA to produce a measurable fluorescent product for detecting NA inhibition of the compound. In brief, 60 μL of WSN was pre-incubated with 20 μL of various concentrations of compound in 32.5 mM MES buffer (containing 4 mM CaCl2, pH 6.5) for 30 min. The mixture was subsequently transferred to 96-well black plates (20 μL per well), mixed with an equal volume of 4-MUNANA, and incubated for 1 h at 37 °C in the dark. The reaction was terminated by the addition of 83 mM NaOH (83% ethanol). The fluorescence value F was measured immediately at an excitation wavelength of 340 nm and an emission wavelength of 440 nm. The inhibition rate was calculated using the following formula: Inhibition rate (%) = (Fvirus − Fsample)/(Fvirus − Fsubstrate) × 100. Fvirus, Fsubstrate and Fsample represent the fluorescence intensities of the influenza virus control, substrate control and test samples, respectively.
Microscale thermophoresis (MST) assay
As previously reported [35], HA protein was labeled with NT647 (NanoTemper Technologies, München, Bavaria, Germany) and applied at a final concentration of 40 nM. The corresponding unlabeled binding partner (J1) was titrated in a two-fold dilution series in PBST containing 5% DMSO (pH 4.5). Subsequently, 10 µL of each dilution point was mixed with 10 µL of labeled HA protein solutions and incubated at RT for 10 min. The samples were filled into hydrophilic capillaries (NanoTemper Technologies) for the measurements. MST experiments were performed using a Monolith NT.115 system (NanoTemper Technologies) with a 100% light-emitting diode (LED) and 20% infrared radiation (IR) laser power. The laser on/off times were set to 30 and 5 s, respectively.
Hemagglutination inhibition (HI) assay
First, J1 (25 µL) was subjected to two-fold serial dilutions and mixed with an equal volume of WSN virus in 96-well V-shaped plates (Solarbio, Beijing, China) at RT for 30 min. Subsequently, the mixture was incubated with 50 µL of chicken 1% red blood cells (cRBCs) diluted for another 30 min at RT. After incubation, the mixture was observed at 4 °C for 1 h. Anti-HA antibody was used as a positive control.
Sandwich enzyme-linked immunosorbent assay (ELISA)
Briefly, 96-well polystyrene plates were pre-coated with HA1 or HA2 monoclonal antibodies (2 μg/mL). Following overnight incubation at 4 °C, the plates were blocked for 1 h at RT with 1% nonfat milk. Mouse monoclonal antibodies IB12 (targeting HA2) or HA-7 antibody (targeting HA1) and diluted J1 were added to 96-well plates and incubated at 37 °C for 1 h. Subsequently, biotin-labeled goat anti-mouse IgG (Sigma-Aldrich, St. Louis, MO, USA), SA-HRP, and TMB were sequentially added. Absorbance was measured at 450 nm using a microplate reader (GENios Pro, TECAN, Bedford, MA, USA).
Surface plasmon resonance (SPR) assay
The HA2 protein (Sino Biological, China) was immobilized onto the surface of an optically cross-linked chip. J1 was diluted to various concentrations using running buffer. Subsequently, J1 was injected as a flowing fluid at a flow rate of 3 mL/s, with a contact time and dissociation time of 300 s. Real-time monitoring of the captured signal was performed using the PlexArray® HT system (Plexera® Bioscience, Beijing, China). The affinity values were calculated using Plexera DE software by fitting the curves using the Langmuir equation.
Polykaryon formation inhibition assay
MDCK cells were seeded in 24-well plates and incubated until reached 50% confluence. MDCK cells were transfected with A/Thailand/Kan353/2004-HA plasmid (2 μg DNA/well) using PEI transfection reagent (4 µL/well) and maintained for 48 h. The cells were washed with PBS and treated with serum-free DMEM containing 5 μg/mL TPCK-trypsin for 15 min at 37 °C. After removing the supernatant, the cells were exposed to a diluted compound in serum-free medium for another 15 min at 37 °C. The compound was then removed, and the cells were exposed to PBS at pH 5.0 or 7.0 for an additional 15 min at 37 °C. The cells were then incubated in DMEM containing 10% FBS for 3 h at 37 °C. Finally, the cells were fixed with 4% paraformaldehyde and observed under a microscope (Nikon, Tokyo, Japan).
Antiviral in vivo experiments
Female and male BALB/c mice (4-5 weeks, 18.0 ± 2.0 g) were randomly assigned to six groups (n = 6 per group). Mice were anesthetized with isoflurane and infected via the intranasal route with 50 μL of the PR8 virus (5×LD50) diluted in PBS. Two hours after inoculation, the mice received oral treatment with oseltamivir phosphate (Ose, 60 mg·kg−1·d−1), J1 (60, 30, or 15 mg·kg−1·d−1), or PBS, and the treatments were repeated once daily for 5 days. The mice were weighed and euthanized on day 5 after inoculation, and mice lungs were weighed and photographed. The lung index was calculated as follows: lung index =lung weight (mg)/body weight (g). The lungs were homogenized in PBS to determine viral titers using Reed-Muench and plaque assays, as mentioned above. Histopathological analysis was performed using hematoxylin and eosin (H&E) staining. In addition, total RNA was extracted from the lung tissue to detect the levels of inflammatory factors and chemokines. In the survival experiments, 10 mice per group were intranasally infected with 50 μL PR8 virus at day 0. Drug administration was repeated once daily for 7 days, and the body weight and mortality of mice were continuously monitored for 14 days. The remaining mice were sacrificed at the end of the experiment on day 14 p.i.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8.0 (San Diego, CA, USA). The results are expressed as the mean ± standard deviation (SD) from triplicate experiments. Differences between groups were analyzed using one-way analysis of variance (ANOVA) or two-way analysis of variance (ANOVA). P < 0.05 was regarded as statistically significant and marked with *.
Results
J1 exhibits low toxicity in vitro and in vivo
MTT assay revealed that J1 exhibited a low cytotoxic effect on MDCK, A549, Vero and HRT18 cells at concentrations up to 200 µM (Fig. 2a). Furthermore, J1 was evaluated for in vivo toxicological properties following the OECD 423 guidelines through a 14-day acute oral toxicity study. No mortality was observed during the experiment. The GHS classification of J1 was category 5, and the LD50 values were cut off at 5000 mg/kg (Fig. 2b). Drug effects can modify behavioral patterns. As previously reported [36], we observed and documented the physical appearance, interactions between mice, functional parameters and mouse activity. These findings demonstrated that J1 did not exert a significant impact on mouse behavior (Supplementary Table S2). Body weight, food intake, and water intake of the mice did not exhibit significant changes compared with the control group (Fig. 2c–e). Histological examination of mice after a single oral dose of J1 revealed no significant damage to the heart, liver, spleen, lungs or kidneys (Fig. 2f). The organ coefficients did not reveal any significant effects of J1 on organ weight, except for a decrease in liver weight in the 300 mg/kg group (Fig. 2g). Mouse peritoneal fluid cells were extracted along with liver and spleen single-cell suspensions to assess cell viability. The results demonstrated that J1 had no effect on cell viability, with a viability rate exceeding 70% (Fig. 2h). Furthermore, serum levels of inflammatory cytokines IL-6 and IL-1β were measured, which indicated that J1 did not induce an increase in inflammation (Supplementary Fig. S4). The cytotoxicity and acute oral toxicity results indicated that compound J1 is safe for use as an antiviral agent.
a The cytotoxicity of J1 in A549, HRT18, Vero-E6 and MDCK cells was determined by MTT assay. b The acute toxicity assessment protocol and LD50 cut-off value of J1 were determined according to OECD 423 guidelines. Body weight (c), food uptake (d) and water uptake (e) of mice were recorded daily for 14 days following a single oral administration of J1 (n = 6). f The heart, liver, spleen, lung and kidney of mice were subjected to histological examination (H&E staining, 100×) following a single administration of J1. scale bar: 100 μm. g The organ coefficient was determined by calculating the ratio of the weight of the organ to the body weight of mice (n = 6), *P < 0.05, ns means not significant vs. control. h Cell viability was evaluated using a single-cell suspension obtained from the peritoneal fluid, liver and spleen of mice (n = 6), ns means not significant vs. control. The data are presented as the mean ±SD.
J1 exhibits broad-spectrum antiviral activity
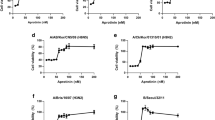
We assessed the inhibitory effect of J1 on various IAV subtypes using the MTT assay. The results demonstrated that J1 was effective against infection by different IAV strains, including H1N1, H3N2, H5N1 and H7N9, with half-maximal inhibitory concentrations (IC50) ranging from 7.81 to 15.65 μM (Table 1). J1 was also active against the oseltamivir-resistant influenza A/Puerto Rico/8/34 (H1N1) virus with NA-H274Y mutation, exhibiting an IC50 of 14.24 μM. More importantly, J1 treatment exhibited inhibitory activity against WSN F10 strains with IC50 values of 11.84 μM, confirming the absence of resistance emergence after 10 passages (Table 1). Oseltamivir and zanamivir (Za) were used as positive controls (Supplementary Figs. S5 and S6). Additionally, the results demonstrated that J1 inhibited infection by two types of coronaviruses. As shown in Fig. 3a, J1 exhibited a dose-dependent inhibitory effect on the wild-type (WT) strain and delta variant of SARS-CoV-2, with IC50 of 6.07 μM and 2.21 μM, respectively. Likewise, J1 inhibited HCoV-OC43 infection and reduced the expression of NP mRNA, chloroquine (20 μM) was used as a positive control (Fig. 3b). J1 also exhibited a significant inhibitory effect on RSV, with ribavirin (50 μM) used as a positive control (Fig. 3c). Interestingly, in addition to inhibiting respiratory viruses, J1 also showed antiviral activity against HSV-induced plaque formation, and acyclovir (2 μM) was used as a positive control (Fig. 3d, e). In summary, compound J1 exhibited broad-spectrum activity against enveloped viruses. The design and modification of J1 will be beneficial for the research and development of broad-spectrum small molecule antiviral drugs.
a The inhibitory effect of J1 on SARS-CoV-2 (MOI = 0.05) infection in Vero-E6 cells was evaluated, and the IC50 value was determined using qRT-PCR. b The inhibitory effect of J1 on the expression of HCoV-OC43 (MOI = 0.1) NP mRNA in HRT18 cells was detected using qRT-PCR. GAPDH was used as an internal control. ***P < 0.001, ns means not significant vs. HCoV-OC43 group. c The inhibitory effect of J1 on the expression of RSV (MOI = 0.1) N and F mRNA in HEp-2 cells was detected using qRT-PCR. GAPDH was used as an internal control. *P < 0.05, ***P < 0.001 vs. RSV group. The inhibitory effect of J1 on (d) HSV1 (MOI = 0.1) and (e) HSV2 (MOI = 0.1) infection in Vero cells was analyzed by plaque assay. The data are presented as the mean ±SD.
J1 shows effective inhibition of IAV infection
Considering the inhibitory effect of J1 on various subtypes of IAV, we further investigated its inhibitory effect and mechanism against IAV using the A/WSN/1933 (H1N1) strain as a representative example because of its high titer and stability. IAV-induced CPE was observed under a microscope, and the infected cells became round, shrunken, and even detached. As shown in Fig. 4a, following treatment with compound J1, IAV-induced CPE was significantly reduced, indicating that J1 may protect host cells from IAV damage. The results of the plaque assay showed that J1 decreased viral plaque formation in MDCK cells in a concentration-dependent manner, confirming the antiviral activity of J1 (Fig. 4b). The expression of influenza NP and PB2 proteins was assessed by Western blotting, whereas the mRNA levels were determined using qRT-PCR. Our findings demonstrated the inhibitory effect of J1 on the expression of viral NP and PB2 at both the protein and mRNA levels (Fig. 4c, d). The expression levels of IAV NP protein were further examined using an indirect immunofluorescence assay. The levels of NP expression (green fluorescence) were significantly reduced in a dose-dependent manner after treatment of infected MDCK cells with J1 (Fig. 4e). Za was used as the positive control. These results demonstrate that J1 can effectively inhibit IAV infection in vitro.
a Microscopic observation (100×) of J1 protecting MDCK cells from WSN-induced cytopathic effects (CPE) at 48 h. Scale bar: 100 μm. b The inhibitory effect of J1 on WSN-induced plaque formation in MDCK cells was assessed by quantifying the number of plaques. ***P < 0.001, ns means not significant vs. virus group. c The expression of NP and PB2 proteins in IAV-infected A549 cells was determined by Western blotting. ***P < 0.001 vs. virus group. d The expression of NP and PB2 mRNA in IAV-infected A549 cells was determined using qRT-PCR. GAPDH was used as an internal control. *P < 0.05, ***P < 0.001, ns means not significant vs. virus group. e The effect of J1 on the level of viral NP protein was detected using indirect immunofluorescence (100×). Scale bar: 100 μm. NP is shown as green fluorescence, and Hoechst marks the location of the nucleus with blue fluorescence. The data are presented as the mean ±SD.
J1 exhibits antiviral activity in the early stages of IAV infection and may target HA
As shown in Supplementary Fig. S7, J1 did not exhibit any inhibitory effect against IAV infection when A549 cells were pre-incubated with J1 for 1 h before IAV infection. However, viral replication was significantly inhibited when the cells were co-incubated with the virus and J1. These findings indicate that the antiviral mechanism of J1 preferentially targets the virus rather than the host cells. Subsequently, we conducted virucidal assays to investigate whether J1 directly impaired IAV infectivity. The results showed that J1 had no significant virucidal efficacy (Supplementary Fig. S8). Therefore, we focused on the role of J1 in viral entry using a time-of-addition assay. As shown in Fig. 5a, the viral life cycle was divided into four stages, J1 was administered at intervals of 0–2 h, 2–5 h, 5–8 h and 8–10 h after IAV infection. The results showed that J1 acted during the early entry stage of IAV (Fig. 5b).
a Schematic of the time-of-addition. b Time-of-addition. A549 cells were infected with WSN virus (MOI = 0.1) for 1 h, followed by treatment with J1 at the time intervals of 0-2 h, 2-5 h, 5-8 h and 0-10 h. NP expression was detected using Western blotting. ***P < 0.001, ns means not significant vs. virus group. c MU-NANA assay was used to assess the inhibitory effect of J1 on neuraminidase (NA). d The binding curve of J1 with NT647 labeled recombinant influenza HA protein was detected using MST assay. NT647 labeled recombinant influenza HA protein was mixed with a two-fold diluted compound in the same volume. MST experiments were performed using a Monolith NT.115 system (NanoTemper Technologies). The data are presented as the mean ±SD.
The inhibitory effect of J1 on viral entry was tested using a pseudovirus entry assay, as previously described [37]. A VSV-G pseudovirus containing an unrelated envelope protein was used as a negative control. As shown in Table 2, J1 effectively inhibited the diverse H5N1 pseudoviruses infection with IC50 ranging from 1.87 to 14.3 µΜ, but did not show detectable inhibition for VSV-G mediated entry. The results highlighted that J1 could specifically interfere with the function of influenza HA or NA proteins because the H5N1 pseudovirus contains two envelope proteins from IAV. We then performed NA inhibition assays using 4-MUNANA as the chemiluminescent substrate and IAV as the enzyme source to investigate the ability of J1 to inhibit NA activity. The NA inhibitor zanamivir was used as a positive control. The results showed that J1 did not affect viral NA activity (Fig. 5c). To further elucidate the binding targets of J1, a solution-based MST assay was performed. It was clear that J1 interacted with the influenza HA protein in a dose-dependent manner, with a dissociation constant Kd values of 0.54 ± 0.18 µΜ (Fig. 5d). Based on these results, we conclude that J1 interferes with viral HA.
The anti-influenza activity of J1 is associated with the HA2 subunit of IAV
Influenza hemagglutinin (HA), a surface glycoprotein, has been extensively characterized and plays a critical role in influenza entry. We first investigated whether J1 interfered with cRBCs agglutination using a hemagglutinin inhibition (HI) assay Fig. 6a demonstrates that even at a concentration as high as 200 μM, J1 was unable to inhibit the agglutination of IAV to cRBCs, suggesting that J1 did not interfere with virus-receptor binding. Anti-HA antibody was used as a positive control. Furthermore, the competitive binding effect of J1 on the HA1-specific monoclonal antibody HA-7 and HA2-specific monoclonal antibody IB12 was determined by ELISA. We found that J1 remarkably inhibited the binding of IB12 to HA2 with an IC50 of 3.23 μM (Fig. 6b), but only background levels of inhibitory activity were seen with HA1, consistent with the result of HI inhibition (Supplementary Fig. S9). To verify the fusion inhibitory effect of J1, MDCK cells transfected with HA were exposed to pH 5.0 conditions to promote membrane fusion and formation of polykaryons. The results in Fig. 6c indicated a significant decrease in the number of polykaryons upon treatment with J1 and the positive control arbidol (10 μM), confirming the ability of J1 to inhibit the function of the HA2 subunit. An SPR assay was used to determine the affinity of J1 for commercially available HA2. It clearly showed that J1 and HA2 proteins bind in a concentration-dependent manner with an equilibrium dissociation constant (KD) of 5.68 × 10−8 M (Fig. 6d). J1 interacts with the HA2 subunit to disrupt the membrane fusion step of IAV entry.
a Inhibition of J1 on WSN-induced aggregation of chicken erythrocytes was evaluated using a hemagglutination inhibition (HI) assay. b Binding of the HA2-specific antibody IB12 to HA2 was inhibited by J1, as demonstrated by ELISA. Absorbance was measured using a microplate reader. c The effect of J1 on membrane fusion was evaluated by detecting polykaryon formation in HA over-expressed MDCK cells at pH 5.0 (100×). Scale bar: 100 μm. d Interactions between influenza HA2 protein and J1 were analyzed using surface plasmon resonance (SPR). The data are presented as the mean ±SD.
Protective effects of J1 against acute lung injury (ALI) induced by IAV
To further investigate whether J1 has an anti-IAV effect in vivo, mouse lung-adapted viral strains A/PR/8/34 (H1N1) were used to infect BALB/c mice and induce ALI. An ALI model was successfully established, as represented by pulmonary edema, atelectasis and hemorrhage. A schematic diagram of the in vivo drug efficacy experiment is shown in Fig. 7a. As shown in Fig. 7b, virus-induced direct tissue damage was significantly alleviated by J1 treatment. Histopathological examination of lung sections after J1 treatment showed that acute lung injury phenotypes (inflammatory cell infiltration, alveolar wall thickening and lung lesions) were remarkably improved compared to the virus group (Fig. 7c). As shown in Fig. 7d, e, J1 treatment significantly reduced viral titers in the lung tissues of mice infected with IAV, particularly in the high-concentration group (60 mg/kg), which was comparable to the efficacy of oseltamivir (Ose). J1 treatment markedly reduced the lung index, suggesting that J1 could alleviate pulmonary edema induced by IVA infection (Fig. 7f). IAV infection leads to a cytokine storm in response to a significant increase in the expression of proinflammatory cytokines and chemokines in the lung tissue. The increased levels of inflammatory mediators, such as IL-6 (Fig. 7g), IL-1β (Fig. 7h), IFN-β (Fig. 7i), MCP-1 (Fig. 7j) and IP-10 (Fig. 7k), were significantly reduced in the J1 treatment group. In summary, our results showed that IAV-induced lung injury was markedly attenuated by J1 treatment.
a Diagram depicts treatment with J1 in BALB/c mice infected with the PR8 virus. b Macroscopic appearance of mouse lungs was examined at 5 d p.i. Three representative mice were selected. c Morphological changes in lung tissues were evaluated by H&E staining (100×). Scale bar: 100 μm. d Viral replication in the lungs of IAV-challenged mice was evaluated by plaque assay. Three representative mice were selected. e Viral titers in the lungs of mice were determined using the Reed–Muench method (n = 6). *P < 0.05, ns means not significant vs. virus group. f Lung index (n = 6). ***P < 0.001, ns means not significant vs. virus group. Effects of J1 on inflammatory factors in lungs of challenged mice, including IL-6 (g), IL-1β (h), IFN-β (i), MCP-1 (j) and IP-10 (k) (n = 6). *P < 0.05, **P < 0.01, ***P < 0.001, ns means not significant vs. virus group. The data are presented as the mean ±SD.
J1 improves the survival of mice infected with IAV
We measured the survival rate and body weight of the mice after IAV infection, according to the schematic diagram (Fig. 8a). As shown in Fig. 8b, the body weight of IAV-infected mice began to decrease at 3 d p.i., reaching a minimum at 9 d p.i., with delayed loss observed in the Ose groups (60 mg/kg), and the high-dose group (60 mg/kg) of J1. The survival rate of the J1-treated group was higher than that of the virus-treated group (Fig. 8c). All infected mice in the virus group died at 10 d p.i. The survival rates of mice treated with J1 at doses of 30 and 60 mg·kg−1·d−1 increased to 30% and 70%, respectively. Additionally, both J1 and Ose effectively alleviated hair shrugs in mice after infection and conferred a protective effect in mice with viral pneumonia.
Discussion
The development of effective prevention and treatment strategies against influenza infection remains a significant challenge, as the emergence of highly virulent strains through antigenic drift/shift, adaptation, and genetic reassortment presents obstacles to timely vaccine production to control influenza pandemics [38]. Antiviral drugs targeting specific steps in the infectious life cycle of IAV have emerged as promising therapeutic options to address these challenges. Entry and fusion inhibitors are important antiviral agents. The anti-HIV-1 fusion inhibitor peptide enfuvirtide, used in the clinical treatment of HIV infection, can be considered successful [39]. During the life cycle of influenza virus, HA mediates the binding of IAV to its cell surface receptor(s) and its subsequent entry, involving endocytosis of the virus and fusion between the viral and host cell membranes [23]. Small molecule entry inhibitors for influenza viruses are currently under development [40]. Given that HA2 is highly conserved, it could effectively reduce the risk of viral escape and present a promising target for the development of inhibitors of viral entry.
Significant attention has been devoted to research on small molecule compounds that target the HA2 subunits. Arbidol, a well-known anti-influenza entry inhibitor that binds to the hydrophobic cavity of the HA trimer stem to stabilize its prefusion conformation and prevent large conformational changes during endosomal membrane fusion at low pH, has been approved by regulatory authorities in Russia (1993) and China (2006) for the treatment and prevention of influenza [41,42,43]. Tert-butyl hydroquinone (TBHQ) was found to bind hydrophobic pockets between HA subunits, thus stabilizing the conformation of neutral pH HA and inhibiting membrane fusion [44]. Furthermore, studies in our laboratory have found that an N-substituted piperidine compound, CL-385319, inhibits viral entry by stabilizing the prefusion state of hemagglutinin [45].
Here, we report a novel fusion inhibitor, J1, which exhibits anti-influenza activity against various IAV subtypes, including H1N1, H7N9, H5N1 and H3N2, with an IC50 of 7.81 to 15.65 μM. J1 was also active against the oseltamivir-resistant strain A/PR/8/34 with the NA-H274Y mutation and maintained an effective inhibitory effect on the WSN virus that had been passaged 10 times in the presence of J1, reflecting its advantage of being less prone to resistance. More importantly, J1 showed antiviral activities against several RNA-enveloped viruses, such as HCoV-OC43, RSV, HSV-1, HSV-2, and the emerging SARS-CoV-2. These findings highlight that J1 has potential broad-spectrum antiviral activity. Following the OECD-423 guidelines, we monitored and documented food and water intake, body weight, and behavioral changes in mice after a single oral administration of J1. Histopathological alterations in vital organs (heart, liver, spleen, lungs, and kidneys) were evaluated using H&E staining. In addition, cell viability, organ coefficients, and serum cytokine levels were measured. Overall, oral administration of J1 did not exhibit any toxic effects in mice, indicating its high safety profile and potential for further investigation.
Time-of-addition studies were performed to investigate the mechanism of action of J1, which indicated that J1 blocks viral entry at an early stage. Next, we conducted H5N1 pseudovirus neutralization assays by transfecting HA and NA plasmids into 293T cells. The results demonstrated that J1 exhibited inhibitory effects on various subtypes of H5N1 pseudoviruses but had no impact on VSV-G, indicating its specific targeting of either HA or NA. The neuraminidase inhibition assay demonstrated that J1 exhibited no inhibitory activity against NA, suggesting that its antiviral effects may be attributed to HA. To validate the binding between J1 and HA, we performed an MST assay, which enabled us to quantitatively analyze the interactions between small molecules and proteins in solution. The MST dose-response curve clearly shows the interaction between HA and J1. However, the addition of compound J1 did not inhibit hemagglutination caused by HA1. Furthermore, MDCK cells transfected with the HA plasmid were exposed to acidic conditions (pH 5.0) for fusion and formation of polykaryons. The addition of J1 resulted in a significant reduction in polykaryon formation, providing compelling evidence for the inhibitory effect of J1 on the functionality of the HA2 subunit. Next, a robust interaction between J1 and the HA2 subunit was detected using SPR. Our findings provide evidence that J1 is an entry inhibitor that effectively inhibits the fusion of the influenza virus with the host cell membrane.
Compound J1 may serve as a promising candidate for the development of novel anti-IAV drugs, thereby necessitating further investigation of its in vivo anti-IAV efficacy. IAV-induced ALI models are frequently used to investigate the efficacy of anti-influenza therapies. Following IAV infection in mice, an abundance of inflammatory mediators induce necrosis in pulmonary tissues, thereby precipitating acute lung injury and subsequent multiorgan injury. In this study, an IAV-induced ALI model was established in BALB/c mice via intranasal infection with PR8. J1 was orally administered, and its impact on viral titers, lung index, histopathological sections, and inflammatory factors in the lungs, as well as its survival-protective effect, were evaluated. These findings demonstrate that J1 effectively reduced viral titers and lung index while mitigating lung inflammation and edema. Moreover, it significantly prolonged the survival time of mice and alleviated weight loss.
Taken together, this study shows that J1 significantly inhibits IAV infection both in vitro and in vivo, with low toxicity. It has been suggested that J1 blocks the membrane fusion process of IAV by interacting with the HA2 subunit. Our findings provide a structural foundation for the development of novel fusion inhibitors, thereby contributing to the development of novel anti-IAV therapeutics. Ongoing efforts in our laboratory are focused on optimizing the structure and synthesizing similar derivatives to enhance anti-influenza activity.
Conclusion
Our study highlights J1 as a promising lead compound for further development as an effective, safe and broad-spectrum viral entry inhibitor.
References
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33.
Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141–54.
Treanor J. Influenza vaccine–outmaneuvering antigenic shift and drift. N Engl J Med. 2004;350:218–20.
Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, et al. Influenza. Nat Rev Dis Prim. 2018;4:3.
Kumar B, Asha K, Khanna M, Ronsard L, Meseko CA, Sanicas M. The emerging influenza virus threat: status and new prospects for its therapy and control. Arch Virol. 2018;163:831–44.
Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12:15–22.
Schäfer JR, Kawaoka Y, Bean WJ, Süss J, Senne D, Webster RG. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–8.
Hsieh YC, Wu TZ, Liu DP, Shao PL, Chang LY, Lu CY, et al. Influenza pandemics: past, present and future. J Formos Med Assoc. 2006;105:1–6.
Kumari R, Sharma SD, Kumar A, Ende Z, Mishina M, Wang Y, et al. Antiviral approaches against influenza virus. Clin Microbiol Rev. 2023;36:e0004022.
Kumar G, Sakharam KA. Tackling influenza A virus by M2 ion channel blockers: latest progress and limitations. Eur J Med Chem. 2024;267:116172.
Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–81.
Toots M, Plemper RK. Next-generation direct-acting influenza therapeutics. Transl Res. 2020;220:33–42.
van der Vries E, Schutten M, Fraaij P, Boucher C, Osterhaus A. Influenza virus resistance to antiviral therapy. Adv Pharmacol. 2013;67:217–46.
Hossain MG, Akter S, Dhole P, Saha S, Kazi T, Majbauddin A, et al. Analysis of the genetic diversity associated with the drug resistance and pathogenicity of influenza A virus isolated in Bangladesh from 2002 to 2019. Front Microbiol. 2021;12:735305.
Lackenby A, Moran Gilad J, Pebody R, Miah S, Calatayud L, Bolotin S, et al. Continued emergence and changing epidemiology of oseltamivir-resistant influenza A(H1N1)2009 virus, United Kingdom, winter 2010/11. Eur Surveill. 2011;16:19784.
Renaud C, Kuypers J, Englund JA. Emerging oseltamivir resistance in seasonal and pandemic influenza A/H1N1. J Clin Virol. 2011;52:70–8.
Shirley M. Baloxavir Marboxil: a review in acute uncomplicated influenza. Drugs. 2020;80:1109–18.
Hickerson BT, Petrovskaya SN, Dickensheets H, Donnelly RP, Ince WL, Ilyushina NA. Impact of baloxavir resistance-associated substitutions on influenza virus growth and drug susceptibility. J Virol. 2023;97:e0015423.
Hayden FG, Sugaya N, Hirotsu N, Lee N, de Jong MD, Hurt AC, et al. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N Engl J Med. 2018;379:913–23.
Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010;285:28403–9.
Russell CJ, Hu M, Okda FA. Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol. 2018;26:841–53.
Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69.
Dou D, Revol R, Östbye H, Wang H, Daniels R. Influenza A virus cell entry, replication, virion assembly and movement. Front Immunol. 2018;9:1581.
Eisfeld AJ, Neumann G, Kawaoka Y. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol. 2015;13:28–41.
Zeng LY, Yang J, Liu S. Investigational hemagglutinin-targeted influenza virus inhibitors. Expert Opin Investig Drugs. 2017;26:63–73.
Vanderlinden E, Naesens L. Emerging antiviral strategies to interfere with influenza virus entry. Med Res Rev. 2014;34:301–39.
Du L, Zhao G, Zhang X, Liu Z, Yu H, Zheng BJ, et al. Development of a safe and convenient neutralization assay for rapid screening of influenza HA-specific neutralizing monoclonal antibodies. Biochem Biophys Res Commun. 2010;397:580–5.
Zhang K, He J, Li C, Bose ME, Henrickson KJ, Zhou J, et al. Complete genome sequences of one human respiratory syncytial antigenic group a virus from China and its four mouse-adapted isolates. Genome Announc. 2015;3:e00062–15.
Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27:493–7.
Chen Y, Wu Y, Chen S, Zhan Q, Wu D, Yang C, et al. Sertraline is an effective SARS-CoV-2 entry inhibitor targeting the spike protein. J Virol. 2022;96:e0124522.
Zhang X, Li A, Li T, Shou Z, Li Y, Qiao X, et al. A potential anti-HIV-1 compound, Q308, inhibits HSV-2 infection and replication in vitro and in vivo. Biomed Pharmacother. 2023;162:114595.
Zwygart AC, Medaglia C, Huber R, Poli R, Marcourt L, Schnee S, et al. Antiviral properties of trans-δ-viniferin derivatives against enveloped viruses. Biomed Pharmacother. 2023;163:114825.
Yu M, Si L, Wang Y, Wu Y, Yu F, Jiao P, et al. Discovery of pentacyclic triterpenoids as potential entry inhibitors of influenza viruses. J Med Chem. 2014;57:10058–71.
Liu AL, Wang HD, Lee SM, Wang YT, Du GH. Structure-activity relationship of flavonoids as influenza virus neuraminidase inhibitors and their in vitro anti-viral activities. Bioorg Med Chem. 2008;16:7141–7.
Wu W, Li R, Li X, He J, Jiang S, Liu S, et al. Quercetin as an antiviral agent inhibits influenza A virus (IAV) entry. Viruses. 2015;8:1999–4915.
Kumar N, Delu V, Shukla A, Singh RK, Ulasov I, Fayzullina D, et al. Safety assessment of a nucleoside analogue FNC (2’-deoxy-2’-β-fluoro-4’-azidocytidine) in Balb/c mice: acute toxicity study. Asian Pac J Cancer Prev. 2023;24:2157–70.
Yang J, Zhang B, Huang Y, Liu T, Zeng B, Chai J, et al. Antiviral activity and mechanism of ESC-1GN from skin secretion of Hylarana guentheri against influenza A virus. J Biochem. 2021;169:757–65.
Pica N, Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med. 2013;64:189–202.
Tilton JC, Doms RW. Entry inhibitors in the treatment of HIV-1 infection. Antivir Res. 2010;85:91–100.
Chen Z, Cui Q, Caffrey M, Rong L, Du R. Small molecule inhibitors of influenza virus entry. Pharmaceuticals. 2021;14:587.
Kadam RU, Wilson IA. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci USA 2017;114:206–14.
Boriskin YS, Leneva IA, Pécheur EI, Polyak SJ. Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr Med Chem. 2008;15:997–1005.
Blaising J, Polyak SJ, Pécheur EI. Arbidol as a broad-spectrum antiviral: an update. Antivir Res. 2014;107:84–94.
Antanasijevic A, Cheng H, Wardrop DJ, Rong L, Caffrey M. Inhibition of influenza H7 hemagglutinin-mediated entry. PLoS One. 2013;8:e76363.
Liu S, Li R, Zhang R, Chan CC, Xi B, Zhu Z, et al. CL-385319 inhibits H5N1 avian influenza A virus infection by blocking viral entry. Eur J Pharmacol. 2011;660:460–7.
Acknowledgements
We thank Shi-bo Jiang at Fudan University for the kindly gift of HA2 antibodies and guidance. This research was funded by the National Natural Science Foundation of China (Grant numbers 82073897, 82373915) and the Science and Technology Innovation Project of Guangdong Medical Products Administration (Grant numbers S2021ZDZ042). We thank BioRender for providing us with the drawing material.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. YYL and JY designed the research study. YYL and GDL conceived and carried out experiments. ZXC, KZ, and LRJ carried out animal experiments. JLL and LRJ coordinated the lung tissue collection of mice. YYL and SZY analyzed data. YYL, JY, FJ and SWL wrote and/or reviewed the manuscript. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Yy., Liang, Gd., Chen, Zx. et al. A small molecule compound targeting hemagglutinin inhibits influenza A virus and exhibits broad-spectrum antiviral activity. Acta Pharmacol Sin (2024). https://doi.org/10.1038/s41401-024-01331-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41401-024-01331-7
- Springer Nature Singapore Pte Ltd.